Targeting Esophageal Squamous Cell Carcinoma by Combining Copper Ionophore Disulfiram and JMJD3/UTX Inhibitor GSK J4
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Compound Library and Drugs
2.2. Sampling
2.3. Cell Lines and Cell Culture
2.4. Screening Bioactive Compound Library
2.5. Drug Synergy Testing
2.6. Annexin V Staining
2.7. Cell Cycle Analysis
2.8. Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
2.9. Western Blot
2.10. Quantitative Proteomics
2.11. In Vivo Tumor Experiments
2.12. Histopathology and IHC Analyses
2.13. TUNEL Assay
2.14. Statistical Analysis
3. Results
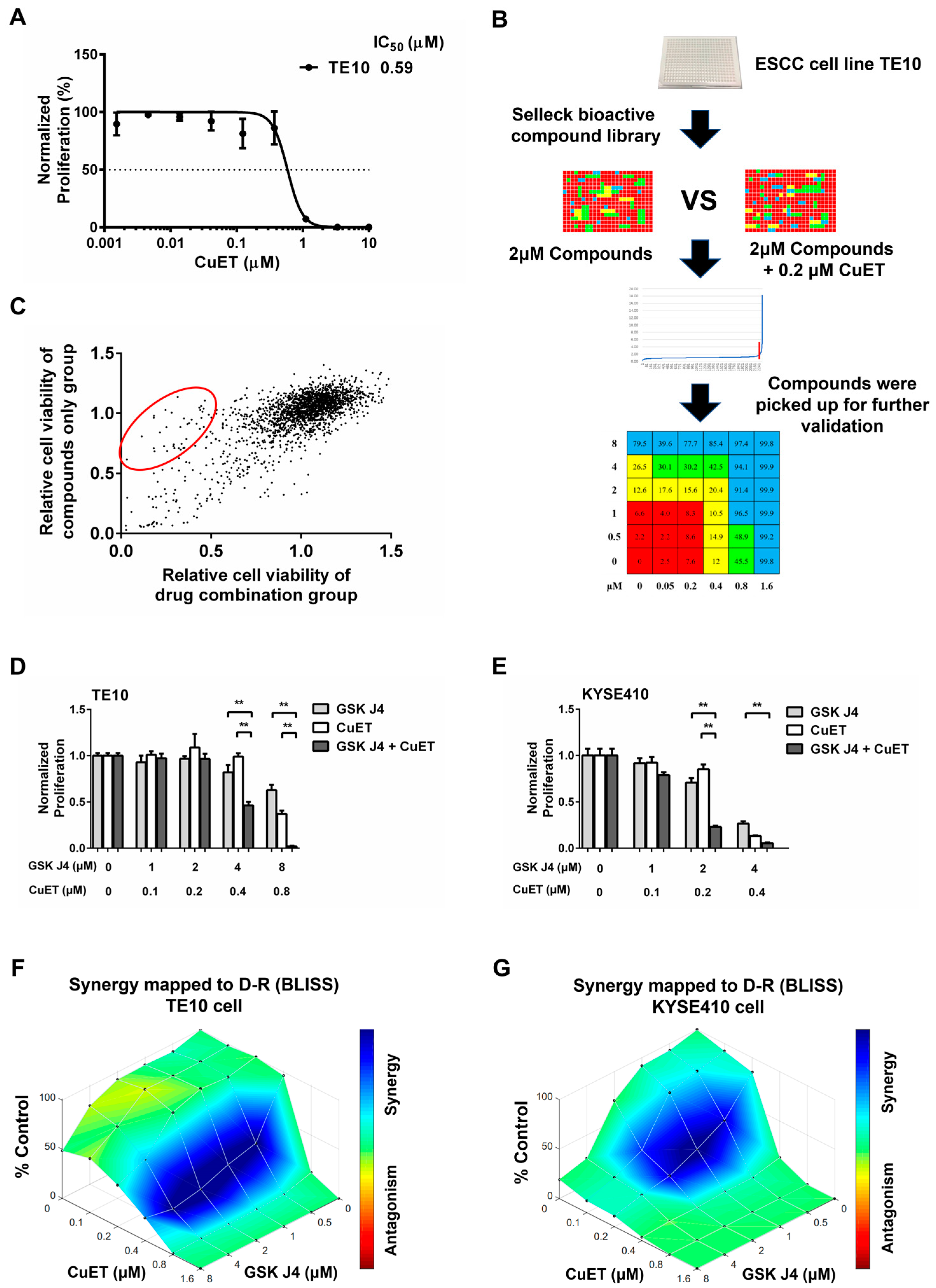
3.1. Identification of Compounds Having Synergistic Effects with CuET
3.2. JMJD3 and UTX Were Highly Expressed in ESCC
3.3. Expression of JMJD3 and UTX Is Associated with the Prognosis of ESCC Patients
3.4. The Combination Effect of CuET with JMJD3/UTX Inhibitor GSK J4
3.5. CuET Combined with GSK J4 Promotes Apoptosis and Cell Cycle Arrest in ESCC Cells
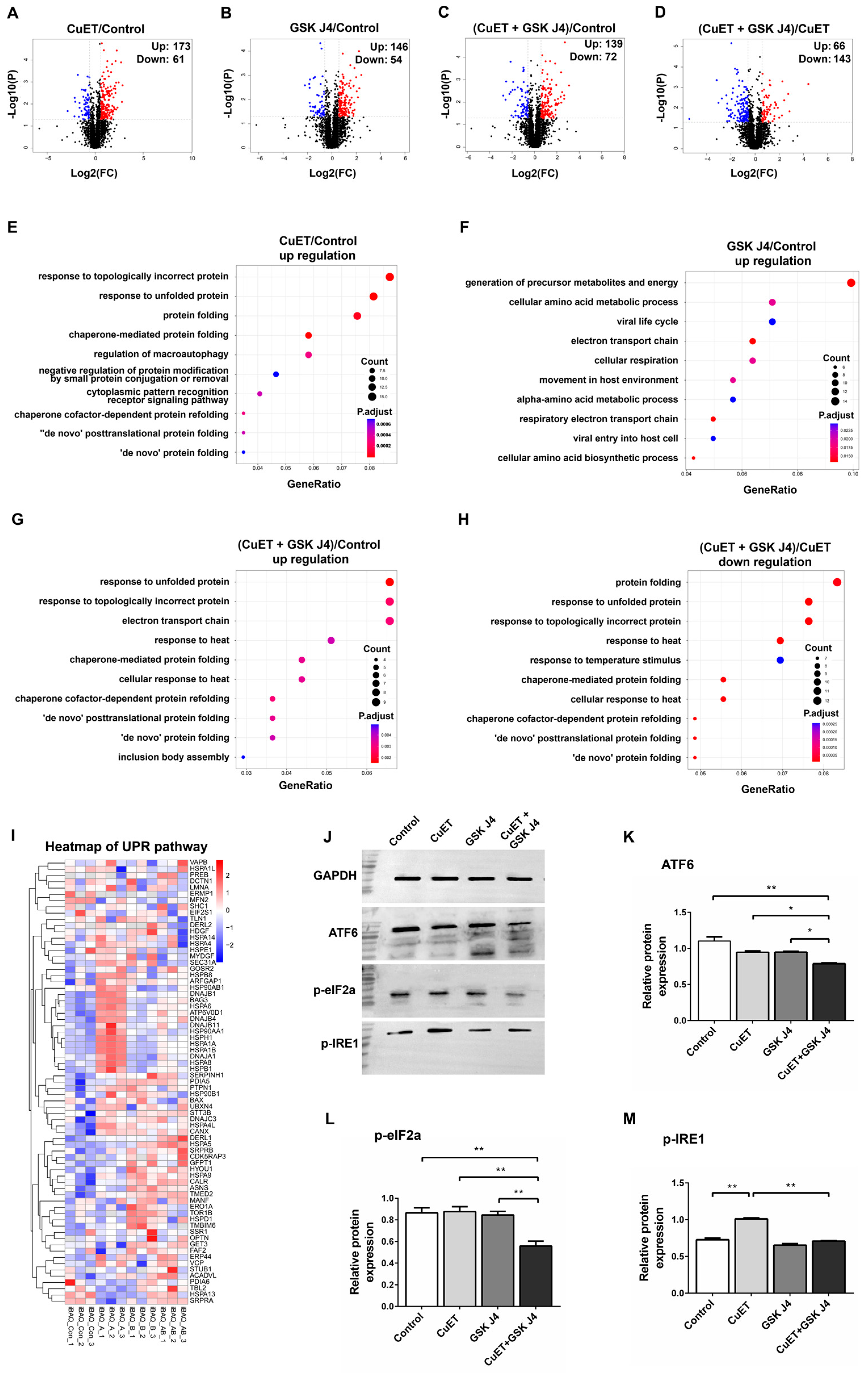
3.6. Proteomic Analysis of ESCC Cells Treated with CuET and GSK J4
3.7. CuET in Combination with GSK J4 Inhibits the Activation of UPR Pathway
3.8. DSF/CuGlu in Combination with GSK J4 Inhibits ESCC Growth In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lagergren, J.; Smyth, E.; Cunningham, D.; Lagergren, P. Oesophageal cancer. Lancet 2017, 390, 2383–2396. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Global Burden of Disease Cancer Collaboration. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar] [CrossRef]
- Zeng, H.; Chen, W.; Zheng, R.; Zhang, S.; Ji, J.S.; Zou, X.; Xia, C.; Sun, K.; Yang, Z.; Li, H.; et al. Changing cancer survival in China during 2003–15: A pooled analysis of 17 population-based cancer registries. Lancet Glob. Health 2018, 6, e555–e567. [Google Scholar] [CrossRef]
- Yang, Y.-M.; Hong, P.; Xu, W.W.; He, Q.-Y.; Li, B. Advances in targeted therapy for esophageal cancer. Signal Transduct. Target. Ther. 2020, 5, 229. [Google Scholar] [CrossRef]
- Shah, M.A.; Kojima, T.; Hochhauser, D.; Enzinger, P.; Raimbourg, J.; Hollebecque, A.; Lordick, F.; Kim, S.-B.; Tajika, M.; Kim, H.T.; et al. Efficacy and Safety of Pembrolizumab for Heavily Pretreated Patients With Advanced, Metastatic Adenocarcinoma or Squamous Cell Carcinoma of the Esophagus: The Phase 2 KEYNOTE-180 Study. JAMA Oncol. 2019, 5, 546–550. [Google Scholar] [CrossRef] [PubMed]
- Cvek, B. Nonprofit drugs as the salvation of the world’s healthcare systems: The case of Antabuse (disulfiram). Drug Discov. Today 2012, 17, 409–412. [Google Scholar] [CrossRef]
- Wiggins, H.L.; Wymant, J.M.; Solfa, F.; Hiscox, S.E.; Taylor, K.M.; Westwell, A.D.; Jones, A.T. Disulfiram-induced cytotoxicity and endo-lysosomal sequestration of zinc in breast cancer cells. Biochem. Pharmacol. 2015, 93, 332–342. [Google Scholar] [CrossRef]
- Lun, X.; Wells, J.C.; Grinshtein, N.; King, J.C.; Hao, X.; Dang, N.-H.; Wang, X.; Aman, A.; Uehling, D.; Datti, A.; et al. Disulfiram when Combined with Copper Enhances the Therapeutic Effects of Temozolomide for the Treatment of Glioblastoma. Clin. Cancer Res. 2016, 22, 3860–3875. [Google Scholar] [CrossRef]
- Jivan, R.; Peres, J.; Damelin, L.H.; Wadee, R.; Veale, R.B.; Prince, S.; Mavri-Damelin, D. Disulfiram with or without metformin inhibits oesophageal squamous cell carcinoma in vivo. Cancer Lett. 2018, 417, 1–10. [Google Scholar] [CrossRef]
- Nechushtan, H.; Hamamreh, Y.; Nidal, S.; Gotfried, M.; Baron, A.; Shalev, Y.I.; Nisman, B.; Peretz, T.; Peylan-Ramu, N. A phase IIb trial assessing the addition of disulfiram to chemotherapy for the treatment of metastatic non-small cell lung cancer. Oncologist 2015, 20, 366–367. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Lu, L.; Luo, J.; Wang, L.; Zhang, Q.; Cao, J.; Jiao, Y. Disulfiram Alone Functions as a Radiosensitizer for Pancreatic Cancer Both In Vitro and In Vivo. Front. Oncol. 2021, 11, 683695. [Google Scholar] [CrossRef] [PubMed]
- Cvek, B. Targeting malignancies with disulfiram (Antabuse): Multidrug resistance, angiogenesis, and proteasome. Curr. Cancer Drug Targets 2011, 11, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Skrott, Z.; Mistrik, M.; Andersen, K.K.; Friis, S.; Majera, D.; Gursky, J.; Ozdian, T.; Bartkova, J.; Turi, Z.; Moudry, P.; et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature 2017, 552, 194–199. [Google Scholar] [CrossRef]
- Ding, N.; Zhu, Q. Disulfiram combats cancer via crippling valosin-containing protein/p97 segregase adaptor NPL4. Transl. Cancer Res. 2018, 7 (Suppl. S4), S495–S499. [Google Scholar] [CrossRef]
- Arcipowski, K.M.; Martinez, C.A.; Ntziachristos, P. Histone demethylases in physiology and cancer: A tale of two enzymes, JMJD3 and UTX. Curr. Opin. Genet. Dev. 2016, 36, 59–67. [Google Scholar] [CrossRef]
- Farzaneh, M.; Kuchaki, Z.; Sheykhahmad, F.R.; Meybodi, S.M.; Abbasi, Y.; Gholami, E.; Ghaedrahmati, F.; Anbiyaee, O. Emerging roles of JMJD3 in cancer. Clin. Transl. Oncol. 2022, 24, 1238–1249. [Google Scholar] [CrossRef]
- Schulz, W.A.; Lang, A.; Koch, J.; Greife, A. The histone demethylase UTX/KDM6A in cancer: Progress and puzzles. Int. J. Cancer 2019, 145, 614–620. [Google Scholar] [CrossRef]
- Bijnsdorp, I.V.; Giovannetti, E.; Peters, G.J. Analysis of drug interactions. Methods Mol. Biol. 2011, 731, 421–434. [Google Scholar]
- Chou, T.-C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef]
- Liu, Q.; Yin, X.; Languino, L.R.; Altieri, D.C. Evaluation of drug combination effect using a Bliss independence dose-response surface model. Stat. Biopharm. Res. 2018, 10, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, A.; Chevet, E. Driving Cancer Tumorigenesis and Metastasis Through UPR Signaling. Coord. Org. Physiol. Through Unfolded Protein Response 2018, 414, 159–192. [Google Scholar] [CrossRef]
- Lin, Y.; Jiang, M.; Chen, W.; Zhao, T.; Wei, Y. Cancer and ER stress: Mutual crosstalk between autophagy, oxidative stress and inflammatory response. Biomed. Pharmacother. 2019, 118, 109249. [Google Scholar] [CrossRef] [PubMed]
- Walczak, A.; Gradzik, K.; Kabzinski, J.; Przybylowska-Sygut, K.; Majsterek, I. The Role of the ER-Induced UPR Pathway and the Efficacy of Its Inhibitors and Inducers in the Inhibition of Tumor Progression. Oxidative Med. Cell. Longev. 2019, 2019, 5729710. [Google Scholar] [CrossRef] [PubMed]
- Lang, B.J.; Guerrero, M.E.; Prince, T.L.; Okusha, Y.; Bonorino, C.; Calderwood, S.K. The functions and regulation of heat shock proteins; key orchestrators of proteostasis and the heat shock response. Arch. Toxicol. 2021, 95, 1943–1970. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-H.; Lu, H.-I.; Chen, Y.-H.; Lo, C.-M.; Huang, W.-T.; Tien, W.-Y.; Lan, Y.-C.; Tsai, H.-T.; Chen, C.-H. JMJD3 expression is an independent prognosticator in patients with esophageal squamous cell carcinoma. Surgery 2019, 165, 946–952. [Google Scholar] [CrossRef]
- Tokunaga, R.; Sakamoto, Y.; Nakagawa, S.; Miyake, K.; Izumi, D.; Kosumi, K.; Taki, K.; Higashi, T.; Imamura, Y.; Ishimoto, T.; et al. The Prognostic Significance of Histone Lysine Demethylase JMJD3/KDM6B in Colorectal Cancer. Ann. Surg. Oncol. 2016, 23, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Xun, J.; Gao, R.; Wang, B.; Li, Y.; Ma, Y.; Guan, J.; Zhang, Q. Histone demethylase KDM6B inhibits breast cancer metastasis by regulating Wnt/β-catenin signaling. FEBS Open Bio 2021, 11, 2273–2281. [Google Scholar] [CrossRef]
- Kim, J.-H.; Sharma, A.; Dhar, S.S.; Lee, S.-H.; Gu, B.; Chan, C.-H.; Lin, H.-K.; Lee, M.G. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014, 74, 1705–1717. [Google Scholar] [CrossRef]
- Kim, G.J.; Kim, D.-H.; Min, K.-W.; Chae, S.W.; Kim, S.H.; Son, B.K.; Moon, K.M.; Kim, Y.H. Expression of UTX Indicates Poor Prognosis in Patients With Luminal Breast Cancer and is Associated With MMP-11 Expression. Appl. Immunohistochem. Mol. Morphol. 2020, 28, 544–550. [Google Scholar] [CrossRef]
- van Haaften, G.; Dalgliesh, G.L.; Davies, H.; Chen, L.; Bignell, G.; Greenman, C.; Edkins, S.; Hardy, C.; O’Meara, S.; Teague, J.; et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 2009, 41, 521–523. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, B.; Nielsen, J.M.; Hudlebusch, H.R.; Lees, M.J.; Larsen, D.V.; Boesen, T.; Labelle, M.; Gerlach, L.-O.; Birk, P.; Helin, K. Inhibition of demethylases by GSK-J1/J4. Nature 2014, 514, E1–E2. [Google Scholar] [CrossRef] [PubMed]
- Lochmann, T.L.; Powell, K.M.; Ham, J.; Floros, K.V.; Heisey, D.A.R.; Kurupi, R.I.J.; Calbert, M.L.; Ghotra, M.S.; Greninger, P.; Dozmorov, M.; et al. Targeted inhibition of histone H3K27 demethylation is effective in high-risk neuroblastoma. Sci. Transl. Med. 2018, 10, eaao4680. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Zhong, L.; Yu, L.; Xiong, L.; Li, J.; Dan, W.; Ye, J.; Liu, C.; Luo, X.; Liu, B. GSK-J4 induces cell cycle arrest and apoptosis via ER stress and the synergism between GSK-J4 and decitabine in acute myeloid leukemia KG-1a cells. Cancer Cell Int. 2020, 20, 209. [Google Scholar] [CrossRef]




| Features | Cases (n) | Expression of JMJD3 | χ2 | p-Value | Expression of UTX | χ2 | p-Value | ||
|---|---|---|---|---|---|---|---|---|---|
| Low | High | Low | High | ||||||
| Gender | |||||||||
| Male | 55 | 27 (49.1%) | 28 (50.9%) | 0.005 | 0.944 | 28 (50.9%) | 27 (49.1%) | 0.005 | 0.944 |
| Female | 20 | 10 (50.0%) | 10 (50.0%) | 10 (50.0%) | 10 (50.0%) | ||||
| Age (years) | |||||||||
| <65 | 32 | 18 (56.3%) | 14 (43.8%) | 1.068 | 0.301 | 18 (56.3%) | 14 (43.8%) | 0.696 | 0.404 |
| ≥65 | 43 | 19 (44.2%) | 24 (55.8%) | 20 (46.5%) | 23 (53.5%) | ||||
| Length of tumor (cm) | |||||||||
| ≤4 | 37 | 20 (50.0%) | 20 (50.0%) | 0.015 | 0.902 | 22 (55.0%) | 18 (45.0%) | 0.644 | 0.422 |
| >4 | 38 | 17 (48.6%) | 18 (51.4%) | 16 (45.7%) | 19 (54.3%) | ||||
| Tumor location | |||||||||
| Upper Middle | 58 | 28 (48.3%) | 30 (51.7%) | 0.114 | 0.735 | 29 (50.0%) | 29 (50.0%) | 0.045 | 0.831 |
| Lower paragraph | 17 | 9 (52.9%) | 8 (47.1%) | 9 (52.9%) | 8 (47.1%) | ||||
| Differentiation | |||||||||
| Well + moderately | 42 | 17 (40.5%) | 25 (59.5%) | 2.996 | 0.083 | 21 (50.0%) | 21 (50.0%) | 0.017 | 0.896 |
| Poorly | 33 | 20 (60.6%) | 13 (39.4%) | 17 (51.5%) | 16 (48.5%) | ||||
| T Stage | |||||||||
| 1–2 | 17 | 13 (76.5%) | 4 (23.5%) | 6.477 | 0.011 * | 13 (76.5%) | 4 (23.5%) | 5.856 | 0.016 * |
| 3–4 | 58 | 24 (41.4%) | 34 (58.6%) | 25 (43.1%) | 33 (56.9%) | ||||
| Lymph node metastasis | |||||||||
| None | 39 | 19 (48.7%) | 20 (51.3%) | 0.012 | 0.912 | 23 (59.0%) | 16 (41.0%) | 2.243 | 0.134 |
| Yes | 36 | 18 (50.0%) | 18 (50.0%) | 15 (41.7%) | 21 (58.3%) | ||||
| TNM Staging | |||||||||
| Phase I–II | 41 | 21 (51.2%) | 20 (48.8%) | 0.129 | 0.72 | 25 (61.0%) | 16 (39.0%) | 3.845 | 0.05 |
| Phase III–IV | 34 | 16 (47.1%) | 18 (52.9%) | 13 (38.2%) | 21 (61.8%) | ||||
| CuET + GSK J4 | Combination Index at | Dose—Reduction Index at | ||||
|---|---|---|---|---|---|---|
| IC50 | IC75 | IC90 | IC50 | IC75 | IC90 | |
| TE10 | 0.21 | 0.08 | 0.03 | 7.78 a | 17.96 a | 41.45 a |
| 12.94 b | 48.78 b | 183.89 b | ||||
| KYSE410 | 0.67 | 0.33 | 0.17 | 2.49 a | 4.49 a | 8.1 a |
| 3.69 b | 9.10 b | 22.49 b | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Li, F.; Ren, Y.; Zhang, Q.; Jiao, B.; Zhang, J.; Huang, J. Targeting Esophageal Squamous Cell Carcinoma by Combining Copper Ionophore Disulfiram and JMJD3/UTX Inhibitor GSK J4. Cancers 2023, 15, 5347. https://doi.org/10.3390/cancers15225347
Yang C, Li F, Ren Y, Zhang Q, Jiao B, Zhang J, Huang J. Targeting Esophageal Squamous Cell Carcinoma by Combining Copper Ionophore Disulfiram and JMJD3/UTX Inhibitor GSK J4. Cancers. 2023; 15(22):5347. https://doi.org/10.3390/cancers15225347
Chicago/Turabian StyleYang, Canlin, Fei Li, Yuanyuan Ren, Qianqian Zhang, Bo Jiao, Jianming Zhang, and Junxing Huang. 2023. "Targeting Esophageal Squamous Cell Carcinoma by Combining Copper Ionophore Disulfiram and JMJD3/UTX Inhibitor GSK J4" Cancers 15, no. 22: 5347. https://doi.org/10.3390/cancers15225347
APA StyleYang, C., Li, F., Ren, Y., Zhang, Q., Jiao, B., Zhang, J., & Huang, J. (2023). Targeting Esophageal Squamous Cell Carcinoma by Combining Copper Ionophore Disulfiram and JMJD3/UTX Inhibitor GSK J4. Cancers, 15(22), 5347. https://doi.org/10.3390/cancers15225347







