A Radiomics Approach to Identify Immunologically Active Tumor in Patients with Head and Neck Squamous Cell Carcinomas
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
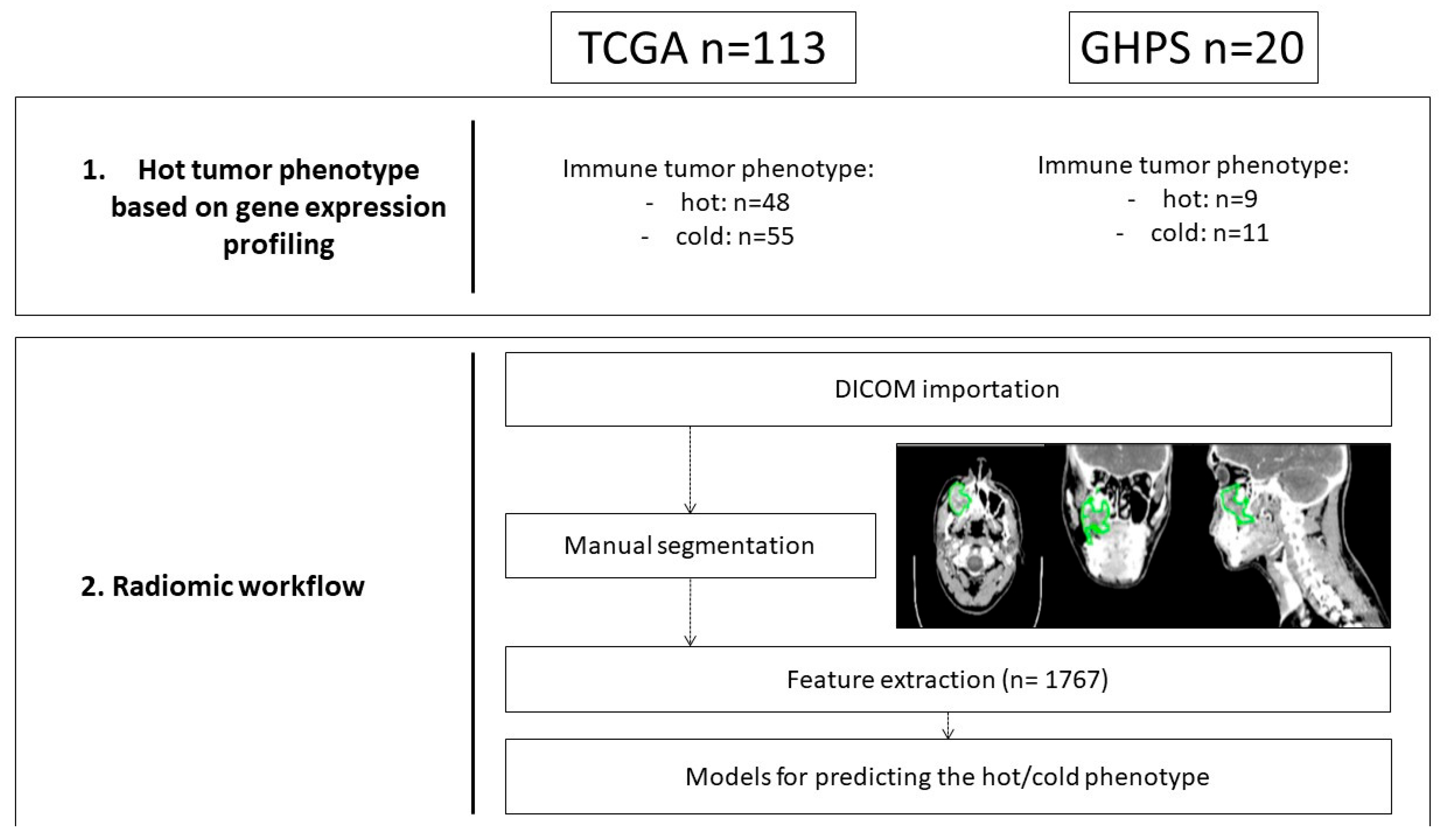
2.1. Datasets (Table 1)
2.1.1. Training and Testing Datasets
2.1.2. External Validating Dataset
| TCGA-HNSCC (N = 113) | GHPS-COSMOS (N = 20) | |
|---|---|---|
| N (%) | N (%) | |
| Age (mean) at diagnosis | 59.69 | 62.15 |
| Gender | ||
| Female | 27 (23.9%) | 8 (40.0%) |
| Male | 86 (76.1%) | 12 (60.0%) |
| Anatomic Site | ||
| Oral cavity | 68 (60.2%) | 20 (100%) |
| Larynx | 31 (27.4%) | 0 |
| Oropharynx | 13 (11.5%) | 0 |
| Hypopharynx | 1 (0.9%) | 0 |
| Alcohol | ||
| Yes | 28 (24.8%) | 8 (40.0%) |
| No | 85 (75.2%) | 10 (50.0%) |
| NA | 0 | 2 (10.0%) |
| Smoking | ||
| Current | 45 (39.8%) | 10 (50.0%) |
| Former | 46 (40.7%) | 2 (10.0%) |
| No | 21 (18.6%) | 8 (40.0%) |
| NA | 1 (0.9%) | 0 |
| Pathological T stage | ||
| T1 | 6 (5.3%) | 1 (5.0%) |
| T2 | 23 (20.3%) | 5 (25.0%) |
| T3 | 23 (30.3%) | 3 (10.0%) |
| T4 | 41 (36.3%) | 11 (55.0%) |
| Tx | 13 (38.0%) | 0 |
| NA | 5 (11.5%) | 0 |
| N stage (%) | ||
| N0 | 47 (41.6%) | 10 (50%) |
| N1 | 13 (11.5%) | 2 (10%) |
| N2 | 32 (28.3%) | 4 (20%) |
| N3 | 1 (0.9%) | 4 (20%) |
| Nx | 15 (13.3%) | 0 |
| NA | 5 (4.4%) | 0 |
| M stage (%) | ||
| M0 | 109 (96.5%) | 20 (100%) |
| M1 | 1 (0.9%) | 0 |
| Mx | 3 (2.6%) | 0 |
| Phenotype (%) | ||
| HOT | 48 (42.5%) | 9 (45%) |
| COLD | 65 (57.5%) | 11 (55%) |
2.2. Radiomics Workflow (Figure 1)
2.2.1. Tumor Volume Segmentation
2.2.2. Quantitative Image Feature Extraction
2.2.3. Feature Selection
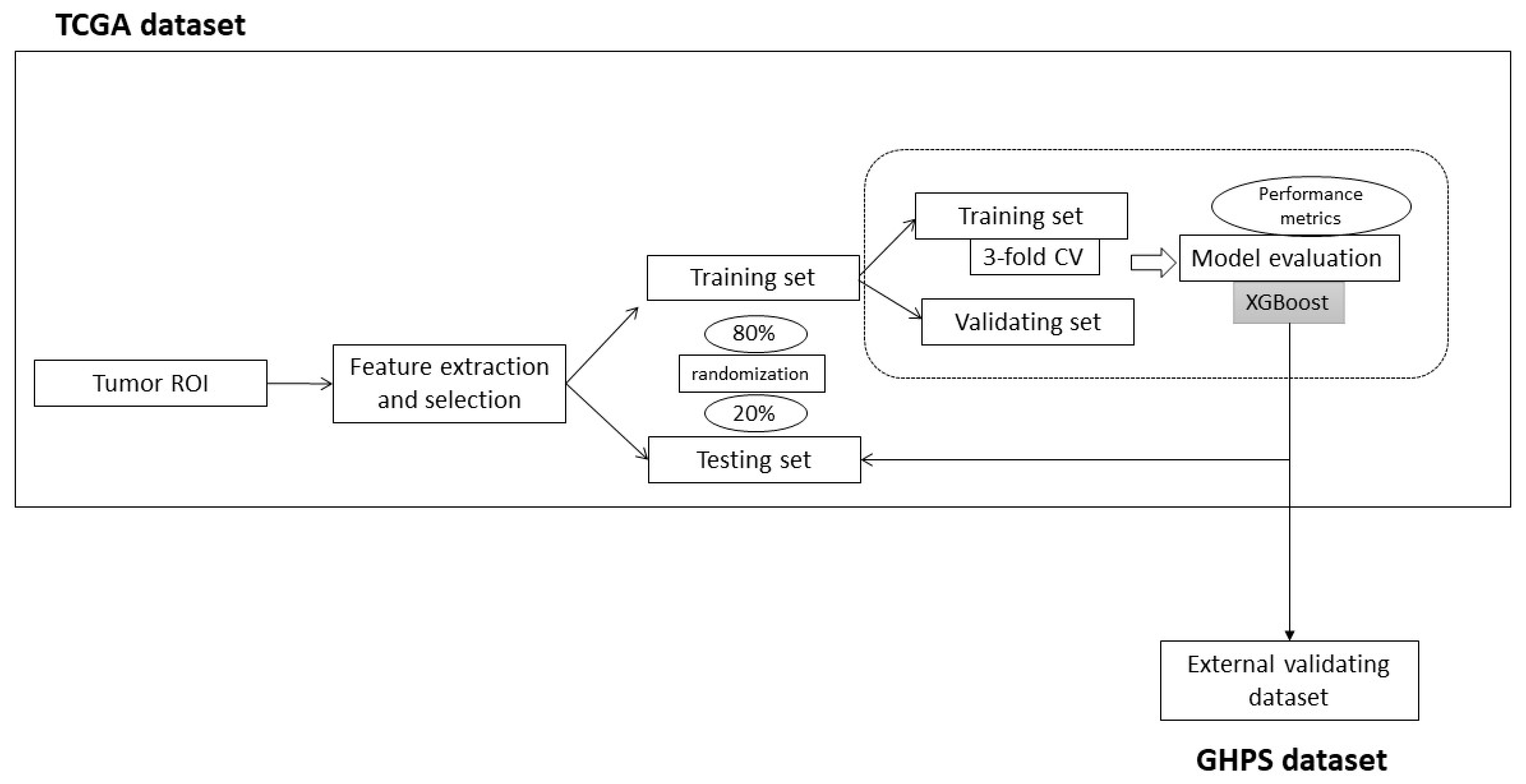
2.3. Radiomic Model (Figure 2)
3. Results
3.1. Identification of the Hot Phenotype and Radiomic Feature Selection
3.2. Predictive Radiomic Model of the Hot Phenotype
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Ferris, R.L.; Blumenschein, G.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Seiwert, T.Y.; Burtness, B.; Mehra, R.; Weiss, J.; Berger, R.; Eder, J.P.; Heath, K.; McClanahan, T.; Lunceford, J.; Gause, C.; et al. Safety and Clinical Activity of Pembrolizumab for Treatment of Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck (KEYNOTE-012): An Open-Label, Multicentre, Phase 1b Trial. Lancet Oncol. 2016, 17, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Haanen, J.B.A.G. Converting Cold into Hot Tumors by Combining Immunotherapies. Cell 2017, 170, 1055–1056. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Approaches to Treat Immune Hot, Altered and Cold Tumours with Combination Immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef]
- De Guillebon, E.; Dardenne, A.; Saldmann, A.; Séguier, S.; Tran, T.; Paolini, L.; Lebbe, C.; Tartour, E. Beyond the Concept of Cold and Hot Tumors for the Development of Novel Predictive Biomarkers and the Rational Design of Immunotherapy Combination. Int. J. Cancer 2020, 147, 1509–1518. [Google Scholar] [CrossRef]
- Foy, J.-P.; Karabajakian, A.; Ortiz-Cuaran, S.; Boussageon, M.; Michon, L.; Bouaoud, J.; Fekiri, D.; Robert, M.; Baffert, K.-A.; Hervé, G.; et al. Immunologically Active Phenotype by Gene Expression Profiling Is Associated with Clinical Benefit from PD-1/PD-L1 Inhibitors in Real-World Head and Neck and Lung Cancer Patients. Eur. J. Cancer 2022, 174, 287–298. [Google Scholar] [CrossRef]
- Gillies, R.J.; Anderson, A.R.; Gatenby, R.A.; Morse, D.L. The Biology Underlying Molecular Imaging in Oncology: From Genome to Anatome and Back Again. Clin. Radiol. 2010, 65, 517–521. [Google Scholar] [CrossRef]
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.W.L.; Dekker, A.; Fenstermacher, D.; et al. Radiomics: The Process and the Challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting More Information from Medical Images Using Advanced Feature Analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Foy, J.P.; Durdux, C.; Giraud, P.; Bibault, J.E. RE: The Rise of Radiomics and Implications for Oncologic Management. J. Natl. Cancer Inst. 2018, 110, 1275–1276. [Google Scholar] [CrossRef] [PubMed]
- Iv, M.; Zhou, M.; Shpanskaya, K.; Perreault, S.; Wang, Z.; Tranvinh, E.; Lanzman, B.; Vajapeyam, S.; Vitanza, N.A.; Fisher, P.G.; et al. MR Imaging-Based Radiomic Signatures of Distinct Molecular Subgroups of Medulloblastoma. Am. J. Neuroradiol. 2019, 40, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Shi, J.; Ye, Z.; Dong, D.; Yu, D.; Zhou, M.; Liu, Y.; Gevaert, O.; Wang, K.; Zhu, Y.; et al. Predicting EGFR Mutation Status in Lung Adenocarcinoma on Computed Tomography Image Using Deep Learning. Eur. Respir. J. 2019, 53, 1800986. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Xie, Z.; Zang, Y.; Zhang, S.; Gu, D.; Zhou, M.; Gevaert, O.; Wei, J.; Li, C.; Chen, H.; et al. Non-Invasive Genotype Prediction of Chromosome 1p/19q Co-Deletion by Development and Validation of an MRI-Based Radiomics Signature in Lower-Grade Gliomas. J. Neuro-Oncol. 2018, 140, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Gevaert, O.; Xu, J.; Hoang, C.D.; Leung, A.N.; Xu, Y.; Quon, A.; Rubin, D.L.; Napel, S.; Plevritis, S.K. Non-Small Cell Lung Cancer: Identifying Prognostic Imaging Biomarkers by Leveraging Public Gene Expression Microarray Data--Methods and Preliminary Results. Radiology 2012, 264, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Carles, M.; Fechter, T.; Grosu, A.L.; Sörensen, A.; Thomann, B.; Stoian, R.G.; Wiedenmann, N.; Rühle, A.; Zamboglou, C.; Ruf, J.; et al. 18 F-FMISO-PET Hypoxia Monitoring for Head-and-Neck Cancer Patients: Radiomics Analyses Predict the Outcome of Chemo-Radiotherapy. Cancers 2021, 13, 3449. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Long, M.; Sun, C.; Yang, Y.; Lin, P.; Shen, Z.; Xia, S.; Shen, W. CT-Based Radiomics Signature Analysis for Evaluation of Response to Induction Chemotherapy and Progression-Free Survival in Locally Advanced Hypopharyngeal Carcinoma. Eur. Radiol. 2022, 32, 7755–7766. [Google Scholar] [CrossRef]
- Joye, I.; Debucquoy, A.; Deroose, C.M.; Vandecaveye, V.; Van Cutsem, E.; Wolthuis, A.; D’Hoore, A.; Sagaert, X.; Zhou, M.; Gevaert, O.; et al. Quantitative Imaging Outperforms Molecular Markers When Predicting Response to Chemoradiotherapy for Rectal Cancer. Radiother. Oncol. 2017, 124, 104–109. [Google Scholar] [CrossRef]
- Bibault, J.E.; Giraud, P.; Housset, M.; Durdux, C.; Taieb, J.; Berger, A.; Coriat, R.; Chaussade, S.; Dousset, B.; Nordlinger, B.; et al. Deep Learning and Radiomics Predict Complete Response after Neo-Adjuvant Chemoradiation for Locally Advanced Rectal Cancer. Sci. Rep. 2018, 8, 12611. [Google Scholar] [CrossRef]
- Itakura, H.; Achrol, A.S.; Mitchell, L.A.; Loya, J.J.; Liu, T.; Westbroek, E.M.; Feroze, A.H.; Rodriguez, S.; Echegaray, S.; Azad, T.D.; et al. Magnetic Resonance Image Features Identify Glioblastoma Phenotypic Subtypes with Distinct Molecular Pathway Activities. Sci. Transl. Med. 2015, 7, 303ra138. [Google Scholar] [CrossRef]
- Gevaert, O.; Mitchell, L.A.; Achrol, A.S.; Xu, J.; Echegaray, S.; Steinberg, G.K.; Cheshier, S.H.; Napel, S.; Zaharchuk, G.; Plevritis, S.K. Glioblastoma Multiforme: Exploratory Radiogenomic Analysis by Using Quantitative Image Features. Radiology 2014, 273, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding Tumour Phenotype by Noninvasive Imaging Using a Quantitative Radiomics Approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Parmar, C.; Leijenaar, R.T.H.; Grossmann, P.; Velazquez, E.R.; Bussink, J.; Rietveld, D.; Rietbergen, M.M.; Haibe-Kains, B.; Lambin, P.; Aerts, H.J.W.L. Radiomic Feature Clusters and Prognostic Signatures Specific for Lung and Head & Neck Cancer. Sci. Rep. 2015, 5, srep11044. [Google Scholar] [CrossRef]
- Parmar, C.; Grossmann, P.; Rietveld, D.; Rietbergen, M.M.; Lambin, P.; Aerts, H.J.W.L. Radiomic Machine-Learning Classifiers for Prognostic Biomarkers of Head and Neck Cancer. Front. Oncol. 2015, 5, 272. [Google Scholar] [CrossRef]
- Bogowicz, M.; Riesterer, O.; Ikenberg, K.; Stieb, S.; Moch, H.; Studer, G.; Guckenberger, M.; Tanadini-Lang, S. Computed Tomography Radiomics Predicts HPV Status and Local Tumor Control After Definitive Radiochemotherapy in Head and Neck Squamous Cell Carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2017, 99, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Ou, D.; Blanchard, P.; Rosellini, S.; Levy, A.; Nguyen, F.; Leijenaar, R.T.H.; Garberis, I.; Gorphe, P.; Bidault, F.; Ferté, C.; et al. Predictive and Prognostic Value of CT Based Radiomics Signature in Locally Advanced Head and Neck Cancers Patients Treated with Concurrent Chemoradiotherapy or Bioradiotherapy and Its Added Value to Human Papillomavirus Status. Oral Oncol. 2017, 71, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Vallières, M.; Kay-Rivest, E.; Perrin, L.J.; Liem, X.; Furstoss, C.; Aerts, H.J.W.L.; Khaouam, N.; Nguyen-Tan, P.F.; Wang, C.S.; Sultanem, K.; et al. Radiomics Strategies for Risk Assessment of Tumour Failure in Head-and-Neck Cancer. Sci. Rep. 2017, 7, 10117. [Google Scholar] [CrossRef]
- Tsai, Y.-L.; Chen, S.-W.; Kao, C.-H.; Cheng, D.-C. Neck Lymph Node Recurrence in HNC Patients Might Be Predicted before Radiotherapy Using Radiomics Extracted from CT Images and XGBoost Algorithm. J. Pers. Med. 2022, 12, 1377. [Google Scholar] [CrossRef]
- Mukherjee, P.; Cintra, M.; Huang, C.; Zhou, M.; Zhu, S.; Colevas, A.D.; Fischbein, N.; Gevaert, O. CT-Based Radiomic Signatures for Predicting Histopathologic Features in Head and Neck Squamous Cell Carcinoma. Radiol. Imaging Cancer 2020, 2, e190039. [Google Scholar] [CrossRef]
- Zhao, X.; Li, W.; Zhang, J.; Tian, S.; Zhou, Y.; Xu, X.; Hu, H.; Lei, D.; Wu, F. Radiomics Analysis of CT Imaging Improves Preoperative Prediction of Cervical Lymph Node Metastasis in Laryngeal Squamous Cell Carcinoma. Eur. Radiol. 2023, 33, 1121–1131. [Google Scholar] [CrossRef]
- Zheng, Y.-M.; Chen, J.; Zhang, M.; Wu, Z.-J.; Tang, G.-Z.; Zhang, Y.; Dong, C. CT Radiomics Nomogram for Prediction of the Ki-67 Index in Head and Neck Squamous Cell Carcinoma. Eur. Radiol. 2023, 33, 2160–2170. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.-M.; Pang, J.; Liu, Z.-J.; Yuan, M.-G.; Li, J.; Wu, Z.-J.; Jiang, Y.; Dong, C. A CT-Based Deep Learning Radiomics Nomogram for the Prediction of EGFR Mutation Status in Head and Neck Squamous Cell Carcinoma. Acad. Radiol. 2023; in press. [Google Scholar] [CrossRef]
- Zwirner, K.; Hilke, F.J.; Demidov, G.; Socarras Fernandez, J.; Ossowski, S.; Gani, C.; Thorwarth, D.; Riess, O.; Zips, D.; Schroeder, C.; et al. Radiogenomics in Head and Neck Cancer: Correlation of Radiomic Heterogeneity and Somatic Mutations in TP53, FAT1 and KMT2D. Strahlenther. Onkol. 2019, 195, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.-M.; Che, J.-Y.; Yuan, M.-G.; Wu, Z.-J.; Pang, J.; Zhou, R.-Z.; Li, X.-L.; Dong, C. A CT-Based Deep Learning Radiomics Nomogram to Predict Histological Grades of Head and Neck Squamous Cell Carcinoma. Acad. Radiol. 2023, 30, 1591–1599. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-H.; Yan, J.-L.; Yap, W.-K.; Kang, C.-J.; Chang, Y.-C.; Tsai, T.-Y.; Chang, K.-P.; Liao, C.-T.; Hsu, C.-L.; Chou, W.-C.; et al. Prognostic Value of Interim CT-Based Peritumoral and Intratumoral Radiomics in Laryngeal and Hypopharyngeal Cancer Patients Undergoing Definitive Radiotherapy. Radiother. Oncol. 2023, 189, 109938. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-T.; Lin, Y.-C.; Yen, C.-H.; Lan, J.; Yu, C.-C.; Lin, W.-C.; Chen, Y.-S.; Wang, C.-K.; Huang, E.-Y.; Ho, S.-Y. Prediction of Extranodal Extension in Head and Neck Squamous Cell Carcinoma by CT Images Using an Evolutionary Learning Model. Cancer Imaging 2023, 23, 84. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Song, L.; Zhou, J.; Yu, M.; Hu, Y.; Zhang, J.; Song, P.; Ye, Y.; Wang, J.; Feng, G.; et al. Prediction of Prognosis of Tongue Squamous Cell Carcinoma Based on Clinical MR Imaging Data Modeling. Technol. Cancer Res. Treat. 2023, 22, 15330338231207006. [Google Scholar] [CrossRef]
- Corti, A.; De Cecco, L.; Cavalieri, S.; Lenoci, D.; Pistore, F.; Calareso, G.; Mattavelli, D.; de Graaf, P.; Leemans, C.R.; Brakenhoff, R.H.; et al. MRI-Based Radiomic Prognostic Signature for Locally Advanced Oral Cavity Squamous Cell Carcinoma: Development, Testing and Comparison with Genomic Prognostic Signatures. Biomark. Res. 2023, 11, 69. [Google Scholar] [CrossRef]
- Jiang, S.; Locatello, L.G.; Maggiore, G.; Gallo, O. Radiomics-Based Analysis in the Prediction of Occult Lymph Node Metastases in Patients with Oral Cancer: A Systematic Review. J. Clin. Med. 2023, 12, 4958. [Google Scholar] [CrossRef]
- Katsoulakis, E.; Yu, Y.; Apte, A.P.; Leeman, J.E.; Katabi, N.; Morris, L.; Deasy, J.O.; Chan, T.A.; Lee, N.Y.; Riaz, N.; et al. Radiomic Analysis Identifies Tumor Subtypes Associated with Distinct Molecular and Microenvironmental Factors in Head and Neck Squamous Cell Carcinoma. Oral Oncol. 2020, 110, 104877. [Google Scholar] [CrossRef]
- Wan, Y.W.; Allen, G.I.; Liu, Z. TCGA2STAT: Simple TCGA Data Access for Integrated Statistical Analysis in R. Bioinformatics 2016, 32, 952–954. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.E.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Foy, J.P.; Karabajakian, A.; Ortiz-Cuaran, S.; Boussageon, M.; Michon, L.; Bouaoud, J.; Fekiri, D.; Robert, M.; Baffert, K.A.; Hervé, G.; et al. Datasets for Gene Expression Profiles of Head and Neck Squamous Cell Carcinoma and Lung Cancer Treated or Not by PD1/PD-L1 Inhibitors. Data Brief. 2022, 44, 108556. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene Set Variation Analysis for Microarray and RNA-Seq Data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Zhang, L.; Fried, D.V.; Fave, X.J.; Hunter, L.A.; Yang, J.; Court, L.E. IBEX: An Open Infrastructure Software Platform to Facilitate Collaborative Work in Radiomics. Med. Phys. 2015, 42, 1341–1353. [Google Scholar] [CrossRef]
- Ger, R.B.; Cardenas, C.E.; Anderson, B.M.; Yang, J.; Mackin, D.S.; Zhang, L.; Court, L.E. Guidelines and Experience Using Imaging Biomarker Explorer (IBEX) for Radiomics. J. Vis. Exp. 2018, 2018, e57132. [Google Scholar] [CrossRef]
- Plaks, V.; Koopman, C.D.; Werb, Z. Cancer. Circulating Tumor Cells. Science 2013, 341, 1186–1188. [Google Scholar] [CrossRef]
- Shrout, P.E.; Fleiss, J.L. Intraclass Correlations: Uses in Assessing Rater Reliability. Psychol. Bull. 1979, 86, 420–428. [Google Scholar] [CrossRef]
- Choy, G.; Khalilzadeh, O.; Michalski, M.; Do, S.; Samir, A.E.; Pianykh, O.S.; Geis, J.R.; Pandharipande, P.V.; Brink, J.A.; Dreyer, K.J. Current Applications and Future Impact of Machine Learning in Radiology. Radiology 2018, 288, 318–328. [Google Scholar] [CrossRef]
- Browne, M.W. Cross-Validation Methods. J. Math. Psychol. 2000, 44, 108–132. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.X.; Huang, J.; Zhang, H. AUC: A Better Measure than Accuracy in Comparing Learning Algorithms. In Proceedings of the Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2003; Volume 2671, pp. 329–341. [Google Scholar]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The Bridge between Medical Imaging and Personalized Medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Mohamed, A.S.R.; Lai, S.Y.; Yang, S.; Kanwar, A.; Wei, L.; Kamal, M.; Sengupta, S.; Elhalawani, H.; Skinner, H.; et al. Imaging-Genomic Study of Head and Neck Squamous Cell Carcinoma: Associations Between Radiomic Phenotypes and Genomic Mechanisms via Integration of The Cancer Genome Atlas and The Cancer Imaging Archive. JCO Clin. Cancer Inform. 2019, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Joye, I.; Debucquoy, A.; Deroose, C.M.; Vandecaveye, V.; Van Cutsem, E.; Wolthuis, A.; D’Hoore, A.; Sagaert, X.; Zhou, M.; Gevaert, O.; et al. Non–Small Cell Lung Cancer Radiogenomics Map Identifies Relationships between Molecular and Imaging Phenotypes with Prognostic Implications. Radiology 2018, 5, 150–155. [Google Scholar] [CrossRef]
- Huang, C.; Cintra, M.; Brennan, K.; Zhou, M.; Colevas, A.D.; Fischbein, N.; Zhu, S.; Gevaert, O. Development and Validation of Radiomic Signatures of Head and Neck Squamous Cell Carcinoma Molecular Features and Subtypes. EBioMedicine 2019, 45, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Caudell, J.J.; Torres-Roca, J.F.; Gillies, R.J.; Enderling, H.; Kim, S.; Rishi, A.; Moros, E.G.; Harrison, L.B. The Future of Personalised Radiotherapy for Head and Neck Cancer. Lancet Oncol. 2017, 18, e266–e273. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.-M.; Zhan, J.-F.; Yuan, M.-G.; Hou, F.; Jiang, G.; Wu, Z.-J.; Dong, C. A CT-Based Radiomics Signature for Preoperative Discrimination between High and Low Expression of Programmed Death Ligand 1 in Head and Neck Squamous Cell Carcinoma. Eur. J. Radiol. 2022, 146, 110093. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, M. Noninvasive Radiomic Analysis of Enhanced CT Predicts CTLA4 Expression and Prognosis in Head and Neck Squamous Cell Carcinoma. Sci. Rep. 2023, 13, 16782. [Google Scholar] [CrossRef]
- Wang, J.H.; Wahid, K.A.; van Dijk, L.V.; Farahani, K.; Thompson, R.F.; Fuller, C.D. Radiomic Biomarkers of Tumor Immune Biology and Immunotherapy Response. Clin. Transl. Radiat. Oncol. 2021, 28, 97–115. [Google Scholar] [CrossRef]
- Tong, H.; Sun, J.; Fang, J.; Zhang, M.; Liu, H.; Xia, R.; Zhou, W.; Liu, K.; Chen, X. A Machine Learning Model Based on PET/CT Radiomics and Clinical Characteristics Predicts Tumor Immune Profiles in Non-Small Cell Lung Cancer: A Retrospective Multicohort Study. Front. Immunol. 2022, 13, 859323. [Google Scholar] [CrossRef]
- Adams, S.; Gatti-Mays, M.E.; Kalinsky, K.; Korde, L.A.; Sharon, E.; Amiri-Kordestani, L.; Bear, H.; McArthur, H.L.; Frank, E.; Perlmutter, J.; et al. Current Landscape of Immunotherapy in Breast Cancer: A Review. JAMA Oncol. 2019, 5, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Hobbs, B.; Amer, A.; Li, X.; Behrens, C.; Canales, J.R.; Cuentas, E.P.; Villalobos, P.; Fried, D.; Chang, J.Y.; et al. Development of an Immune-Pathology Informed Radiomics Model for Non-Small Cell Lung Cancer. Sci. Rep. 2018, 8, 1922. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Li, H.; Liu, Q.; Duan, J.; Zhou, W.; Yu, X.; Chen, Q.; Liu, Z.; Wang, W.; Rong, P. CT Radiomics to Predict Macrotrabecular-Massive Subtype and Immune Status in Hepatocellular Carcinoma. Radiology 2023, 307, e221291. [Google Scholar] [CrossRef] [PubMed]
- Khalili, N.; Kazerooni, A.F.; Familiar, A.; Haldar, D.; Kraya, A.; Foster, J.; Koptyra, M.; Storm, P.B.; Resnick, A.C.; Nabavizadeh, A. Radiomics for Characterization of the Glioma Immune Microenvironment. npj Precis. Oncol. 2023, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Henry, T.; Laville, A.; Carré, A.; Hamaoui, A.; Bockel, S.; Chaffai, I.; Levy, A.; Chargari, C.; Robert, C.; et al. Imaging Approaches and Radiomics: Toward a New Era of Ultraprecision Radioimmunotherapy? J. Immunother. Cancer 2022, 10, e004848. [Google Scholar] [CrossRef]
- Kang, C.Y.; Duarte, S.E.; Kim, H.S.; Kim, E.; Park, J.; Lee, A.D.; Kim, Y.; Kim, L.; Cho, S.; Oh, Y.; et al. Artificial Intelligence-Based Radiomics in the Era of Immuno-Oncology. Oncologist 2022, 27, e471–e483. [Google Scholar] [CrossRef]
- Tanadini-Lang, S.; Balermpas, P.; Guckenberger, M.; Pavic, M.; Riesterer, O.; Vuong, D.; Bogowicz, M. Radiomic Biomarkers for Head and Neck Squamous Cell Carcinoma. Strahlenther. Onkol. 2020, 196, 868–878. [Google Scholar] [CrossRef]
- Sun, R.; Limkin, E.J.; Vakalopoulou, M.; Dercle, L.; Champiat, S.; Han, S.R.; Verlingue, L.; Brandao, D.; Lancia, A.; Ammari, S.; et al. A Radiomics Approach to Assess Tumour-Infiltrating CD8 Cells and Response to Anti-PD-1 or Anti-PD-L1 Immunotherapy: An Imaging Biomarker, Retrospective Multicohort Study. Lancet Oncol. 2018, 19, 1180–1191. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, W.; Chai, Y.; Wang, H.; Liu, Z.; He, Y. Constrast-Enhanced Computed Tomography Radiomics Predicts CD27 Expression and Clinical Prognosis in Head and Neck Squamous Cell Carcinoma. Front. Immunol. 2022, 13, 1015436. [Google Scholar] [CrossRef]
- Chen, R.-Y.; Lin, Y.-C.; Shen, W.-C.; Hsieh, T.-C.; Yen, K.-Y.; Chen, S.-W.; Kao, C.-H. Associations of Tumor PD-1 Ligands, Immunohistochemical Studies, and Textural Features in 18F-FDG PET in Squamous Cell Carcinoma of the Head and Neck. Sci. Rep. 2018, 8, 105. [Google Scholar] [CrossRef]
- Tramm, T.; Di Caterino, T.; Jylling, A.-M.B.; Lelkaitis, G.; Lænkholm, A.-V.; Ragó, P.; Tabor, T.P.; Talman, M.-L.M.; Vouza, E. Scientific Committee of Pathology, Danish Breast Cancer Group (DBCG) Standardized Assessment of Tumor-Infiltrating Lymphocytes in Breast Cancer: An Evaluation of Inter-Observer Agreement between Pathologists. Acta Oncol. 2018, 57, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Huang, H.; Tong, Q.; Cao, M.; Ming, W.; Zhang, R.; Zhu, W.; Wang, Y.; Sun, X. Multi-View Radiomics Feature Fusion Reveals Distinct Immuno-Oncological Characteristics and Clinical Prognoses in Hepatocellular Carcinoma. Cancers 2023, 15, 2338. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Zhang, Z.; Chen, Y.; Zhao, Y.; Sun, Q.; Wang, W.; Zheng, H.; Liang, D.; Cheng, J.; Yan, J.; et al. Imaging Phenotypes from MRI for the Prediction of Glioma Immune Subtypes from RNA Sequencing: A Multicenter Study. Mol. Oncol. 2023, 17, 629–646. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mayer, A.T.; Li, R. Integrated Imaging and Molecular Analysis to Decipher Tumor Microenvironment in the Era of Immunotherapy. Semin. Cancer Biol. 2022, 84, 310–328. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. arXiv 2016, arXiv:1603.02754. [Google Scholar]
- Shi, Y.; Zou, Y.; Liu, J.; Wang, Y.; Chen, Y.; Sun, F.; Yang, Z.; Cui, G.; Zhu, X.; Cui, X.; et al. Ultrasound-Based Radiomics XGBoost Model to Assess the Risk of Central Cervical Lymph Node Metastasis in Patients with Papillary Thyroid Carcinoma: Individual Application of SHAP. Front. Oncol. 2022, 12, 897596. [Google Scholar] [CrossRef]
- Li, J.; Shi, Z.; Liu, F.; Fang, X.; Cao, K.; Meng, Y.; Zhang, H.; Yu, J.; Feng, X.; Li, Q.; et al. XGBoost Classifier Based on Computed Tomography Radiomics for Prediction of Tumor-Infiltrating CD8+ T-Cells in Patients with Pancreatic Ductal Adenocarcinoma. Front. Oncol. 2021, 11, 671333. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; You, X.; Zhang, L.; Huang, D.; Aramini, B.; Shabaturov, L.; Jiang, G.; Fan, J. A Radiomics Model Combined with XGBoost May Improve the Accuracy of Distinguishing between Mediastinal Cysts and Tumors: A Multicenter Validation Analysis. Ann. Transl. Med. 2021, 9, 1737. [Google Scholar] [CrossRef]
- Song, B.-I. A Machine Learning-Based Radiomics Model for the Prediction of Axillary Lymph-Node Metastasis in Breast Cancer. Breast Cancer 2021, 28, 664–671. [Google Scholar] [CrossRef]
- Fournier, L.; Costaridou, L.; Bidaut, L.; Michoux, N.; Lecouvet, F.E.; de Geus-Oei, L.-F.; Boellaard, R.; Oprea-Lager, D.E.; Obuchowski, N.A.; Caroli, A.; et al. Incorporating Radiomics into Clinical Trials: Expert Consensus Endorsed by the European Society of Radiology on Considerations for Data-Driven Compared to Biologically Driven Quantitative Biomarkers. Eur. Radiol. 2021, 31, 6001–6012. [Google Scholar] [CrossRef]
- Narang, S.; Kim, D.; Aithala, S.; Heimberger, A.B.; Ahmed, S.; Rao, D.; Rao, G.; Rao, A. Tumor Image-Derived Texture Features Are Associated with CD3 T-Cell Infiltration Status in Glioblastoma. Oncotarget 2017, 8, 101244–101254. [Google Scholar] [CrossRef]
- Mackin, D.; Fave, X.; Zhang, L.; Fried, D.; Yang, J.; Taylor, B.; Rodriguez-Rivera, E.; Dodge, C.; Jones, A.K.; Court, L. Measuring Computed Tomography Scanner Variability of Radiomics Features. Investig. Radiol. 2015, 50, 757–765. [Google Scholar] [CrossRef]
- Kocak, B.; Durmaz, E.S.; Kaya, O.K.; Ates, E.; Kilickesmez, O. Reliability of Single-Slice-Based 2D CT Texture Analysis of Renal Masses: Influence of Intra- and Interobserver Manual Segmentation Variability on Radiomic Feature Reproducibility. Am. J. Roentgenol. 2019, 213, 377–383. [Google Scholar] [CrossRef]
- Larue, R.T.H.M.; Defraene, G.; De Ruysscher, D.; Lambin, P.; Van Elmpt, W. Quantitative Radiomics Studies for Tissue Characterization: A Review of Technology and Methodological Procedures. Br. J. Radiol. 2017, 90, 20160665. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, T.M.; Bertolus, C.; Giraud, P.; Burgun, A.; Saintigny, P.; Bibault, J.-E.; Foy, J.-P. A Radiomics Approach to Identify Immunologically Active Tumor in Patients with Head and Neck Squamous Cell Carcinomas. Cancers 2023, 15, 5369. https://doi.org/10.3390/cancers15225369
Nguyen TM, Bertolus C, Giraud P, Burgun A, Saintigny P, Bibault J-E, Foy J-P. A Radiomics Approach to Identify Immunologically Active Tumor in Patients with Head and Neck Squamous Cell Carcinomas. Cancers. 2023; 15(22):5369. https://doi.org/10.3390/cancers15225369
Chicago/Turabian StyleNguyen, Tan Mai, Chloé Bertolus, Paul Giraud, Anita Burgun, Pierre Saintigny, Jean-Emmanuel Bibault, and Jean-Philippe Foy. 2023. "A Radiomics Approach to Identify Immunologically Active Tumor in Patients with Head and Neck Squamous Cell Carcinomas" Cancers 15, no. 22: 5369. https://doi.org/10.3390/cancers15225369
APA StyleNguyen, T. M., Bertolus, C., Giraud, P., Burgun, A., Saintigny, P., Bibault, J.-E., & Foy, J.-P. (2023). A Radiomics Approach to Identify Immunologically Active Tumor in Patients with Head and Neck Squamous Cell Carcinomas. Cancers, 15(22), 5369. https://doi.org/10.3390/cancers15225369







