PDZ and LIM Domain-Encoding Genes: Their Role in Cancer Development
Simple Summary
Abstract
1. Introduction
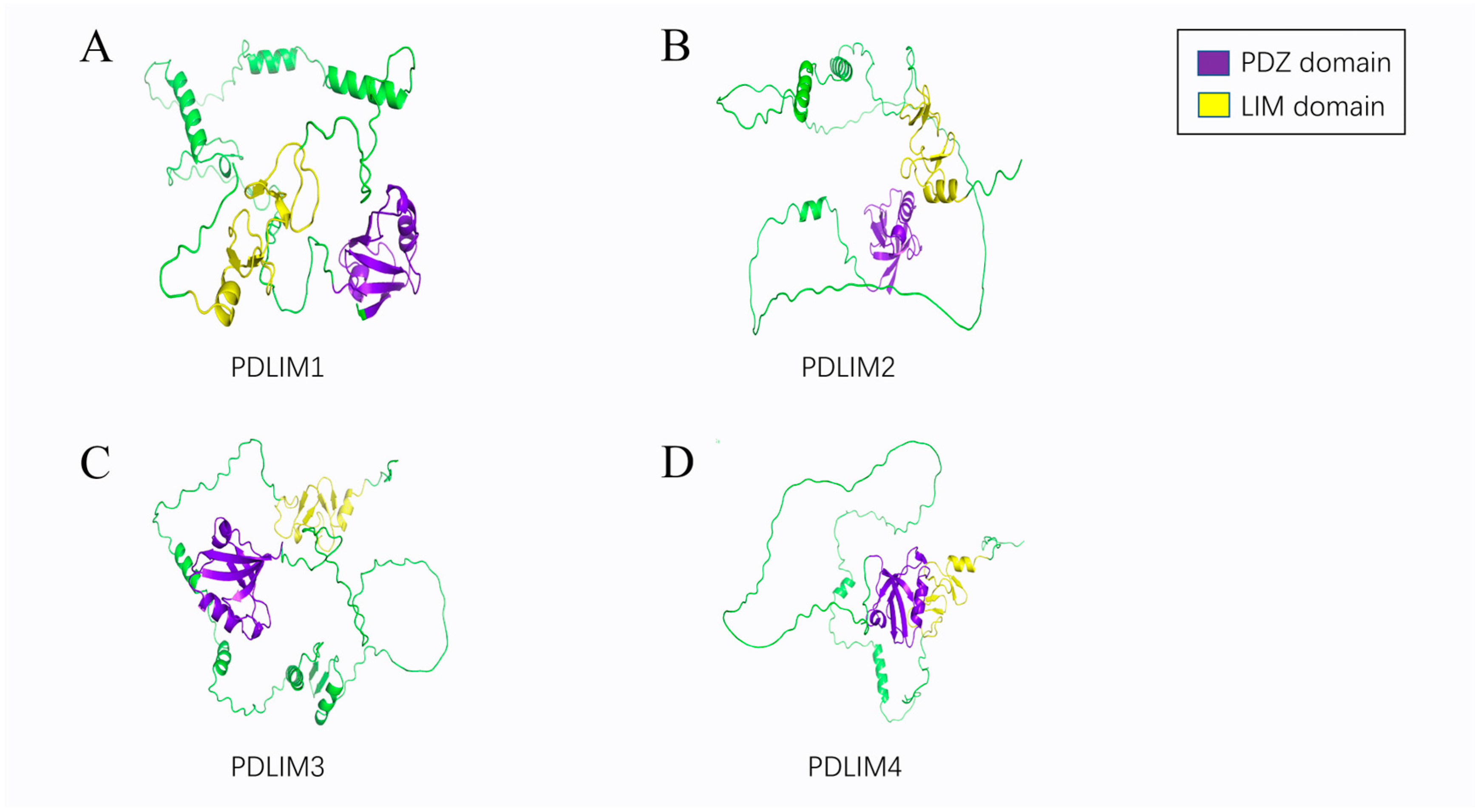
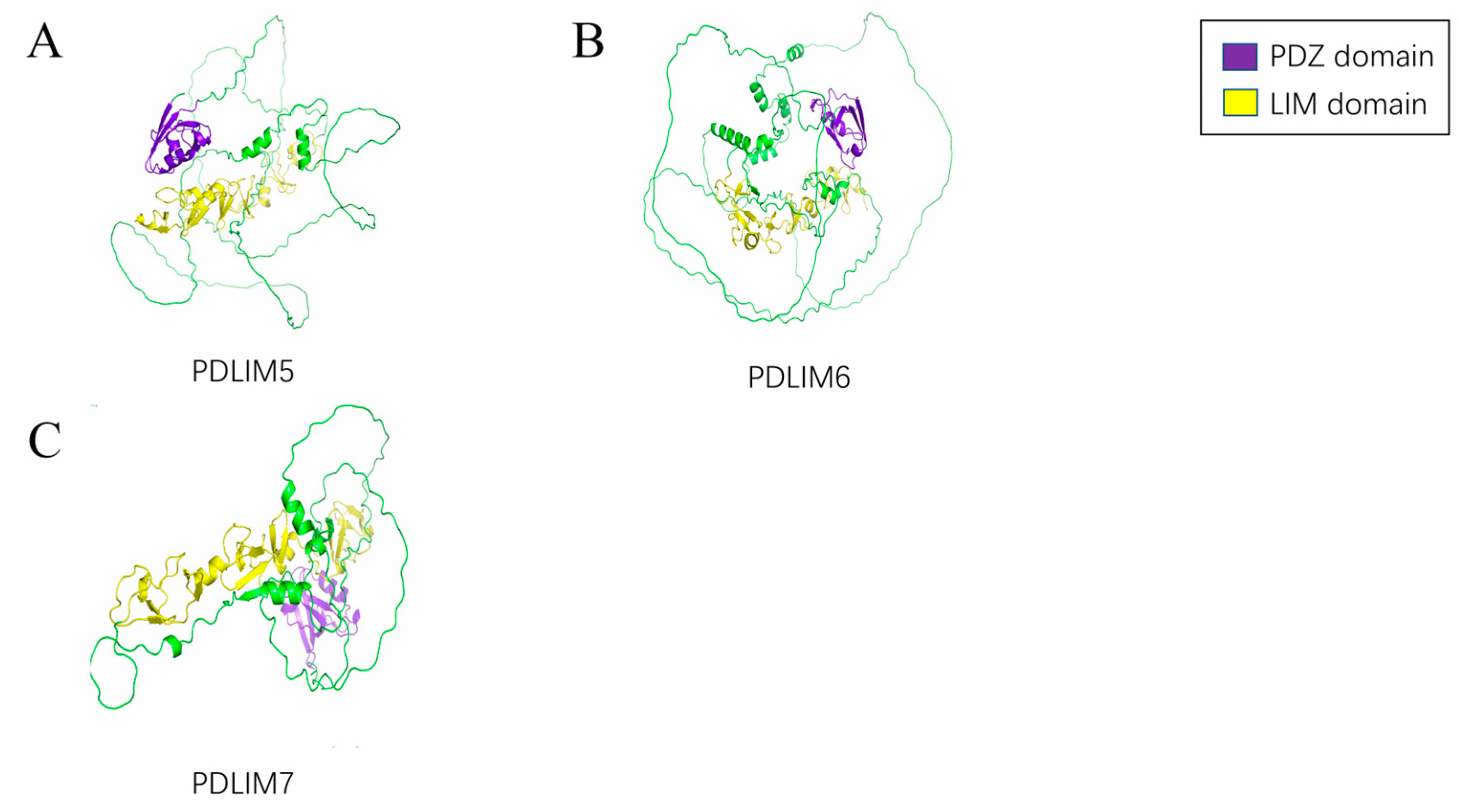
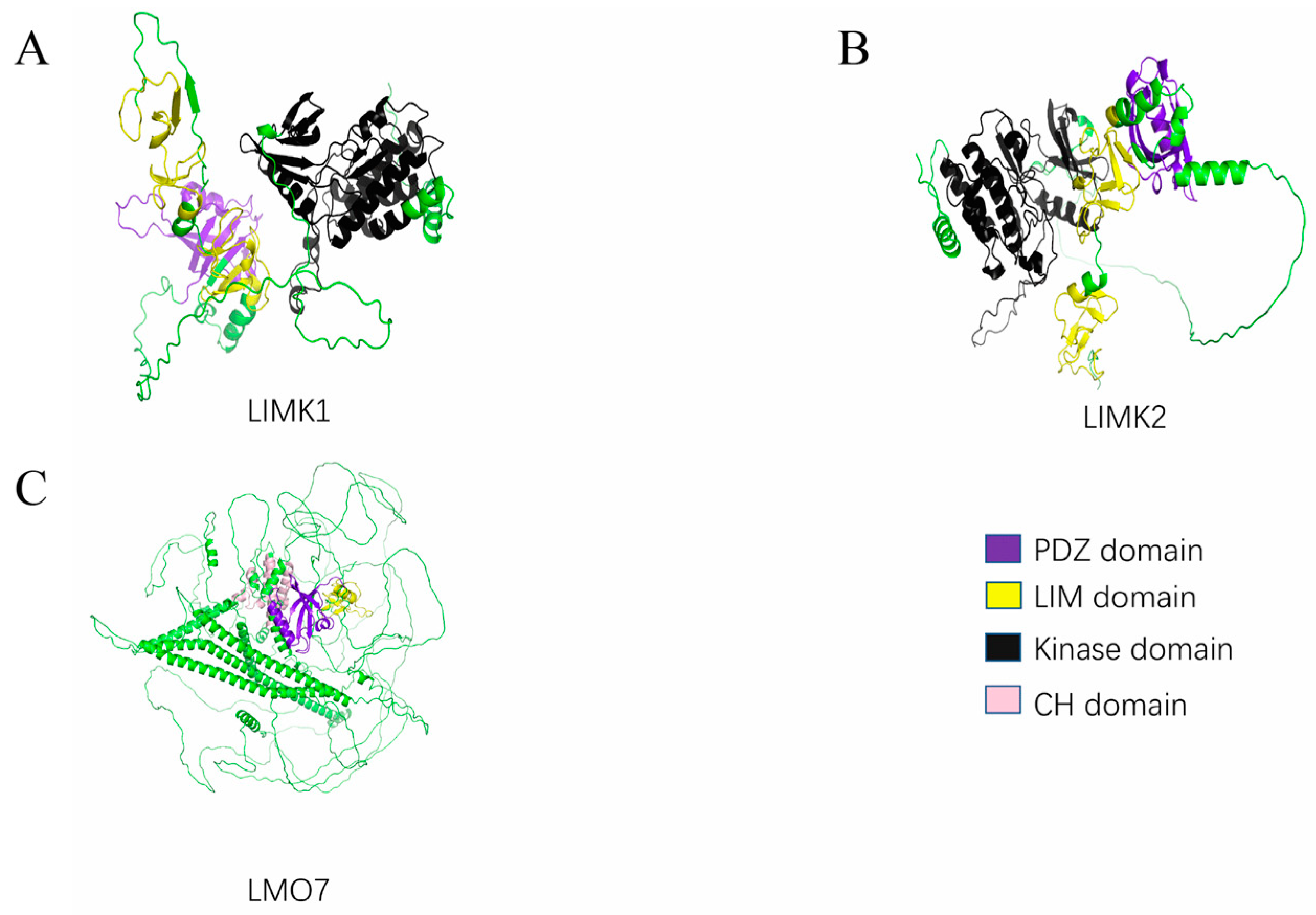
2. Structural Features of the PDLIMs
2.1. PDZ Domain
2.2. LIM Domain
2.3. Other Domains
3. PDLIMs and Signaling Pathways
3.1. Integrin Signaling Pathway
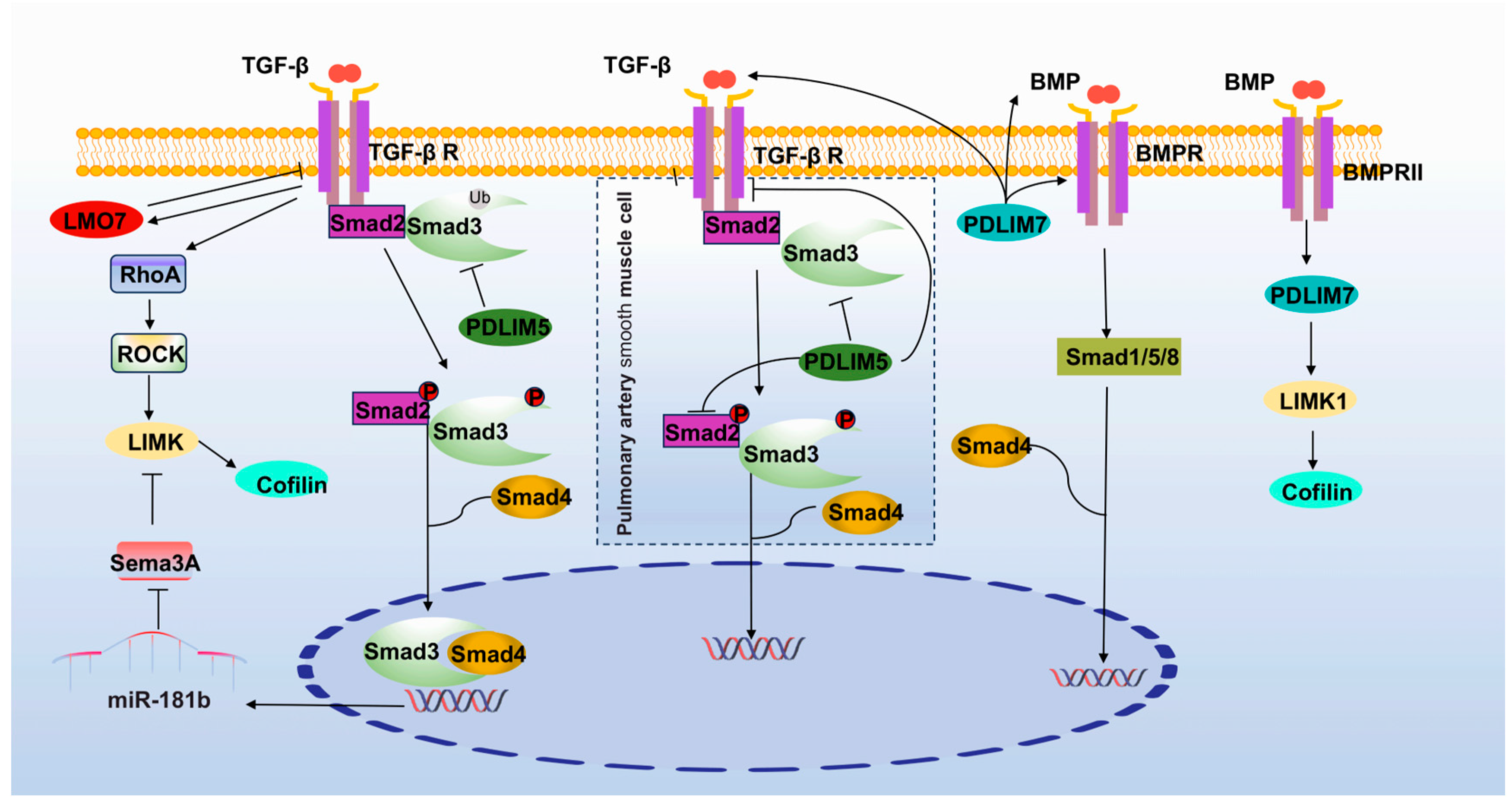
3.2. TGF-β Signaling Pathway
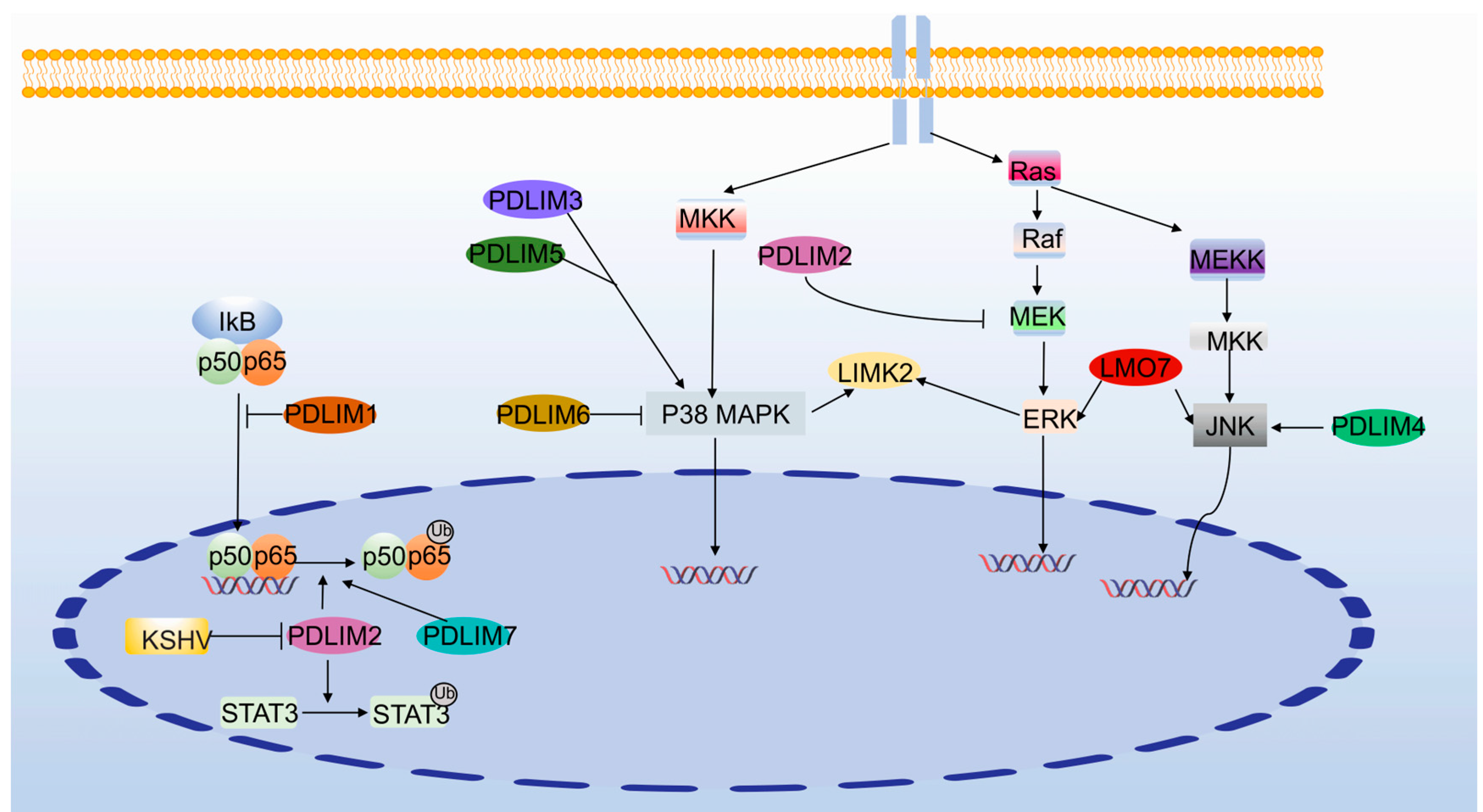
3.3. NF-κB Signaling Pathway
3.4. MAPK Signaling Pathway
4. PDLIMs and Tumor
4.1. ALP Subfamily
4.1.1. PDLIM1
4.1.2. PDLIM2
4.1.3. PDLIM3
4.1.4. PDLIM4
4.2. Enigma Subfamily
4.2.1. PDLIM5
4.2.2. PDLIM6
4.2.3. PDLIM7
4.3. LMO7
4.4. LIM Kinase
4.4.1. LIMK1
LIMK1 in GC
LIMK1 in CRC
LIMK1 in BC
LIMK1 in PC
LIMK1 in LC
LIMK1 in OS
LIMK1 in Cervical Cancer (CC)
LIMK1 in HCC
LIMK1 in Other Tumors
4.4.2. LIMK2
LIMK2 in CRC
LIMK2 in PC
LIMK2 in BC
LIMK2 in LC
LIMK2 in OS
LIMK2 in Neuroblastoma
LIMK2 in Other Tumors
5. Conclusions and Future Directions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DiRusso, C.J.; Dashtiahangar, M.; Gilmore, T.D. Scaffold Proteins as Dynamic Integrators of Biological Processes. J. Biol. Chem. 2022, 298, 102628. [Google Scholar] [CrossRef]
- Shaw, A.S.; Filbert, E.L. Scaffold Proteins and Immune-Cell Signalling. Nat. Rev. Immunol. 2009, 9, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Velthuis, A.J.W.T.; Bagowski, C.P. PDZ and LIM Domain-Encoding Genes: Molecular Interactions and Their Role in Development. Sci. World J. 2007, 7, 1470–1492. [Google Scholar] [CrossRef]
- Vallenius, T.; Luukko, K.; Mäkelä, T.P. CLP-36 PDZ-LIM Protein Associates with Nonmuscle Alpha-Actinin-1 and Alpha-Actinin-4. J. Biol. Chem. 2000, 275, 11100–11105. [Google Scholar] [CrossRef]
- Zhou, Q.; Ruiz-Lozano, P.; Martone, M.E.; Chen, J. Cypher, a Striated Muscle-Restricted PDZ and LIM Domain-Containing Protein, Binds to Alpha-Actinin-2 and Protein Kinase C. J. Biol. Chem. 1999, 274, 19807–19813. [Google Scholar] [CrossRef] [PubMed]
- Krcmery, J.; Camarata, T.; Kulisz, A.; Simon, H.-G. Nucleocytoplasmic Functions of the PDZ-LIM Protein Family: New Insights into Organ Development. Bioessays 2010, 32, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, V.C.; Kyle, W.B.; Kojic, S.; Vitulo, N.; Li, Z.; Belgrano, A.; Maiuri, P.; Banks, L.; Vatta, M.; Valle, G.; et al. ZASP Interacts with the Mechanosensing Protein Ankrd2 and P53 in the Signalling Network of Striated Muscle. PLoS ONE 2014, 9, e92259. [Google Scholar] [CrossRef]
- Lasorella, A.; Iavarone, A. The Protein ENH Is a Cytoplasmic Sequestration Factor for Id2 in Normal and Tumor Cells from the Nervous System. Proc. Natl. Acad. Sci. USA 2006, 103, 4976–4981. [Google Scholar] [CrossRef]
- Piao, S.; Zheng, L.; Zheng, H.; Zhou, M.; Feng, Q.; Zhou, S.; Ke, M.; Yang, H.; Wang, X. High Expression of PDLIM2 Predicts a Poor Prognosis in Prostate Cancer and Is Correlated with Epithelial-Mesenchymal Transition and Immune Cell Infiltration. J. Immunol. Res. 2022, 2022, 2922832. [Google Scholar] [CrossRef]
- Kundu, J.; Bakshi, S.; Joshi, H.; Bhadada, S.K.; Verma, I.; Sharma, S. Proteomic Profiling of Peripheral Blood Mononuclear Cells Isolated from Patients with Tuberculosis and Diabetes Copathogenesis—A Pilot Study. PLoS ONE 2020, 15, e0233326. [Google Scholar] [CrossRef]
- Loughran, G.; Healy, N.C.; Kiely, P.A.; Huigsloot, M.; Kedersha, N.L.; O’Connor, R. Mystique Is a New Insulin-like Growth Factor-I-Regulated PDZ-LIM Domain Protein That Promotes Cell Attachment and Migration and Suppresses Anchorage-Independent Growth. Mol. Biol. Cell 2005, 16, 1811–1822. [Google Scholar] [CrossRef] [PubMed]
- Ooshio, T.; Irie, K.; Morimoto, K.; Fukuhara, A.; Imai, T.; Takai, Y. Involvement of LMO7 in the Association of Two Cell-Cell Adhesion Molecules, Nectin and E-Cadherin, through Afadin and Alpha-Actinin in Epithelial Cells. J. Biol. Chem. 2004, 279, 31365–31373. [Google Scholar] [CrossRef] [PubMed]
- Healy, N.C.; O’connor, R. Sequestration of PDLIM2 in the Cytoplasm of Monocytic/Macrophage Cells Is Associated with Adhesion and Increased Nuclear Activity of NF-kappaB. J. Leukoc. Biol. 2009, 85, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Itoh, M.; Nagafuchi, A.; Yonemura, S.; Kitani-Yasuda, T.; Tsukita, S. The 220-kD Protein Colocalizing with Cadherins in Non-Epithelial Cells Is Identical to ZO-1, a Tight Junction-Associated Protein in Epithelial Cells: cDNA Cloning and Immunoelectron Microscopy. J. Cell Biol. 1993, 121, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.-O.; Hunt, C.A.; Kennedy, M.B. The Rat Brain Postsynaptic Density Fraction Contains a Homolog of the Drosophila Discs-Large Tumor Suppressor Protein. Neuron 1992, 9, 929–942. [Google Scholar] [CrossRef] [PubMed]
- Woods, D.F.; Bryant, P.J. The Discs-Large Tumor Suppressor Gene of Drosophila Encodes a Guanylate Kinase Homolog Localized at Septate Junctions. Cell 1991, 66, 451–464. [Google Scholar] [CrossRef]
- Dev, K.K. Making Protein Interactions Druggable: Targeting PDZ Domains. Nat. Rev. Drug Discov. 2004, 3, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Zhang, F.; Chen, X.; Lin, J.; Shi, J. PDZ Protein Mediated Activity-Dependent LTP/LTD Developmental Switch at Rat Retinocollicular Synapses. Am. J. Physiol. Cell Physiol. 2010, 298, C1572–C1582. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.-X.; Johns, R.A. PDZ Domains at Excitatory Synapses: Potential Molecular Targets for Persistent Pain Treatment. Curr. Neuropharmacol. 2006, 4, 217–223. [Google Scholar] [CrossRef]
- Gallardo, R.; Ivarsson, Y.; Schymkowitz, J.; Rousseau, F.; Zimmermann, P. Structural Diversity of PDZ-Lipid Interactions. Chembiochem 2010, 11, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Lenfant, N.; Polanowska, J.; Bamps, S.; Omi, S.; Borg, J.-P.; Reboul, J. A Genome-Wide Study of PDZ-Domain Interactions in C. Elegans Reveals a High Frequency of Non-Canonical Binding. BMC Genom. 2010, 11, 671. [Google Scholar] [CrossRef]
- Tonikian, R.; Zhang, Y.; Sazinsky, S.L.; Currell, B.; Yeh, J.-H.; Reva, B.; A Held, H.; A Appleton, B.; Evangelista, M.; Wu, Y.; et al. A Specificity Map for the PDZ Domain Family. PLoS Biol. 2008, 6, e239. [Google Scholar] [CrossRef] [PubMed]
- Fanning, A.S.; Anderson, J.M. PDZ Domains: Fundamental Building Blocks in the Organization of Protein Complexes at the Plasma Membrane. J. Clin. Investig. 1999, 103, 767–772. [Google Scholar] [CrossRef]
- Jeleń, F.; Oleksy, A.; Smietana, K.; Otlewski, J. PDZ Domains—Common Players in the Cell Signaling. Acta Biochim. Pol. 2003, 50, 985–1017. [Google Scholar] [CrossRef] [PubMed]
- E Brenman, J.; Chao, D.S.; Gee, S.H.; McGee, A.W.; E Craven, S.; Santillano, D.R.; Wu, Z.; Huang, F.; Xia, H.; Peters, M.F.; et al. Interaction of Nitric Oxide Synthase with the Postsynaptic Density Protein PSD-95 and Alpha1-Syntrophin Mediated by PDZ Domains. Cell 1996, 84, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Ernst, A.; Gfeller, D.; Kan, Z.; Seshagiri, S.; Kim, P.M.; Bader, G.D.; Sidhu, S.S. Coevolution of PDZ Domain-Ligand Interactions Analyzed by High-Throughput Phage Display and Deep Sequencing. Mol. Biosyst. 2010, 6, 1782–1790. [Google Scholar] [CrossRef]
- Grootjans, J.J.; Reekmans, G.; Ceulemans, H.; David, G. Syntenin-Syndecan Binding Requires Syndecan-Synteny and the Co-Operation of Both PDZ Domains of Syntenin. J. Biol. Chem. 2000, 275, 19933–19941. [Google Scholar] [CrossRef]
- Kornau, H.-C.; Schenker, L.T.; Kennedy, M.B.; Seeburg, P.H. Domain Interaction between NMDA Receptor Subunits and the Postsynaptic Density Protein PSD-95. Science 1995, 269, 1737–1740. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Feng, W.; Chen, J.; Chan, L.-N.; Huang, S.; Zhang, M. PDZ Domains of Par-3 as Potential Phosphoinositide Signaling Integrators. Mol. Cell 2007, 28, 886–898. [Google Scholar] [CrossRef]
- Perroy, J.; El Far, O.; Bertaso, F.; Pin, J.; Betz, H.; Bockaert, J.; Fagni, L. PICK1 Is Required for the Control of Synaptic Transmission by the Metabotropic Glutamate Receptor 7. EMBO J. 2002, 21, 2990–2999. [Google Scholar] [CrossRef]
- Gardiol, D. PDZ-Containing Proteins as Targets in Human Pathologies. FEBS J. 2012, 279, 3529. [Google Scholar] [CrossRef]
- Sheng, M.; Sala, C. PDZ Domains and the Organization of Supramolecular Complexes. Annu. Rev. Neurosci. 2001, 24, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Tran, Y.H.; Xu, Z.; Kato, A.; Mistry, A.C.; Goya, Y.; Taira, M.; Brandt, S.J.; Hirose, S. Spliced Isoforms of LIM-Domain-Binding Protein (CLIM/NLI/Ldb) Lacking the LIM-Interaction Domain. J. Biochem. 2006, 140, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Kadrmas, J.L.; Beckerle, M.C. The LIM Domain: From the Cytoskeleton to the Nucleus. Nat. Rev. Mol. Cell Biol. 2004, 5, 920–931. [Google Scholar] [CrossRef]
- Pérez-Alvarado, G.C.; Miles, C.; Michelsen, J.W.; Louis, H.A.; Winge, D.R.; Beckerle, M.C.; Summers, M.F. Structure of the Carboxy-Terminal LIM Domain from the Cysteine Rich Protein CRP. Nat. Struct. Biol. 1994, 1, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Fan, Z.; Liang, C.; Li, L.; Wang, L.; Liang, Y.; Wu, J.; Chang, S.; Yan, Z.; Lv, Z.; et al. A Signature Motif in LIM Proteins Mediates Binding to Checkpoint Proteins and Increases Tumour Radiosensitivity. Nat. Commun. 2017, 8, 14059. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.A.; Kovar, D.R.; Gardel, M.L.; Winkelman, J.D. LIM Domain Proteins in Cell Mechanobiology. Cytoskeleton 2021, 78, 303–311. [Google Scholar] [CrossRef]
- Wu, R.Y.; Gill, G.N. LIM Domain Recognition of a Tyrosine-Containing Tight Turn. J. Biol. Chem. 1994, 269, 25085–25090. [Google Scholar] [CrossRef]
- Dawid, I.B.; Breen, J.J.; Toyama, R. LIM Domains: Multiple Roles as Adapters and Functional Modifiers in Protein Interactions. Trends Genet. 1998, 14, 156–162. [Google Scholar] [CrossRef]
- Schiller, H.B.; Friedel, C.C.; Boulegue, C.; Fässler, R. Quantitative Proteomics of the Integrin Adhesome Show a Myosin II-Dependent Recruitment of LIM Domain Proteins. EMBO Rep. 2011, 12, 259–266. [Google Scholar] [CrossRef]
- Germain, P.; Delalande, A.; Pichon, C. Role of Muscle LIM Protein in Mechanotransduction Process. Int. J. Mol. Sci. 2022, 23, 9785. [Google Scholar] [CrossRef] [PubMed]
- Bouaouina, M.; Jani, K.; Long, J.Y.; Czerniecki, S.; Morse, E.M.; Ellis, S.J.; Tanentzapf, G.; Schöck, F.; Calderwood, D.A. Zasp Regulates Integrin Activation. J. Cell Sci. 2012, 125 Pt 23, 5647–5657. [Google Scholar] [CrossRef]
- Matthews, J.M.; Lester, K.; Joseph, S.; Curtis, D.J. LIM-Domain-Only Proteins in Cancer. Nat. Rev. Cancer 2013, 13, 111–122. [Google Scholar] [CrossRef] [PubMed]
- González-Morales, N.; Xiao, Y.S.; Schilling, M.A.; Marescal, O.; Liao, K.A.; Schöck, F. Myofibril Diameter Is Set by a Finely Tuned Mechanism of Protein Oligomerization in Drosophila. eLife 2019, 8, e50496. [Google Scholar] [CrossRef]
- She, M.; Tang, M.; Jiang, T.; Zeng, Q. The Roles of the LIM Domain Proteins in Drosophila Cardiac and Hematopoietic Morphogenesis. Front. Cardiovasc. Med. 2021, 8, 616851. [Google Scholar] [CrossRef]
- Jani, K.; Schöck, F. Zasp Is Required for the Assembly of Functional Integrin Adhesion Sites. J. Cell Biol. 2007, 179, 1583–1597. [Google Scholar] [CrossRef] [PubMed]
- Klaavuniemi, T.; Kelloniemi, A.; Ylänne, J. The ZASP-like Motif in Actinin-Associated LIM Protein Is Required for Interaction with the Alpha-Actinin Rod and for Targeting to the Muscle Z-Line. J. Biol. Chem. 2004, 279, 26402–26410. [Google Scholar] [CrossRef]
- Yang, N.; Higuchi, O.; Ohashi, K.; Nagata, K.; Wada, A.; Kangawa, K.; Nishida, E.; Mizuno, K. Cofilin Phosphorylation by LIM-Kinase 1 and Its Role in Rac-Mediated Actin Reorganization. Nature 1998, 393, 809–812. [Google Scholar] [CrossRef]
- Manetti, F. LIM Kinases Are Attractive Targets with Many Macromolecular Partners and Only a Few Small Molecule Regulators. Med. Res. Rev. 2012, 32, 968–998. [Google Scholar] [CrossRef]
- Yin, L.-M.; Schnoor, M.; Jun, C.-D. Structural Characteristics, Binding Partners and Related Diseases of the Calponin Homology (CH) Domain. Front. Cell Dev. Biol. 2020, 8, 342. [Google Scholar] [CrossRef]
- Cox, D.; Brennan, M.; Moran, N. Integrins as Therapeutic Targets: Lessons and Opportunities. Nat. Rev. Drug Discov. 2010, 9, 804–820. [Google Scholar] [CrossRef]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in Cancer: Biological Implications and Therapeutic Opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef]
- Campbell, I.D.; Humphries, M.J. Integrin Structure, Activation, and Interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a004994. [Google Scholar] [CrossRef] [PubMed]
- Horton, E.R.; Astudillo, P.; Humphries, M.J.; Humphries, J.D. Mechanosensitivity of Integrin Adhesion Complexes: Role of the Consensus Adhesome. Exp. Cell Res. 2016, 343, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Yao, Y.; Yue, Q.-X.; Zhou, X.-W.; Yang, P.-Y.; Wu, W.-Y.; Guan, S.-H.; Jiang, B.-H.; Yang, M.; Liu, X.; et al. Differential Proteomic Analysis of Platelets Suggested Possible Signal Cascades Network in Platelets Treated with Salvianolic Acid B. PLoS ONE 2011, 6, e14692. [Google Scholar] [CrossRef]
- Lv, J.; Pan, Z.; Chen, J.; Xu, R.; Wang, D.; Huang, J.; Dong, Y.; Jiang, J.; Yin, X.; Cheng, H.; et al. Phosphoproteomic Analysis Reveals Downstream PKA Effectors of AKAP Cypher/ZASP in the Pathogenesis of Dilated Cardiomyopathy. Front. Cardiovasc. Med. 2021, 8, 753072. [Google Scholar] [CrossRef] [PubMed]
- Cox, O.T.; O’shea, S.; Tresse, E.; Bustamante-Garrido, M.; Kiran-Deevi, R.; O’connor, R. iGF-1 Receptor and Adhesion Signaling: An Important Axis in Determining Cancer Cell Phenotype and Therapy Resistance. Front. Endocrinol. 2015, 6, 106. [Google Scholar] [CrossRef] [PubMed]
- Elbediwy, A.; Vanyai, H.; Diaz-De-La-Loza, M.-D.; Frith, D.; Snijders, A.P.; Thompson, B.J. Enigma Proteins Regulate YAP Mechanotransduction. J. Cell Sci. 2018, 131, jcs221788. [Google Scholar] [CrossRef]
- Wozniak, M.A.; Baker, B.M.; Chen, C.S.; Wilson, K.L. The Emerin-Binding Transcription Factor Lmo7 Is Regulated by Association with p130Cas at Focal Adhesions. PeerJ 2013, 1, e134. [Google Scholar] [CrossRef]
- Chen, M.; Sinha, M.; Luxon, B.A.; Bresnick, A.R.; O’Connor, K.L. Integrin Alpha6beta4 Controls the Expression of Genes Associated with Cell Motility, Invasion, and Metastasis, Including S100A4/Metastasin. J. Biol. Chem. 2009, 284, 1484–1494. [Google Scholar] [CrossRef]
- Shibue, T.; Brooks, M.W.; Weinberg, R.A. An Integrin-Linked Machinery of Cytoskeletal Regulation That Enables Experimental Tumor Initiation and Metastatic Colonization. Cancer Cell 2013, 24, 481–498. [Google Scholar] [CrossRef] [PubMed]
- Lui, W.-Y.; Lee, W.M.; Cheng, C.Y. Sertoli-Germ Cell Adherens Junction Dynamics in the Testis Are Regulated by RhoB GTPase via the ROCK/LIMK Signaling Pathway. Biol. Reprod. 2003, 68, 2189–2206. [Google Scholar] [CrossRef] [PubMed]
- Loubet, D.; Dakowski, C.; Pietri, M.; Pradines, E.; Bernard, S.; Callebert, J.; Kellermann, O.; Schneider, B.; Ardila-Osorio, H.; Mouillet-Richard, S.; et al. Neuritogenesis: The Prion Protein Controls Β1 Integrin Signaling Activity. FASEB J. 2012, 26, 678–690. [Google Scholar] [CrossRef]
- Peng, D.; Fu, M.; Wang, M.; Wei, Y.; Wei, X. Targeting TGF-β Signal Transduction for Fibrosis and Cancer Therapy. Mol. Cancer 2022, 21, 104. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.; Wickman, G.; Munro, J.; Inman, G.J.; Olson, M.F. Differing Contributions of LIMK and ROCK to TGFβ-Induced Transcription, Motility and Invasion. Eur. J. Cell Biol. 2011, 90, 13–25. [Google Scholar] [CrossRef]
- Lee, J.; Ko, M.; Joo, C.-K. Rho Plays a Key Role in TGF-Beta1-Induced Cytoskeletal Rearrangement in Human Retinal Pigment Epithelium. J. Cell Physiol. 2008, 216, 520–526. [Google Scholar] [CrossRef]
- Lai, Y.-J.; Tsai, F.-C.; Chang, G.-J.; Chang, S.-H.; Huang, C.-C.; Chen, W.-J.; Yeh, Y.-H. miR-181b Targets Semaphorin 3A to Mediate TGF-β-Induced Endothelial-Mesenchymal Transition Related to Atrial Fibrillation. J. Clin. Investig. 2022, 132, e142548. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, X.; Xu, Z.; He, Y.; Guo, C.; He, L.; Huan, C.; Cai, C.; Huang, J.; Zhang, J.; et al. PDLIM5 Inhibits STUB1-Mediated Degradation of SMAD3 and Promotes the Migration and Invasion of Lung Cancer Cells. J. Biol. Chem. 2020, 295, 13798–13811. [Google Scholar] [CrossRef]
- Cheng, H.; Chen, T.; Tor, M.; Park, D.; Zhou, Q.; Huang, J.B.; Khatib, N.; Rong, L.; Zhou, G. A High-Throughput Screening Platform Targeting PDLIM5 for Pulmonary Hypertension. J. Biomol. Screen. 2016, 21, 333–341. [Google Scholar] [CrossRef]
- Xie, Y.; Ostriker, A.C.; Jin, Y.; Hu, H.; Sizer, A.J.; Peng, G.; Morris, A.H.; Ryu, C.; Herzog, E.L.; Kyriakides, T.; et al. LMO7 Is a Negative Feedback Regulator of Transforming Growth Factor β Signaling and Fibrosis. Circulation 2019, 139, 679–693. [Google Scholar] [CrossRef]
- Nakamura, H.; Mukai, M.; Komatsu, K.; Tanaka-Okamoto, M.; Itoh, Y.; Ishizaki, H.; Tatsuta, M.; Inoue, M.; Miyoshi, J. Transforming Growth Factor-Beta1 Induces LMO7 While Enhancing the Invasiveness of Rat Ascites Hepatoma Cells. Cancer Lett. 2005, 220, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, X.; Chen, Z.; Liu, G.; Chen, Z. Semi-Quantitative RT-PCR Analysis of LIM Mineralization Protein 1 and Its Associated Molecules in Cultured Human Dental Pulp Cells. Arch. Oral. Biol. 2007, 52, 720–726. [Google Scholar] [CrossRef]
- Minamide, A.; Boden, S.D.; Viggeswarapu, M.; Hair, G.A.; Oliver, C.; Titus, L. Mechanism of Bone Formation with Gene Transfer of the cDNA Encoding for the Intracellular Protein LMP-1. J. Bone Jt. Surg. Am. 2003, 85, 1030–1039. [Google Scholar] [CrossRef]
- Ma, J.; Guo, W.; Gao, M.; Huang, B.; Qi, Q.; Ling, Z.; Chen, Y.; Hu, H.; Zhou, H.; Yu, F.; et al. Biomimetic Matrix Fabricated by LMP-1 Gene-Transduced MC3T3-E1 Cells for Bone Regeneration. Biofabrication 2017, 9, 045010. [Google Scholar] [CrossRef]
- Boden, S.D. Biology of Lumbar Spine Fusion and Use of Bone Graft Substitutes: Present, Future, and next Generation. Tissue Eng. 2000, 6, 383–399. [Google Scholar] [CrossRef]
- Jiang, X.; Chen, Y.; Fan, X.; Zhang, H.; Kun, L. Osteogenesis and Mineralization in a Rabbit Mandibular Distraction Osteogenesis Model Is Promoted by the Human LMP-1 Gene. J. Orthop. Res. 2015, 33, 521–526. [Google Scholar] [CrossRef]
- Yoon, S.T.; Park, J.S.; Kim, K.S.; Li, J.; Attallah-Wasif, E.S.; Hutton, W.C.; Boden, S.D. ISSLS Prize Winner: LMP-1 Upregulates Intervertebral Disc Cell Production of Proteoglycans and BMPs in Vitro and in Vivo. Spine (Phila. Pa. 1976) 2004, 29, 2603–2611. [Google Scholar] [CrossRef]
- Park, J.S.; Nagata, K. [BMP and LMP-1 for intervertebral disc regeneration]. Clin. Calcium 2004, 14, 76–78. [Google Scholar] [PubMed]
- Mu, R.; Chen, B.; Bi, B.; Yu, H.; Liu, J.; Li, J.; He, M.; Rong, L.; Liu, B.; Liu, K.; et al. LIM Mineralization Protein-1 Enhances the Committed Differentiation of Dental Pulp Stem Cells through the ERK1/2 and P38 MAPK Pathways and BMP Signaling. Int. J. Med. Sci. 2022, 19, 1307–1319. [Google Scholar] [CrossRef]
- Matsuura, I.; Endo, M.; Hata, K.; Kubo, T.; Yamaguchi, A.; Saeki, N.; Yamashita, T. BMP Inhibits Neurite Growth by a Mechanism Dependent on LIM-Kinase. Biochem. Biophys. Res. Commun. 2007, 360, 868–873. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Ben-Neriah, Y. Phosphorylation Meets Ubiquitination: The Control of NF- [Kappa]B Activity. Annu. Rev. Immunol. 2000, 18, 621–663. [Google Scholar] [CrossRef] [PubMed]
- Ono, R.; Kaisho, T.; Tanaka, T. PDLIM1 Inhibits NF-κB-Mediated Inflammatory Signaling by Sequestering the P65 Subunit of NF-κB in the Cytoplasm. Sci. Rep. 2015, 5, 18327. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Grusby, M.J.; Kaisho, T. PDLIM2-Mediated Termination of Transcription Factor NF-kappaB Activation by Intranuclear Sequestration and Degradation of the P65 Subunit. Nat. Immunol. 2007, 8, 584–591. [Google Scholar] [CrossRef]
- Jodo, A.; Shibazaki, A.; Onuma, A.; Kaisho, T.; Tanaka, T. PDLIM7 Synergizes With PDLIM2 and P62/Sqstm1 to Inhibit Inflammatory Signaling by Promoting Degradation of the P65 Subunit of NF-κB. Front. Immunol. 2020, 11, 1559. [Google Scholar] [CrossRef]
- Guo, Q.; Xu, J.; Shi, Q.; Wu, S. PDLIM2 Protects Articular Chondrocytes from Lipopolysaccharide-Induced Apoptosis, Degeneration and Inflammatory Injury through down-Regulation of Nuclear Factor (NF)-κB Signaling. Int. Immunopharmacol. 2020, 88, 106883. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.-R.; Tang, F.-J.; Zhang, X.; Wang, H. Suppression of NF-κB Activation by PDLIM2 Restrains Hepatic Lipogenesis and Inflammation in High Fat Diet Induced Mice. Biochem. Biophys. Res. Commun. 2018, 503, 564–571. [Google Scholar] [CrossRef]
- Sun, F.; Li, L.; Yan, P.; Zhou, J.; Shapiro, S.D.; Xiao, G.; Qu, Z. Causative Role of PDLIM2 Epigenetic Repression in Lung Cancer and Therapeutic Resistance. Nat. Commun. 2019, 10, 5324. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Xiao, Y.; Qu, Z. Oncovirus Kaposi Sarcoma Herpesvirus (KSHV) Represses Tumor Suppressor PDLIM2 to Persistently Activate Nuclear Factor κB (NF-κB) and STAT3 Transcription Factors for Tumorigenesis and Tumor Maintenance. J. Biol. Chem. 2015, 290, 7362–7368. [Google Scholar] [CrossRef]
- Fang, J.Y.; Richardson, B.C. The MAPK Signalling Pathways and Colorectal Cancer. Lancet Oncol. 2005, 6, 322–327. [Google Scholar] [CrossRef]
- He, H.; Li, W.; Yan, P.; Bundschuh, R.; Killian, J.A.; Labanowska, J.; Brock, P.; Shen, R.; Heerema, N.A.; de la Chapelle, A. Identification of a Recurrent LMO7-BRAF Fusion in Papillary Thyroid Carcinoma. Thyroid 2018, 28, 748–754. [Google Scholar] [CrossRef]
- Kang, M.; Lee, K.-H.; Lee, H.S.; Park, Y.H.; Jeong, C.W.; Ku, J.H.; Kim, H.H.; Kwak, C. PDLIM2 Suppression Efficiently Reduces Tumor Growth and Invasiveness of Human Castration-Resistant Prostate Cancer-like Cells. Prostate 2016, 76, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Zhao, J.; He, H.; Chen, Y.; Wang, Y.; Li, D.; Zhu, Q. Gga-miR-3525 Targets PDLIM3 through the MAPK Signaling Pathway to Regulate the Proliferation and Differentiation of Skeletal Muscle Satellite Cells. Int. J. Mol. Sci. 2020, 21, 5573. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Yin, H.; Yu, X.; Zhang, Y.; Ma, M.; Li, D.; Zhu, Q. PDLIM5 Affects Chicken Skeletal Muscle Satellite Cell Proliferation and Differentiation via the P38-MAPK Pathway. Animals 2021, 11, 1016. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.-Y.; Jung, N.-C.; Lee, J.-H.; Choi, S.-Y.; Choi, H.-J.; Park, S.-Y.; Jang, J.-S.; Byun, S.-H.; Hwang, S.-U.; Noh, K.-E.; et al. Pdlim4 Is Essential for CCR7-JNK-Mediated Dendritic Cell Migration and F-Actin-Related Dendrite Formation. FASEB J. 2019, 33, 11035–11044. [Google Scholar] [CrossRef]
- Xuan, T.; Wang, D.; Lv, J.; Pan, Z.; Fang, J.; Xiang, Y.; Cheng, H.; Wang, X.; Guo, X. Downregulation of Cypher Induces Apoptosis in Cardiomyocytes via Akt/P38 MAPK Signaling Pathway. Int. J. Med. Sci. 2020, 17, 2328–2337. [Google Scholar] [CrossRef]
- Bongalon, S.; Dai, Y.-P.; Singer, C.A.; Yamboliev, I.A. PDGF and IL-1beta Upregulate Cofilin and LIMK2 in Canine Cultured Pulmonary Artery Smooth Muscle Cells. J. Vasc. Res. 2004, 41, 412–421. [Google Scholar] [CrossRef]
- Zhou, J.-K.; Fan, X.; Cheng, J.; Liu, W.; Peng, Y. PDLIM1: Structure, Function and Implication in Cancer. Cell Stress 2021, 5, 119–127. [Google Scholar] [CrossRef]
- Ahn, B.Y.; Saldanha-Gama, R.F.G.; Rahn, J.J.; Hao, X.; Zhang, J.; Dang, N.-H.; Alshehri, M.; Robbins, S.M.; Senger, D.L. Glioma Invasion Mediated by the P75 Neurotrophin Receptor (P75(NTR)/CD271) Requires Regulated Interaction with PDLIM1. Oncogene 2016, 35, 1411–1422. [Google Scholar] [CrossRef]
- Liu, Z.; Zhan, Y.; Tu, Y.; Chen, K.; Wu, C. PDZ and LIM Domain Protein 1(PDLIM1)/CLP36 Promotes Breast Cancer Cell Migration, Invasion and Metastasis through Interaction with α-Actinin. Oncogene 2015, 34, 1300–1311. [Google Scholar] [CrossRef]
- Gupta, P.; Suman, S.; Mishra, M.; Mishra, S.; Srivastava, N.; Kumar, V.; Singh, P.K.; Shukla, Y. Autoantibodies against TYMS and PDLIM1 Proteins Detected as Circulatory Signatures in Indian Breast Cancer Patients. Proteom. Clin. Appl. 2016, 10, 564–573. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, J.; Wang, K.; Chen, H.; Qin, S.; Liu, J.; Luo, M.; Chen, Y.; Jiang, J.; Zhou, L.; et al. PDLIM1 Inhibits Tumor Metastasis Through Activating Hippo Signaling in Hepatocellular Carcinoma. Hepatology 2020, 71, 1643–1659. [Google Scholar] [CrossRef]
- Tan, Y.; Li, Y.; Zhu, H.; Wu, X.; Mei, K.; Li, P.; Yang, Q. miR-187/PDLIM1 Gets Involved in Gastric Cancer Progression and Cisplatin Sensitivity of Cisplatin by Mediating the Hippo-YAP Signaling Pathway. J. Oncol. 2022, 2022, 5456016. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-N.; Yuan, K.; Xie, N.; Wang, K.; Huang, Z.; Chen, Y.; Dou, Q.; Wu, M.; Nice, E.C.; Zhou, Z.-G.; et al. PDLIM1 Stabilizes the E-Cadherin/β-Catenin Complex to Prevent Epithelial-Mesenchymal Transition and Metastatic Potential of Colorectal Cancer Cells. Cancer Res. 2016, 76, 1122–1134. [Google Scholar] [CrossRef] [PubMed]
- Tamura, N.; Ohno, K.; Katayama, T.; Kanayama, N.; Sato, K. The PDZ-LIM Protein CLP36 Is Required for Actin Stress Fiber Formation and Focal Adhesion Assembly in BeWo Cells. Biochem. Biophys. Res. Commun. 2007, 364, 589–594. [Google Scholar] [CrossRef][Green Version]
- Hong, S.-H. Identification of CLP36 as a Tumor Antigen That Induces an Antibody Response in Pancreatic Cancer. Cancer Res. Treat. 2005, 37, 71–77. [Google Scholar] [CrossRef]
- Macartney-Coxson, D.P.; A Hood, K.; Shi, H.-J.; Ward, T.; Wiles, A.; O’Connor, R.; A Hall, D.; A Lea, R.; A Royds, J.; Stubbs, R.S.; et al. Metastatic Susceptibility Locus, an 8p Hot-Spot for Tumour Progression Disrupted in Colorectal Liver Metastases: 13 Candidate Genes Examined at the DNA, mRNA and Protein Level. BMC Cancer 2008, 8, 187. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Xu, Z.; Zhang, Y.; Tan, C.; Hu, W.; Wang, M.; Xu, Y.; Tang, J. Exosome-Mediated miR-222 Transferring: An Insight into NF-κB-Mediated Breast Cancer Metastasis. Exp. Cell Res. 2018, 369, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Fu, J.; Yan, P.; Hu, J.; Cheng, S.-Y.; Xiao, G. Epigenetic Repression of PDZ-LIM Domain-Containing Protein 2: Implications for the Biology and Treatment of Breast Cancer. J. Biol. Chem. 2010, 285, 11786–11792. [Google Scholar] [CrossRef] [PubMed]
- Deevi, R.K.; Cox, O.T.; O’Connor, R. Essential Function for PDLIM2 in Cell Polarization in Three-Dimensional Cultures by Feedback Regulation of the Β1-Integrin-RhoA Signaling Axis. Neoplasia 2014, 16, 422–431. [Google Scholar] [CrossRef]
- Bowe, R.A.; Cox, O.T.; Ayllón, V.; Tresse, E.; Healy, N.C.; Edmunds, S.J.; Huigsloot, M.; O’Connor, R. PDLIM2 Regulates Transcription Factor Activity in Epithelial-to-Mesenchymal Transition via the COP9 Signalosome. Mol. Biol. Cell 2014, 25, 184–195. [Google Scholar] [CrossRef]
- Shi, H.; Ji, Y.; Li, W.; Zhong, Y.; Ming, Z. PDLIM2 Acts as a Cancer Suppressor Gene in Non-Small Cell Lung Cancer via the down Regulation of NF-κB Signaling. Mol. Cell. Probes 2020, 53, 101628. [Google Scholar] [CrossRef]
- Song, G.; Xu, J.; He, L.; Sun, X.; Xiong, R.; Luo, Y.; Hu, X.; Zhang, R.; Yue, Q.; Liu, K.; et al. Systematic Profiling Identifies PDLIM2 as a Novel Prognostic Predictor for Oesophageal Squamous Cell Carcinoma (ESCC). J. Cell. Mol. Med. 2019, 23, 5751–5761. [Google Scholar] [CrossRef]
- Jiang, X.; Chu, Z.; Cao, Y.; Tang, Y.; Shi, Y.; Shi, X. PDLIM2 Prevents the Malignant Phenotype of Hepatocellular Carcinoma Cells by Negatively Regulating β-Catenin. Cancer Gene Ther. 2021, 28, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, G.-Y.; Zhou, L.; Jiang, H.-L.; Yang, Y.; Wu, H.-T. Exosomes from M2 Macrophages Promoted Glycolysis in FaDu Cells by Inhibiting PDLIM2 Expression to Stabilize PFKL. Neoplasma 2022, 69, 1041–1053. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yu, C.; Zhou, S.; Lau, W.B.; Lau, B.; Luo, Z.; Lin, Q.; Yang, H.; Xuan, Y.; Yi, T.; et al. Epigenetic Repression of PDZ-LIM Domain-Containing Protein 2 Promotes Ovarian Cancer via NOS2-Derived Nitric Oxide Signaling. Oncotarget 2016, 7, 1408–1420. [Google Scholar] [CrossRef]
- Oh, B.Y.; Cho, J.; Hong, H.K.; Bae, J.S.; Park, W.-Y.; Joung, J.-G.; Cho, Y.B. Exome and Transcriptome Sequencing Identifies Loss of PDLIM2 in Metastatic Colorectal Cancers. Cancer Manag. Res. 2017, 9, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Yang, Z.; Zhi, Q.; Wang, D.; Guo, L.; Li, G.; Miao, R.; Shi, Y.; Kuang, Y. Long Noncoding RNA OR3A4 Promotes Metastasis and Tumorigenicity in Gastric Cancer. Oncotarget 2016, 7, 30276–30294. [Google Scholar] [CrossRef]
- Shou, Y.; Robinson, D.M.; Amakye, D.D.; Rose, K.L.; Cho, Y.-J.; Ligon, K.L.; Sharp, T.; Haider, A.S.; Bandaru, R.; Ando, Y.; et al. A Five-Gene Hedgehog Signature Developed as a Patient Preselection Tool for Hedgehog Inhibitor Therapy in Medulloblastoma. Clin. Cancer Res. 2015, 21, 585–593. [Google Scholar] [CrossRef]
- Feng, Y.; Jiang, Y.; Wen, T.; Meng, F.; Shu, X. Identifying Potential Prognostic Markers for Muscle-Invasive Bladder Urothelial Carcinoma by Weighted Gene Co-Expression Network Analysis. Pathol. Oncol. Res. 2020, 26, 1063–1072. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, P.; Wen, W.; Grubbs, C.J.; Townsend, R.R.; Malone, J.P.; Lubet, R.A.; You, M. Cross-Species Comparison of Orthologous Gene Expression in Human Bladder Cancer and Carcinogen-Induced Rodent Models. Am. J. Transl. Res. 2010, 3, 8–27. [Google Scholar]
- Stein, L.; Rothschild, J.; Luce, J.; Cowell, J.K.; Thomas, G.A.; Bogdanova, T.I.; Tronko, M.D.; Hawthorn, L.; Richter, H.; Braselmann, H.; et al. Copy Number and Gene Expression Alterations in Radiation-Induced Papillary Thyroid Carcinoma from Chernobyl Pediatric Patients. Thyroid 2010, 20, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Shi, H.; Cao, Y.; Feng, W.; Li, M.; Li, X. PDZ and LIM Domain Protein 4 Suppresses the Growth and Invasion of Ovarian Cancer Cells via Inactivation of STAT3 Signaling. Life Sci. 2019, 233, 116715. [Google Scholar] [CrossRef] [PubMed]
- Vanaja, D.K.; Grossmann, M.E.; Cheville, J.C.; Gazi, M.H.; Gong, A.; Zhang, J.S.; Ajtai, K.; Burghardt, T.P.; Young, C.Y.F. Pdlim4, an Actin Binding Protein, Suppresses Prostate Cancer Cell Growth. Cancer Investig. 2009, 27, 264–272. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vanaja, D.K.; Ballman, K.V.; Morlan, B.W.; Cheville, J.C.; Neumann, R.M.; Lieber, M.M.; Tindall, D.J.; Young, C.Y.F. PDLIM4 Repression by Hypermethylation as a Potential Biomarker for Prostate Cancer. Clin. Cancer Res. 2006, 12, 1128–1136. [Google Scholar] [CrossRef] [PubMed]
- Kolluru, V.; Tyagi, A.; Chandrasekaran, B.; Damodaran, C. Profiling of Differentially Expressed Genes in Cadmium-Induced Prostate Carcinogenesis. Toxicol. Appl. Pharmacol. 2019, 375, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Vasiljević, N.; Wu, K.; Brentnall, A.R.; Kim, D.C.; Thorat, M.A.; Kudahetti, S.C.; Mao, X.; Xue, L.; Yu, Y.; Shaw, G.L.; et al. Absolute Quantitation of DNA Methylation of 28 Candidate Genes in Prostate Cancer Using Pyrosequencing. Dis. Markers 2011, 30, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.R.; Ricketts, C.; Gentle, D.; Abdulrahman, M.; Clarke, N.; Brown, M.; Kishida, T.; Yao, M.; Latif, F.; Maher, E.R. Identification of Candidate Tumour Suppressor Genes Frequently Methylated in Renal Cell Carcinoma. Oncogene 2010, 29, 2104–2117. [Google Scholar] [CrossRef]
- Patai, V.; Barták, B.K.; Péterfia, B.; Micsik, T.; Horváth, R.; Sumánszki, C.; Péter, Z.; Valcz, G.; Kalmár, A.; Tóth, K.; et al. Comprehensive DNA Methylation and Mutation Analyses Reveal a Methylation Signature in Colorectal Sessile Serrated Adenomas. Pathol. Oncol. Res. 2017, 23, 589–594. [Google Scholar] [CrossRef]
- Feng, W.; Orlandi, R.; Zhao, N.; Carcangiu, M.L.; Tagliabue, E.; Xu, J.; Bast, R.C.; Yu, Y. Tumor Suppressor Genes Are Frequently Methylated in Lymph Node Metastases of Breast Cancers. BMC Cancer 2010, 10, 378. [Google Scholar] [CrossRef]
- Xu, J.; Shetty, P.B.; Feng, W.; Chenault, C.; Bast, R.C.; Issa, J.-P.J.; Hilsenbeck, S.G.; Yu, Y. Methylation of HIN-1, RASSF1A, RIL and CDH13 in Breast Cancer Is Associated with Clinical Characteristics, but Only RASSF1A Methylation Is Associated with Outcome. BMC Cancer 2012, 12, 243. [Google Scholar] [CrossRef]
- Kravchenko, D.S.; Ivanova, A.E.; Podshivalova, E.S.; Chumakov, S.P. PDLIM4/RIL-Mediated Regulation of Src and Malignant Properties of Breast Cancer Cells. Oncotarget 2020, 11, 22–30. [Google Scholar] [CrossRef]
- Liu, X.; Chen, L.; Huang, H.; Lv, J.-M.; Chen, M.; Qu, F.-J.; Pan, X.-W.; Li, L.; Yin, L.; Cui, X.-G.; et al. High Expression of PDLIM5 Facilitates Cell Tumorigenesis and Migration by Maintaining AMPK Activation in Prostate Cancer. Oncotarget 2017, 8, 98117–98134. [Google Scholar] [CrossRef]
- Shui, I.M.; Lindström, S.; Kibel, A.S.; Berndt, S.I.; Campa, D.; Gerke, T.; Penney, K.L.; Albanes, D.; Berg, C.; Bueno-De-Mesquita, H.B.; et al. Prostate Cancer (PCa) Risk Variants and Risk of Fatal PCa in the National Cancer Institute Breast and Prostate Cancer Cohort Consortium. Eur. Urol. 2014, 65, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Tsukamoto, O.; Nakano, A.; Kato, H.; Kioka, H.; Ito, N.; Higo, S.; Yamazaki, S.; Shintani, Y.; Matsuoka, K.; et al. Augmented AMPK Activity Inhibits Cell Migration by Phosphorylating the Novel Substrate Pdlim5. Nat. Commun. 2015, 6, 6137. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zhang, Y.; Yu, S.; Li, S.; Jiang, W.; Zhu, Y.; Xu, Y.; Yang, C.; Tian, G.; Mi, J.; et al. PDLIM5 Identified by Label-Free Quantitative Proteomics as a Potential Novel Biomarker of Papillary Thyroid Carcinoma. Biochem. Biophys. Res. Commun. 2018, 499, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lv, W.; Li, H.; Dong, T.; Wu, H.; Su, C.; Shu, H.; Nie, F. Exploring the Relationship between Autophagy and Gefitinib Resistance in NSCLC by Silencing PDLIM5 Using Ultrasound-Targeted Microbubble Destruction Technology. Cancer Cell Int. 2022, 22, 293. [Google Scholar] [CrossRef]
- Edlund, K.; Lindskog, C.; Saito, A.; Berglund, A.; Pontén, F.; Göransson-Kultima, H.; Isaksson, A.; Jirström, K.; Planck, M.; Johansson, L.; et al. CD99 Is a Novel Prognostic Stromal Marker in Non-Small Cell Lung Cancer. Int. J. Cancer 2012, 131, 2264–2273. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Lv, W.-H.; Zhu, Y.-Y.; Jia, Y.-Y.; Nie, F. Ultrasound-Mediated Mesoporous Silica Nanoparticles Loaded with PDLIM5 siRNA Inhibit Gefitinib Resistance in NSCLC Cells by Attenuating EMT. Eur. J. Pharm. Sci. 2023, 182, 106372. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, Y.; Matsuura, T.; Kurosaki, T.; Amakusa, Y.; Kinoshita, M.; Ibi, T.; Sahashi, K.; Ohno, K. LDB3 Splicing Abnormalities Are Specific to Skeletal Muscles of Patients with Myotonic Dystrophy Type 1 and Alter Its PKC Binding Affinity. Neurobiol. Dis. 2014, 69, 200–205. [Google Scholar] [CrossRef]
- Yu, H.; Yuan, C.; Westenbroek, R.E.; Catterall, W.A. The AKAP Cypher/Zasp Contributes to β-Adrenergic/PKA Stimulation of Cardiac CaV1.2 Calcium Channels. J. Gen. Physiol. 2018, 150, 883–889. [Google Scholar] [CrossRef]
- Leung, M.-C.; Hitchen, P.G.; Ward, D.G.; Messer, A.E.; Marston, S.B. Z-Band Alternatively Spliced PDZ Motif Protein (ZASP) Is the Major O-Linked β-N-Acetylglucosamine-Substituted Protein in Human Heart Myofibrils. J. Biol. Chem. 2013, 288, 4891–4898. [Google Scholar] [CrossRef]
- Cui, L.; Cheng, Z.; Hu, K.; Pang, Y.; Liu, Y.; Qian, T.; Quan, L.; Dai, Y.; Pang, Y.; Ye, X.; et al. Prognostic Value of the PDLIM Family in Acute Myeloid Leukemia. Am. J. Transl. Res. 2019, 11, 6124–6131. [Google Scholar] [PubMed]
- Kales, S.C.; Nau, M.M.; Merchant, A.S.; Lipkowitz, S. Enigma Prevents Cbl-c-Mediated Ubiquitination and Degradation of RETMEN2A. PLoS ONE 2014, 9, e87116. [Google Scholar] [CrossRef]
- Tabariès, S.; McNulty, A.; Ouellet, V.; Annis, M.G.; Dessureault, M.; Vinette, M.; Hachem, Y.; Lavoie, B.; Omeroglu, A.; Simon, H.-G.; et al. Afadin Cooperates with Claudin-2 to Promote Breast Cancer Metastasis. Genes Dev. 2019, 33, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.-R.; Lim, J.H.; Choi, Y.; Kim, D.-G.; Kang, K.J.; Noh, S.-M.; Im, D.-S. Enigma Negatively Regulates P53 through MDM2 and Promotes Tumor Cell Survival in Mice. J. Clin. Investig. 2010, 120, 4493–4506. [Google Scholar] [CrossRef]
- Firek, A.A.; Perez, M.C.; Gonda, A.; Lei, L.; Munir, I.; Simental, A.A.; Carr, F.E.; Becerra, B.J.; De Leon, M.; Khan, S. Pathologic Significance of a Novel Oncoprotein in Thyroid Cancer Progression. Head Neck 2017, 39, 2459–2469. [Google Scholar] [CrossRef] [PubMed]
- Borrello, M.G.; Mercalli, E.; Perego, C.; Degl’Innocenti, D.; Ghizzoni, S.; Arighi, E.; Eroini, B.; Rizzetti, M.G.; A Pierotti, M. Differential Interaction of Enigma Protein with the Two RET Isoforms. Biochem. Biophys. Res. Commun. 2002, 296, 515–522. [Google Scholar] [CrossRef]
- Kim, Y.J.; Hwang, H.-J.; Kang, J.G.; Kim, C.S.; Ihm, S.-H.; Choi, M.G.; Lee, S.J. Enigma Plays Roles in Survival of Thyroid Carcinoma Cells through PI3K/AKT Signaling and Survivin. Anticancer Res. 2018, 38, 3515–3525. [Google Scholar] [CrossRef]
- Lu, Y.; Jin, Z.; Hou, J.; Wu, X.; Yu, Z.; Yao, L.; Pan, T.; Chang, X.; Yu, B.; Li, J.; et al. Calponin 1 Increases Cancer-Associated Fibroblasts-Mediated Matrix Stiffness to Promote Chemoresistance in Gastric Cancer. Matrix Biol. 2023, 115, 1–15. [Google Scholar] [CrossRef]
- Liu, H.; Huang, L.; Zhang, Z.; Zhang, Z.; Yu, Z.; Chen, X.; Chen, Z.; Zen, Y.; Yang, D.; Han, Z.; et al. LIM Mineralization Protein-1 Inhibits the Malignant Phenotypes of Human Osteosarcoma Cells. Int. J. Mol. Sci. 2014, 15, 7037–7048. [Google Scholar] [CrossRef]
- Hu, Q.; Guo, C.; Li, Y.; Aronow, B.J.; Zhang, J. LMO7 Mediates Cell-Specific Activation of the Rho-Myocardin-Related Transcription Factor-Serum Response Factor Pathway and Plays an Important Role in Breast Cancer Cell Migration. Mol. Cell. Biol. 2011, 31, 3223–3240. [Google Scholar] [CrossRef]
- Nakamura, H.; Hori, K.; Tanaka-Okamoto, M.; Higashiyama, M.; Itoh, Y.; Inoue, M.; Morinaka, S.; Miyoshi, J. Decreased Expression of LMO7 and Its Clinicopathological Significance in Human Lung Adenocarcinoma. Exp. Ther. Med. 2011, 2, 1053–1057. [Google Scholar] [CrossRef]
- Tanaka-Okamoto, M.; Hori, K.; Ishizaki, H.; Hosoi, A.; Itoh, Y.; Wei, M.; Wanibuchi, H.; Mizoguchi, A.; Nakamura, H.; Miyoshi, J. Increased Susceptibility to Spontaneous Lung Cancer in Mice Lacking LIM-Domain Only 7. Cancer Sci. 2009, 100, 608–616. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhou, J.; Mei, S.; Wu, D.; Mu, Z.; Chen, B.; Xie, Y.; Ye, Y.; Liu, J. Circulating Exosomal microRNA-96 Promotes Cell Proliferation, Migration and Drug Resistance by Targeting LMO7. J. Cell. Mol. Med. 2017, 21, 1228–1236. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zheng, H.; Li, Z.; Shi, S.; Zhong, L.; Gong, L.; Lan, B. LMO7-ALK Fusion in a Lung Adenocarcinoma Patient With Crizotinib: A Case Report. Front. Oncol. 2022, 12, 841493. [Google Scholar] [CrossRef]
- Mixed Responses to First-Line Alectinib in Non-Small Cell Lung Cancer Patients with Rare ALK Gene Fusions: A Case Series and Literature Review—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/34541785/ (accessed on 4 May 2023).
- Tzeng, Y.-W.; Li, D.-Y.; Chen, Y.; Yang, C.-H.; Chang, C.-Y.; Juang, Y.-L. LMO7 Exerts an Effect on Mitosis Progression and the Spindle Assembly Checkpoint. Int. J. Biochem. Cell Biol. 2018, 94, 22–30. [Google Scholar] [CrossRef]
- Liu, X.; Yuan, H.; Zhou, J.; Wang, Q.; Qi, X.; Bernal, C.; Avella, D.; Kaifi, J.T.; Kimchi, E.T.; Timothy, P.; et al. LMO7 as an Unrecognized Factor Promoting Pancreatic Cancer Progression and Metastasis. Front. Cell Dev. Biol. 2021, 9, 647387. [Google Scholar] [CrossRef]
- Takahashi, M.; Lio, C.-W.J.; Campeau, A.; Steger, M.; Ay, F.; Mann, M.; Gonzalez, D.J.; Jain, M.; Sharma, S. The Tumor Suppressor Kinase DAPK3 Drives Tumor-Intrinsic Immunity through the STING-IFN-β Pathway. Nat. Immunol. 2021, 22, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Amano, T.; Tanabe, K.; Eto, T.; Narumiya, S.; Mizuno, K. LIM-Kinase 2 Induces Formation of Stress Fibres, Focal Adhesions and Membrane Blebs, Dependent on Its Activation by Rho-Associated Kinase-Catalysed Phosphorylation at Threonine-505. Biochem. J. 2001, 354 Pt 1, 149–159. [Google Scholar] [CrossRef]
- Maekawa, M.; Ishizaki, T.; Boku, S.; Watanabe, N.; Fujita, A.; Iwamatsu, A.; Obinata, T.; Ohashi, K.; Mizuno, K.; Narumiya, S. Signaling from Rho to the Actin Cytoskeleton through Protein Kinases ROCK and LIM-Kinase. Science 1999, 285, 895–898. [Google Scholar] [CrossRef]
- Ohashi, K.; Nagata, K.; Maekawa, M.; Ishizaki, T.; Narumiya, S.; Mizuno, K. Rho-Associated Kinase ROCK Activates LIM-Kinase 1 by Phosphorylation at Threonine 508 within the Activation Loop. J. Biol. Chem. 2000, 275, 3577–3582. [Google Scholar] [CrossRef]
- Suyama, E.; Wadhwa, R.; Kawasaki, H.; Yaguchi, T.; Kaul, S.C.; Nakajima, M.; Taira, K. LIM Kinase-2 Targeting as a Possible Anti-Metastasis Therapy. J. Gene Med. 2004, 6, 357–363. [Google Scholar] [CrossRef]
- Yoshioka, K.; Foletta, V.; Bernard, O.; Itoh, K. A Role for LIM Kinase in Cancer Invasion. Proc. Natl. Acad. Sci. USA 2003, 100, 7247–7252. [Google Scholar] [CrossRef]
- You, T.; Gao, W.; Wei, J.; Jin, X.; Zhao, Z.; Wang, C.; Li, Y. Overexpression of LIMK1 Promotes Tumor Growth and Metastasis in Gastric Cancer. Biomed. Pharmacother. 2015, 69, 96–101. [Google Scholar] [CrossRef]
- Li, X.; Ke, Q.; Li, Y.; Liu, F.; Zhu, G.; Li, F. DGCR6L, a Novel PAK4 Interaction Protein, Regulates PAK4-Mediated Migration of Human Gastric Cancer Cell via LIMK1. Int. J. Biochem. Cell Biol. 2010, 42, 70–79. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.-Y.; Wang, J.; You, L.-H.; Zhou, R.-Z.; Zhao, D.-M.; Cheng, M.-S.; Li, F. GL-1196 Suppresses the Proliferation and Invasion of Gastric Cancer Cells via Targeting PAK4 and Inhibiting PAK4-Mediated Signaling Pathways. Int. J. Mol. Sci. 2016, 17, 470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Zhang, J.; Hao, C.Z.; Zhou, Y.; Wang, J.; Cheng, M.S.; Zhao, D.M.; Li, F. LC-0882 Targets PAK4 and Inhibits PAK4-Related Signaling Pathways to Suppress the Proliferation and Invasion of Gastric Cancer Cells. Am. J. Transl. Res. 2017, 9, 2736–2747. [Google Scholar] [PubMed]
- Zhang, J.; Wang, J.; Guo, Q.; Wang, Y.; Zhou, Y.; Peng, H.; Cheng, M.; Zhao, D.; Li, F. LCH-7749944, a Novel and Potent P21-Activated Kinase 4 Inhibitor, Suppresses Proliferation and Invasion in Human Gastric Cancer Cells. Cancer Lett. 2012, 317, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Su, B.; Su, J.; Zeng, Y.; Liu, F.; Xia, H.; Ma, Y.-H.; Zhou, Z.-G.; Zhang, S.; Yang, B.-M.; Wu, Y.-H.; et al. Diallyl Disulfide Suppresses Epithelial-Mesenchymal Transition, Invasion and Proliferation by Downregulation of LIMK1 in Gastric Cancer. Oncotarget 2016, 7, 10498–10512. [Google Scholar] [CrossRef]
- Guo, B.; Zhang, T.; Su, J.; Wang, K.; Li, X. Oxymatrine Targets EGFR(p-Tyr845) and Inhibits EGFR-Related Signaling Pathways to Suppress the Proliferation and Invasion of Gastric Cancer Cells. Cancer Chemother. Pharmacol. 2015, 75, 353–363. [Google Scholar] [CrossRef]
- Li, H.; Chen, C. Quercetin Has Antimetastatic Effects on Gastric Cancer Cells via the Interruption of uPA/uPAR Function by Modulating NF-Κb, PKC-δ, ERK1/2, and AMPKα. Integr. Cancer Ther. 2018, 17, 511–523. [Google Scholar] [CrossRef]
- Zeng, Y.; Ren, M.; Li, Y.; Liu, Y.; Chen, C.; Su, J.; Su, B.; Xia, H.; Liu, F.; Jiang, H.; et al. Knockdown of RhoGDI2 Represses Human Gastric Cancer Cell Proliferation, Invasion and Drug Resistance via the Rac1/Pak1/LIMK1 Pathway. Cancer Lett. 2020, 492, 136–146. [Google Scholar] [CrossRef]
- Kang, X.; Li, W.; Liu, W.; Liang, H.; Deng, J.; Wong, C.C.; Zhao, S.; Kang, W.; To, K.F.; Chiu, P.W.Y.; et al. LIMK1 Promotes Peritoneal Metastasis of Gastric Cancer and Is a Therapeutic Target. Oncogene 2021, 40, 3422–3433. [Google Scholar] [CrossRef]
- Liu, X.; Song, Q.; Wang, D.; Liu, Y.; Zhang, Z.; Fu, W. LIMK1: A Promising Prognostic and Immune Infiltration Indicator in Colorectal Cancer. Oncol. Lett. 2022, 24, 234. [Google Scholar] [CrossRef]
- Su, J.; Zhou, Y.; Pan, Z.; Shi, L.; Yang, J.; Liao, A.; Liao, Q.; Su, Q. Downregulation of LIMK1-ADF/Cofilin by DADS Inhibits the Migration and Invasion of Colon Cancer. Sci. Rep. 2017, 7, 45624. [Google Scholar] [CrossRef]
- Liao, Q.; Li, R.; Zhou, R.; Pan, Z.; Xu, L.; Ding, Y.; Zhao, L. LIM Kinase 1 Interacts with Myosin-9 and Alpha-Actinin-4 and Promotes Colorectal Cancer Progression. Br. J. Cancer 2017, 117, 563–571. [Google Scholar] [CrossRef]
- Sun, X.; Li, S.; Lin, H. LIMK1 Interacts with STK25 to Regulate EMT and Promote the Proliferation and Metastasis of Colorectal Cancer. J. Oncol. 2022, 2022, 3963883. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Su, J.; Shi, L.; Liao, Q.; Su, Q. DADS Downregulates the Rac1-ROCK1/PAK1-LIMK1-ADF/Cofilin Signaling Pathway, Inhibiting Cell Migration and Invasion. Oncol. Rep. 2013, 29, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Sheng, N.; Tan, G.; You, W.; Chen, H.; Gong, J.; Chen, D.; Zhang, H.; Wang, Z. MiR-145 Inhibits Human Colorectal Cancer Cell Migration and Invasion via PAK4-Dependent Pathway. Cancer Med. 2017, 6, 1331–1340. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, G.; Zhang, B.; Liu, C.; Yu, Y.; Jin, Y. miR-27b-3p Suppresses Cell Proliferation, Migration and Invasion by Targeting LIMK1 in Colorectal Cancer. Int. J. Clin. Exp. Pathol. 2017, 10, 9251–9261. [Google Scholar] [PubMed]
- Zhu, Q.; Wu, Y.; Yang, M.; Wang, Z.; Zhang, H.; Jiang, X.; Chen, M.; Jin, T.; Wang, T. IRX5 Promotes Colorectal Cancer Metastasis by Negatively Regulating the Core Components of the RHOA Pathway. Mol. Carcinog. 2019, 58, 2065–2076. [Google Scholar] [CrossRef]
- Hu, Y.H.; Lu, Y.X.; Zhang, Z.Y.; Zhang, J.M.; Zhang, W.J.; Zheng, L.; Lin, W.H.; Zhang, W.; Li, X.N. SSH3 Facilitates Colorectal Cancer Cell Invasion and Metastasis by Affecting Signaling Cascades Involving LIMK1/Rac1. Am. J. Cancer Res. 2019, 9, 1061–1073. [Google Scholar]
- Croft, D.R.; Coleman, M.L.; Li, S.; Robertson, D.; Sullivan, T.; Stewart, C.L.; Olson, M.F. Actin-Myosin–Based Contraction Is Responsible for Apoptotic Nuclear Disintegration. J. Cell Biol. 2005, 168, 245–255. [Google Scholar] [CrossRef] [PubMed]
- McConnell, B.V.; Koto, K.; Gutierrez-Hartmann, A. Nuclear and Cytoplasmic LIMK1 Enhances Human Breast Cancer Progression. Mol. Cancer 2011, 10, 75. [Google Scholar] [CrossRef] [PubMed]
- Bagheri-Yarmand, R.; Mazumdar, A.; Sahin, A.A.; Kumar, R. LIM Kinase 1 Increases Tumor Metastasis of Human Breast Cancer Cells via Regulation of the Urokinase-Type Plasminogen Activator System. Int. J. Cancer 2006, 118, 2703–2710. [Google Scholar] [CrossRef] [PubMed]
- Lagoutte, E.; Villeneuve, C.; Lafanechère, L.; Wells, C.M.; Jones, G.E.; Chavrier, P.; Rossé, C. LIMK Regulates Tumor-Cell Invasion and Matrix Degradation through Tyrosine Phosphorylation of MT1-MMP. Sci. Rep. 2016, 6, 24925. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Li, H.; An, W.; Wei, W.; Zhang, X.; Wang, L. Mex-3 RNA Binding MEX3A Promotes the Proliferation and Migration of Breast Cancer Cells via Regulating RhoA/ROCK1/LIMK1 Signaling Pathway. Bioengineered 2021, 12, 5850–5858. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Ma, D.; Cao, Y.; Hu, L.; Liu, S.; Yan, D.; Zhang, S.; Zhang, G.; Wang, Z.; Wu, J.; et al. SphK2/S1P Promotes Metastasis of Triple-Negative Breast Cancer through the PAK1/LIMK1/Cofilin1 Signaling Pathway. Front. Mol. Biosci. 2021, 8, 598218. [Google Scholar] [CrossRef]
- Li, H.; Zhang, B.; Liu, Y.; Yin, C. EBP50 Inhibits the Migration and Invasion of Human Breast Cancer Cells via LIMK/Cofilin and the PI3K/Akt/mTOR/MMP Signaling Pathway. Med. Oncol. 2014, 31, 162. [Google Scholar] [CrossRef]
- Shahi, P.; Wang, C.-Y.; Chou, J.; Hagerling, C.; Velozo, H.G.; Ruderisch, A.; Yu, Y.; Lai, M.-D.; Werb, Z. GATA3 Targets Semaphorin 3B in Mammary Epithelial Cells to Suppress Breast Cancer Progression and Metastasis. Oncogene 2017, 36, 5567–5575. [Google Scholar] [CrossRef]
- Fu, J.; Yu, J.; Chen, J.; Xu, H.; Luo, Y.; Lu, H. In Vitro Inhibitory Properties of Sesquiterpenes from Chloranthus Serratus on Cell Motility via Down-Regulation of LIMK1 Activation in Human Breast Cancer. Phytomedicine 2018, 49, 23–31. [Google Scholar] [CrossRef]
- Zhao, J.; Li, D.; Fang, L. MiR-128-3p Suppresses Breast Cancer Cellular Progression via Targeting LIMK1. Biomed. Pharmacother. 2019, 115, 108947. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, H.; Song, H.; Xu, H.; Zhao, B.; Wu, C.; Hu, J.; Wu, T.; Xie, D.; Zhao, J.; et al. The microRNAs miR-200b-3p and miR-429-5p Target the LIMK1/CFL1 Pathway to Inhibit Growth and Motility of Breast Cancer Cells. Oncotarget 2017, 8, 85276–85289. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Song, H.; Wu, T.; Xie, D.; Hu, J.; Zhao, J.; Shen, Q.; Fang, L. MiR-519d-3p Suppresses Breast Cancer Cell Growth and Motility via Targeting LIM Domain Kinase 1. Mol. Cell. Biochem. 2018, 444, 169–178. [Google Scholar] [CrossRef]
- Li, D.; Hu, J.; Song, H.; Xu, H.; Wu, C.; Zhao, B.; Xie, D.; Wu, T.; Zhao, J.; Fang, L. miR-143-3p Targeting LIM Domain Kinase 1 Suppresses the Progression of Triple-Negative Breast Cancer Cells. Am. J. Transl. Res. 2017, 9, 2276–2285. [Google Scholar] [PubMed]
- Davila, M.; Jhala, D.; Ghosh, D.; E Grizzle, W.; Chakrabarti, R. Expression of LIM Kinase 1 Is Associated with Reversible G1/S Phase Arrest, Chromosomal Instability and Prostate Cancer. Mol. Cancer 2007, 6, 40. [Google Scholar] [CrossRef]
- Limonta, P.; Moretti, R.M.; Mai, S.; Marelli, M.M.; Rizzi, F.; Bettuzzi, S. Molecular Mechanisms of the Antimetastatic Activity of Nuclear Clusterin in Prostate Cancer Cells. Int. J. Oncol. 2011, 39, 225–234. [Google Scholar] [CrossRef]
- Cai, S.; Chen, R.; Li, X.; Cai, Y.; Ye, Z.; Li, S.; Li, J.; Huang, H.; Peng, S.; Wang, J.; et al. Downregulation of microRNA-23a Suppresses Prostate Cancer Metastasis by Targeting the PAK6-LIMK1 Signaling Pathway. Oncotarget 2015, 6, 3904–3917. [Google Scholar] [CrossRef]
- Ngalame, N.N.; Makia, N.L.; Waalkes, M.P.; Tokar, E.J. Mitigation of Arsenic-Induced Acquired Cancer Phenotype in Prostate Cancer Stem Cells by miR-143 Restoration. Toxicol. Appl. Pharmacol. 2016, 312, 11–18. [Google Scholar] [CrossRef][Green Version]
- Bhardwaj, A.; Srivastava, S.K.; Singh, S.; Arora, S.; Tyagi, N.; Andrews, J.; McClellan, S.; Carter, J.E.; Singh, A.P. CXCL12/CXCR4 Signaling Counteracts Docetaxel-Induced Microtubule Stabilization via P21-Activated Kinase 4-Dependent Activation of LIM Domain Kinase 1. Oncotarget 2014, 5, 11490–11500. [Google Scholar] [CrossRef]
- Mardilovich, K.; Gabrielsen, M.; McGarry, L.; Orange, C.; Patel, R.; Shanks, E.; Edwards, J.; Olson, M.F. Elevated LIM Kinase 1 in Nonmetastatic Prostate Cancer Reflects Its Role in Facilitating Androgen Receptor Nuclear Translocation. Mol. Cancer Ther. 2015, 14, 246–258. [Google Scholar] [CrossRef]
- Lu, G.; Zhou, Y.; Zhang, C.; Zhang, Y. Upregulation of LIMK1 Is Correlated with Poor Prognosis and Immune Infiltrates in Lung Adenocarcinoma. Front. Genet. 2021, 12, 671585. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Ye, Z.; Wang, X.; Pan, Y.; Weng, Y.; Lao, S.; Wei, H.; Li, L. Overexpression of P21-Activated Kinase 4 Is Associated with Poor Prognosis in Non-Small Cell Lung Cancer and Promotes Migration and Invasion. J. Exp. Clin. Cancer Res. 2015, 34, 48. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Li, X.; Song, S.; Chen, M.; Cheng, M.; Zhao, D.; Li, F. (-)-β-Hydrastine Suppresses the Proliferation and Invasion of Human Lung Adenocarcinoma Cells by Inhibiting PAK4 Kinase Activity. Oncol. Rep. 2016, 35, 2246–2256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, R.; Tian, J.; Song, M.; Zhao, R.; Liu, K.; Zhu, F.; Shim, J.; Dong, Z.; Lee, M. Targeting LIMK1 with Luteolin Inhibits the Growth of Lung Cancer In Vitro and In Vivo. J. Cell. Mol. Med. 2021, 25, 5560–5571. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.G.; Im, E.; Lee, H.-J.; Lee, E.-O. Plumbagin Reduces Osteopontin-Induced Invasion through Inhibiting the Rho-Associated Kinase Signaling Pathway in A549 Cells and Suppresses Osteopontin-Induced Lung Metastasis in BalB/c Mice. Bioorg Med. Chem. Lett. 2017, 27, 1914–1918. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Zhang, L.; Fan, K.; Wang, J. MiR-27b Targets LIMK1 to Inhibit Growth and Invasion of NSCLC Cells. Mol. Cell. Biochem. 2014, 390, 85–91. [Google Scholar] [CrossRef]
- Chen, Q.; Jiao, D.; Hu, H.; Song, J.; Yan, J.; Wu, L.; Xu, L.-Q. Downregulation of LIMK1 Level Inhibits Migration of Lung Cancer Cells and Enhances Sensitivity to Chemotherapy Drugs. Oncol. Res. 2013, 20, 491–498. [Google Scholar] [CrossRef]
- Yang, J.Z.; Huang, L.H.; Chen, R.; Meng, L.J.; Gao, Y.Y.; Ji, Q.Y.; Wang, Y. LIM Kinase 1 Serves an Important Role in the Multidrug Resistance of Osteosarcoma Cells. Oncol. Lett. 2018, 15, 250–256. [Google Scholar] [CrossRef]
- Zhang, H.-S.; Zhao, J.-W.; Wang, H.; Zhang, H.-Y.; Ji, Q.-Y.; Meng, L.-J.; Xing, F.-J.; Yang, S.-T.; Wang, Y. LIM Kinase 1 Is Required for Insulin-dependent Cell Growth of Osteosarcoma Cell Lines. Mol. Med. Rep. 2014, 9, 103–108. [Google Scholar] [CrossRef]
- Yoshizawa, M.; Nakamura, S.; Sugiyama, Y.; Tamai, S.; Ishida, Y.; Sueyoshi, M.; Toda, Y.; Hosogi, S.; Yano, Y.; Ashihara, E. 6-Hydroxythiobinupharidine Inhibits Migration of LM8 Osteosarcoma Cells by Decreasing Expression of LIM Domain Kinase 1. Anticancer Res. 2019, 39, 6507–6513. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Y.; Xing, F.; Wang, J.; Wang, H.; Yang, Y.; Gao, Z.; Wang, Y. Overexpression of LIMK1 Promotes Migration Ability of Multidrug-Resistant Osteosarcoma Cells. Oncol. Res. 2011, 19, 501–509. [Google Scholar] [CrossRef]
- Chhavi; Saxena, M.; Singh, S.; Negi, M.; Srivastava, A.; Trivedi, R.; Singh, U.; Pant, M.; Bhatt, M. Expression Profiling of G2/M Phase Regulatory Proteins in Normal, Premalignant and Malignant Uterine Cervix and Their Correlation with Survival of Patients. J. Cancer Res. Ther. 2010, 6, 167–171. [Google Scholar] [CrossRef]
- Yang, X.; Du, H.; Bian, W.; Li, Q.; Sun, H. FOXD3-AS1/miR-128-3p/LIMK1 Axis Regulates Cervical Cancer Progression. Oncol. Rep. 2021, 45, 62. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zheng, Y.; Duan, Y.; Ma, L.; Nan, P. MicroRNA-125a-5p Targets LIM Kinase 1 to Inhibit Cisplatin Resistance of Cervical Cancer Cells. Oncol. Lett. 2021, 21, 392. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Lou, W.; Fan, W.; Pan, J. Exosomal miR-374c-5p derived from mesenchymal stem cells suppresses epithelial-mesenchymal transition of hepatocellular carcinoma via the LIMK1-Wnt/β-catenin axis. Environ. Toxicol. 2023, 38, 1038–1052. [Google Scholar] [CrossRef]
- Wang, D.; Xing, N.; Yang, T.; Liu, J.; Zhao, H.; He, J.; Ai, Y.; Yang, J. Exosomal lncRNA H19 Promotes the Progression of Hepatocellular Carcinoma Treated with Propofol via miR-520a-3p/LIMK1 Axis. Cancer Med. 2020, 9, 7218–7230. [Google Scholar] [CrossRef]
- Pan, Z.; Liu, C.; Zhi, Y.; Xie, Z.; Wu, L.; Jiang, M.; Zhang, Y.; Zhou, R.; Zhao, L. LIMK1 Nuclear Translocation Promotes Hepatocellular Carcinoma Progression by Increasing P-ERK Nuclear Shuttling and by Activating c-Myc Signalling upon EGF Stimulation. Oncogene 2021, 40, 2581–2595. [Google Scholar] [CrossRef]
- Lee, C.; Siu, A.; Chang, J.; Dang, D.; Ramos, D.M. Alphavbeta3 Suppresses the RhoA-LIMK1 Pathway in K1735 Melanoma. J. Calif. Dent. Assoc. 2012, 40, 921–927. [Google Scholar] [CrossRef]
- Shi, B.; Ma, C.; Liu, G.; Guo, Y. MiR-106a Directly Targets LIMK1 to Inhibit Proliferation and EMT of Oral Carcinoma Cells. Cell Mol. Biol. Lett. 2019, 24, 1. [Google Scholar] [CrossRef]
- Peng, T.; Wang, T.; Liu, G.; Zhou, L.; Teng, Z. Effects of miR-373 Inhibition on Glioblastoma Growth by Reducing Limk1 In Vitro. J. Immunol. Res. 2020, 2020, 7671502. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhang, L.; Kebebew, E. MiR-20a Is Upregulated in Anaplastic Thyroid Cancer and Targets LIMK1. PLoS ONE 2014, 9, e96103. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, R.; Luo, C.; Zhou, X.; Xia, K.; Chen, X.; Zhou, M.; Zou, Q.; Cao, P.; Cao, K. MiR-20a Inhibits Cutaneous Squamous Cell Carcinoma Metastasis and Proliferation by Directly Targeting LIMK1. Cancer Biol. Ther. 2014, 15, 1340–1349. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; He, Z.; Xu, J.; Chen, P.; Jiang, J. Long Noncoding RNA LINC00941 Promotes Pancreatic Cancer Progression by Competitively Binding miR-335-5p to Regulate ROCK1-Mediated LIMK1/Cofilin-1 Signaling. Cell Death Dis. 2021, 12, 36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gan, N.; Zhou, J. Immunohistochemical Investigation of the Correlation between LIM Kinase 1 Expression and Development and Progression of Human Ovarian Carcinoma. J. Int. Med. Res. 2012, 40, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zeng, M.; Zhao, Y.; Fang, X. Upregulation of Limk1 Caused by microRNA-138 Loss Aggravates the Metastasis of Ovarian Cancer by Activation of Limk1/Cofilin Signaling. Oncol. Rep. 2014, 32, 2070–2076. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, A.; Shi, J.; Fang, Y.; Gu, C.; Cai, J.; Lin, C.; Zhao, L.; Liu, S. Imbalanced LIMK1 and LIMK2 Expression Leads to Human Colorectal Cancer Progression and Metastasis via Promoting β-Catenin Nuclear Translocation. Cell Death Dis. 2018, 9, 749. [Google Scholar] [CrossRef]
- Lourenço, F.C.; Munro, J.; Brown, J.; Cordero, J.; Stefanatos, R.; Strathdee, K.; Orange, C.; Feller, S.M.; Sansom, O.J.; Vidal, M.; et al. Reduced LIMK2 Expression in Colorectal Cancer Reflects Its Role in Limiting Stem Cell Proliferation. Gut 2014, 63, 480–493. [Google Scholar] [CrossRef]
- Aggelou, H.; Chadla, P.; Nikou, S.; Karteri, S.; Maroulis, I.; Kalofonos, H.P.; Papadaki, H.; Bravou, V. LIMK/Cofilin Pathway and Slingshot Are Implicated in Human Colorectal Cancer Progression and Chemoresistance. Virchows Arch. 2018, 472, 727–737. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Li, Q.; Zhang, Y. lncRNA LINC00460 Promoted Colorectal Cancer Cells Metastasis via miR-939-5p Sponging. Cancer Manag. Res. 2019, 11, 1779–1789. [Google Scholar] [CrossRef]
- Nikhil, K.; Chang, L.; Viccaro, K.; Jacobsen, M.; McGuire, C.; Satapathy, S.R.; Tandiary, M.; Broman, M.M.; Cresswell, G.; He, Y.J.; et al. Identification of LIMK2 as a Therapeutic Target in Castration Resistant Prostate Cancer. Cancer Lett. 2019, 448, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Ma, F.; Huang, W.; Fang, S.; Li, M.; Wei, T.; Guo, L. Long Non-Coding RNA TUG1 Is Involved in Cell Growth and Chemoresistance of Small Cell Lung Cancer by Regulating LIMK2b via EZH2. Mol. Cancer 2017, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Xu, B.; Shen, Q.; Lei, Z.; Zhang, W.; Hu, T. LIMK2 Is a Novel Prognostic Biomarker and Correlates With Tumor Immune Cell Infiltration in Lung Squamous Cell Carcinoma. Front. Immunol. 2022, 13, 788375. [Google Scholar] [CrossRef]
- Ren, T.; Zheng, B.; Huang, Y.; Wang, S.; Bao, X.; Liu, K.; Guo, W. Osteosarcoma Cell Intrinsic PD-L2 Signals Promote Invasion and Metastasis via the RhoA-ROCK-LIMK2 and Autophagy Pathways. Cell Death Dis. 2019, 10, 261. [Google Scholar] [CrossRef]
- Sooreshjani, M.A.; Nikhil, K.; Kamra, M.; Nguyen, D.N.; Kumar, D.; Shah, K. LIMK2-NKX3.1 Engagement Promotes Castration-Resistant Prostate Cancer. Cancers 2021, 13, 2324. [Google Scholar] [CrossRef] [PubMed]
- Malvi, P.; Janostiak, R.; Chava, S.; Manrai, P.; Yoon, E.; Singh, K.; Harigopal, M.; Gupta, R.; Wajapeyee, N. LIMK2 Promotes the Metastatic Progression of Triple-Negative Breast Cancer by Activating SRPK1. Oncogenesis 2020, 9, 77. [Google Scholar] [CrossRef]
- Xu, M.; Wang, F.; Li, G.; Wang, X.; Fang, X.; Jin, H.; Chen, Z.; Zhang, J.; Fu, L. MED12 Exerts an Emerging Role in Actinmediated Cytokinesis via LIMK2/Cofilin Pathway in NSCLC. Mol. Cancer 2019, 18, 93. [Google Scholar] [CrossRef]
- Wang, S.; Ren, T.; Jiao, G.; Huang, Y.; Bao, X.; Zhang, F.; Liu, K.; Zheng, B.; Sun, K.; Guo, W. BMPR2 Promotes Invasion and Metastasis via the RhoA-ROCK-LIMK2 Pathway in Human Osteosarcoma Cells. Oncotarget 2017, 8, 58625–58641. [Google Scholar] [CrossRef]
- Po’Uha, S.T.; Shum, M.S.Y.; Goebel, A.; Bernard, O.; Kavallaris, M. LIM-Kinase 2, a Regulator of Actin Dynamics, Is Involved in Mitotic Spindle Integrity and Sensitivity to Microtubule-Destabilizing Drugs. Oncogene 2010, 29, 597–607. [Google Scholar] [CrossRef]
- Gamell, C.; Schofield, A.V.; Suryadinata, R.; Sarcevic, B.; Bernard, O. LIMK2 Mediates Resistance to Chemotherapeutic Drugs in Neuroblastoma Cells through Regulation of Drug-Induced Cell Cycle Arrest. PLoS ONE 2013, 8, e72850. [Google Scholar] [CrossRef]
- Vlecken, D.H.; Bagowski, C.P. LIMK1 and LIMK2 Are Important for Metastatic Behavior and Tumor Cell-Induced Angiogenesis of Pancreatic Cancer Cells. Zebrafish 2009, 6, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Rak, R.; Haklai, R.; Elad-Tzfadia, G.; Wolfson, H.J.; Carmeli, S.; Kloog, Y. Novel LIMK2 Inhibitor Blocks Panc-1 Tumor Growth in a Mouse Xenograft Model. Oncoscience 2014, 1, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, C.; Nie, H.; Qiu, X.; Zhang, L.; Xiao, Y.; Zhou, W.; Zeng, Q.; Zhang, X.; Wu, Y.; et al. LIMK2 Acts as an Oncogene in Bladder Cancer and Its Functional SNP in the microRNA-135a Binding Site Affects Bladder Cancer Risk. Int. J. Cancer 2019, 144, 1345–1355. [Google Scholar] [CrossRef]
- Hsu, F.-F.; Lin, T.-Y.; Chen, J.-Y.; Shieh, S.-Y. P53-Mediated Transactivation of LIMK2b Links Actin Dynamics to Cell Cycle Checkpoint Control. Oncogene 2010, 29, 2864–2876. [Google Scholar] [CrossRef]
- Nikhil, K.; Haymour, H.S.; Kamra, M.; Shah, K. Phosphorylation-Dependent Regulation of SPOP by LIMK2 Promotes Castration-Resistant Prostate Cancer. Br. J. Cancer 2021, 124, 995–1008. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.O.; Chang, K.-H.; Ghosh, S.; Venkatesh, C.; Giger, K.; Low, P.S.; Shah, K. LIMK2 Is a Crucial Regulator and Effector of Aurora-A-Kinase-Mediated Malignancy. J. Cell Sci. 2012, 125 Pt 5, 1204–1216. [Google Scholar] [CrossRef] [PubMed]
- Nikhil, K.; Kamra, M.; Raza, A.; Shah, K. Negative Cross Talk between LIMK2 and PTEN Promotes Castration Resistant Prostate Cancer Pathogenesis in Cells and in Vivo. Cancer Lett. 2021, 498, 1–18. [Google Scholar] [CrossRef]
- The Cause of Cancer. JAMA 2021, 325, 311. [CrossRef]
- Lasorsa, F.; Rutigliano, M.; Milella, M.; Ferro, M.; Pandolfo, S.D.; Crocetto, F.; Autorino, R.; Battaglia, M.; Ditonno, P.; Lucarelli, G. Cancer Stem Cells in Renal Cell Carcinoma: Origins and Biomarkers. Int. J. Mol. Sci. 2023, 24, 13179. [Google Scholar] [CrossRef]
- Aveta, A.; Cilio, S.; Contieri, R.; Spena, G.; Napolitano, L.; Manfredi, C.; Franco, A.; Crocerossa, F.; Cerrato, C.; Ferro, M.; et al. Urinary MicroRNAs as Biomarkers of Urological Cancers: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 10846. [Google Scholar] [CrossRef]
- Gao, J.; Hou, B.; Zhu, Q.; Yang, L.; Jiang, X.; Zou, Z.; Li, X.; Xu, T.; Zheng, M.; Chen, Y.-H.; et al. Engineered Bioorthogonal POLY-PROTAC Nanoparticles for Tumour-Specific Protein Degradation and Precise Cancer Therapy. Nat. Commun. 2022, 13, 4318. [Google Scholar] [CrossRef] [PubMed]

| PDLIMs | Tumor Type | Expression | Effect | Reference |
|---|---|---|---|---|
| PDLIM1 | Glioma | Exogenous suppression | Inhibits tumor invasion | Ahn et al., 2016 [98] |
| Breast cancer | Increased | Promotes tumor migration and invasion | Liu et al., 2015 [99]; Gupta et al., 2016 [100] | |
| Pancreatic cancer | Increased | As a tumor antigen that induces antibody response | Hong 2005 [105] | |
| Hepatocellular carcinoma | Reduced | Promotes tumor migration | Huang et al., 2020 [101] | |
| Gastric cancer | Reduced | Promotes tumor progression and cisplatin sensitivity | Tan et al., 2022 [102] | |
| Colorectal Cancer | Reduced | Inhibits tumor metastasis formation and EMT | Chen et al., 2016 [103] | |
| Choriocarcinoma | Exogenous suppression | Inhibits tumor cell actin stress fiber formation and focal adhesion assembly | Tamura et al., 2007 [104] | |
| PDLIM2 | Breast cancer | Exogenous suppression | Promotes tumor progression and lymphatic metastasis | Ding 2018 [107]; Qu et al., 2010 [108] |
| Breast cancer | Increased | Promotes cell migration, cytoskeletal polarization, and EMT | Deevi, Cox, and O’Connor 2014 [109]; Bowe et al., 2014 [110] | |
| Lung cancer | Reduced | Promotes tumor progression and therapeutic resistance | Sun et al., 2019 [87]; Shi et al., 2020 [111] | |
| Esophageal squamous cell carcinoma | High PDLIM2 expression group has longer overall survival | Song et al., 2019 [112] | ||
| Hepatocellular carcinoma | Exogenous overexpression | Inhibits tumor malignant phenotype | Jiang et al., 2021 [113] | |
| Laryngeal squamous cell carcinoma | Reduced | Promotes tumor cell proliferation | Wang et al., 2022 [114] | |
| Ovarian cancer | Reduced | Promotes tumor pathogenesis | Zhao et al., 2016 [115] | |
| Metastatic colorectal cancer | Reduced | Promotes tumor metastasis | Oh et al., 2017 [116] | |
| Castration-Resistant Prostate Cancer | Increased | Promotes tumor growth and invasion | Kang et al., 2016 [91] | |
| Gastric cancer | Exogenous activation | Promotes tumorigenicity and metastasis | Guo et al., 2016 [117] | |
| PDLIM3 | Medulloblastoma | Increased | Unknown | Shou et al., 2015 [118] |
| Invasive bladder urothelium carcinoma | Increased | Associated with the unfavorable survival | Feng et al., 2020 [119] | |
| Bladder cancer | Reduced | Unknown | Lu et al., 2010 [120] | |
| Thyroid Papillary carcinoma | Reduced | Unknown | Stein et al., 2010 [121] | |
| PDLIM4 | Ovarian cancer | Reduced | Associated with aggressive tumor features and poor prognosis | Jia et al., 2019 [122] |
| Prostatic carcinoma | Reduced | Promotes carcinogenesis | Vanaja et al., 2009 [123]; Vanaja et al., 2006 [124]; Kolluru et al., 2019 [125]; Vasiljević et al., 2011 [126] | |
| Thyroid carcinoma | Reduced | Unknown | Patai et al., 2017 [128] | |
| Kidney cancer | Reduced | Unknown | Morris et al., 2010 [127] | |
| Breast cancer | Reduced | Promotes tumor progression | Feng et al., 2010 [129] Xu et al., 2012 [130] | |
| Breast cancer | Exogenous overexpression | Promotes tumor metastasis | Kravchenko et al., 2020 [131] | |
| PDLIMs | Tumor Type | Expression | Effect | Reference |
|---|---|---|---|---|
| PDLIM5 | Prostatic carcinoma | Increased | Promotes tumorigenesis and migration | Liu et al., 2017 [132]; Shui et al., 2014 [133] |
| Thyroid Papillary carcinoma | Increased | Promotes tumor migration, invasion and proliferation | Wei et al., 2018 [135] | |
| Lung cancer | Increased | Promotes tumor migration and invasion | Shi et al., 2020 [68] Edlund et al., 2012 [137] Zhang et al., 2022 [136] Wu et al., 2023 [138] | |
| PDLIM6 | Unknown | |||
| PDLIM7 | Acute myeloid leukemia | Increased | An independent risk factor for EFS and OS | Cui et al., 2019 [142] |
| Breast cancer | Increased | Correlates with a poor outcome | Kales et al., 2014 [143] Tabariès et al., 2019 [144] | |
| Colorectal cancer | Exogenous overexpression | Promotes tumor cell survival | Jung et al., 2010 [145] | |
| Hepatocellular carcinoma | Exogenous overexpression | Promotes tumor cell survival | Jung et al., 2010 [145] | |
| Thyroid carcinoma | Increased | Promotes carcinogenesis | Firek et al., 2017 [146] Borrello et al., 2002 [147] Kim et al., 2018 [148] | |
| Gastric cancer | Degradation Inhibited | Contributes to 5-Fu resistance in GC cells | Lu et al., 2023 [149] | |
| Osteosarcoma | Reduced | Promotes tumor malignant phenotypes | Liu et al., 2014 [150] | |
| PDLIMs | Tumor Type | Expression | Effect | Reference |
|---|---|---|---|---|
| LIMK1 | Gastric cancer | Increased | Promotes tumor growth and metastasis | You et al., 2015 [165]; Li et al., 2010 [166]; Zhang et al., 2016 [167]; H.-Y. Zhang et al., 2017 [168]; J. Zhang et al., 2012 [169]; Su et al., 2016 [170]; Guo et al., 2015 [171]; Li and Chen 2018 [172]; Zeng et al., 2020 [173]; Kang et al., 2021 [174] |
| Colorectal cancer | Increased | Promoted tumor development | Liu et al., 2022 [175]; Su et al., 2017 [176]; Liao et al., 2017 [177]; Sun, Li, and Lin 2022 [178]; Zhou et al., 2013 [179]; Sheng et al., 2017 [180]; Chen et al., 2017 [181] | |
| Colorectal cancer | Exogenous suppression | Promoted tumor development | Zhu et al., 2019 [182]; Hu et al., 2019 [183] | |
| Breast cancer | Increased | Promoted tumor development | Croft et al., 2005 [184]; McConnell, Koto, and Gutierrez-Hartmann 2011 [185]; Bagheri-Yarmand et al., 2006 [186]; Lagoutte et al., 2016 [187]; Yan et al., 2021 [188]; Shi et al., 2021 [189]; Li et al., 2014 [190]; Shahi et al., [191]; Fu et al., 2018 [192]; Zhao, Li, and Fang 2019 [193]; Li, Hu, et al., 2017 [194]; Li, Wang, et al., 2017 [195]; Li et al., 2018, 1 [196] J. | |
| Prostatic carcinoma | Exogenous overexpression | Promoted tumor development | Davila et al., 2007 [197]; Moretti et al., 2011 [198]; Cai et al., 2015 [199]; Ngalame et al., 2016 [200]; Bhardwaj et al., 2014, 1 [201]; Mardilovich et al., 2015 [202] | |
| Lung cancer | Increased | Promoted tumor development | Lu et al., 2021 [203]; Cai et al., 2015 [204]; Guo et al., 2016 [205]; Zhang et al., 2021 [206]; Kang et al., 2017 [207]; Wan et al., 2014 [208]; Chen et al., 2013 [209] | |
| Osteosarcoma | Increased | Promoted tumor development | Yang et al., 2018 [210]; H.-S. Zhang et al., 2014 [211]; Yoshizawa et al., 2019, 1 [212]; H. Zhang et al., 2011 [213] | |
| Uterine cervix carcinoma | Increased | Promoted tumor development | Chhavi et al., 2010 [214]; Yang et al., 2021 [215]; Xu et al., 2021 [216] | |
| Hepatocellular carcinoma | Exogenous overexpression | Promoted tumor development | Ding et al., 2023 [217]; Wang et al., 2020 [218]; Pan et al., 2021 [219] | |
| Melanoma | Exogenous suppression | Promotes tumor invasion | Lee et al., 2012 [220] | |
| Oral cancer | Exogenous suppression | Inhibits tumor proliferation, migration and EMT | Shi et al., 2019 [221] | |
| Thyroid Papillary carcinoma | Exogenous suppression | Inhibits tumor progression | Xiong, Zhang, and Kebebew 2014 [223] | |
| Cutaneous squamous cell carcinoma | Exogenous suppression | Inhibits tumor progression | Zhou et al., 2014 [224] | |
| Glioblastoma | Exogenous suppression | Inhibits tumor progression | Peng et al., 2020 [222] | |
| Pancreatic adenocarcinoma | Exogenous activation | Promotes tumor growth and metastasis | Wang et al., 2021 [225] | |
| Ovarian cancer | Increased | Promotes tumor migration | Zhang, Gan, and Zhou 2012 [226]; Chen et al., 2014 [227] | |
| LIMK2 | Colorectal cancer | Exogenous overexpression | Inhibits tumor cell proliferation | Zhang et al., 2019 [231] |
| Colorectal cancer | Increased | Promotes tumor development | Aggelou et al., 2018 [230] | |
| Colorectal cancer | Reduced | Promotes tumor development | Yue Zhang et al., 2018 [228]; Lourenço et al., 2014 [229] | |
| Castration resistant prostate cancer | Increased | Promotes tumor initiation, progression, and poor prognosis | Nikhil et al., 2019 [232] | |
| Prostate cancer | LIMK regulates other molecules | Promotes tumor progression and drug resistance | Nikhil, Kamra, et al., 2021 [246]; Nikhil et al., 2021 [247]; Sooreshjani et al., 2021 [248] | |
| Triple negative breast cancer | LIMK regulates other molecules | Promotes tumor migration | Malvi et al., 2020 [237] | |
| Breast cancer | Exogenous suppression | Inhibits tumor migration | Shahi et al., 2017 [191]; Malvi et al., 2020 [237] | |
| Non-small-cell lung cancer | Exogenous activation | Inhibits tumor proliferation | Xu et al., 2019 [238] | |
| Non-small-cell lung cancer | Exogenous overexpression | Promotes tumor proliferation and drug resistance | Niu et al., 2017 [233]; Su et al., 2022 [234] | |
| Osteosarcoma | Exogenous activation | Promotes tumor migration | Ren et al., 2019 [235]; Wang et al., 2017 [239] | |
| Neuroblastoma | Increased | Promotes tumor drug resistance | Po’uha et al., 2010 [240]; Gamell et al., 2013 [241] | |
| Pancreatic adenocarcinoma | Exogenous suppression | Inhibits tumor development | Rak et al., 2014 [243] | |
| Bladder cancer | Increased | Promotes tumor proliferation, migration, and invasion | Wang et al., 2019 [244] | |
| Thyroid carcinoma | Reduced | Promotes tumor migration | Hsu et al., 2010 [245] | |
| Esophageal cancer | Reduced | Promotes tumor migration | Hsu et al., 2010 [245] | |
| LMO7 | Breast cancer | Increased | Promotes tumor migration | Hu et al., 2011 [151]; Tanaka-Okamoto et al., 2009, 7 [152]; Nakamura et al., 2011 [153] |
| Lung cancer | Exogenous suppression | Promoted tumor development | Wu et al., 2017 [154]; Yang et al., 2022 [155]; Li et al., 2021 [156] | |
| Cervical carcinoma | Exogenous overexpression | Inhibits tumor cell proliferation | Tzeng et al., 2018 [157] | |
| Colorectal cancer | Exogenous overexpression | Inhibits tumor cell proliferation | Tzeng et al., 2018 [157] | |
| Pancreatic cancer | Increased | Promotes tumor progression and metastasis | Lin et al., 2021 [158] | |
| Hepatocellular carcinoma | Exogenous overexpression | Promotes tumor invasion | Nakamura et al., 2005 [72] |
| ATC | Undifferentiated thyroid cancer |
| BC | Breast cancer |
| BCA | Bladder carcinoma |
| BRCA | Breast invasive carcinoma |
| CC | Choriocarcinoma |
| CCA | Cervical carcinoma |
| CRC | Colorectal cancer |
| EMT | Epithelial–mesenchymal transition |
| EC | Esophageal cancer |
| FVPTC | Follicular papillary thyroid carcinoma |
| GC | Gastric cancer |
| GCC | Hepatocarcinoma |
| LSCC | Laryngeal squamous cell carcinoma |
| LUAD | Lung adenocarcinoma |
| LC | Lung cancer |
| NSCLC | Non-small-cell lung cancer |
| OS | Osteosarcoma |
| OV | Ovarian Cancer |
| PAAD | Pancreatic adenocarcinoma |
| PTC | Papillary thyroid carcinoma |
| PDTC | Poorly differentiated thyroid carcinoma |
| PC | Prostatic carcinoma |
| THCA | Thyroid carcinoma |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, X.; Xu, Z.; Jiang, S.; Wang, H.; Xiao, M.; Shi, Y.; Wang, K. PDZ and LIM Domain-Encoding Genes: Their Role in Cancer Development. Cancers 2023, 15, 5042. https://doi.org/10.3390/cancers15205042
Jiang X, Xu Z, Jiang S, Wang H, Xiao M, Shi Y, Wang K. PDZ and LIM Domain-Encoding Genes: Their Role in Cancer Development. Cancers. 2023; 15(20):5042. https://doi.org/10.3390/cancers15205042
Chicago/Turabian StyleJiang, Xinyuan, Zhiyong Xu, Sujing Jiang, Huan Wang, Mingshu Xiao, Yueli Shi, and Kai Wang. 2023. "PDZ and LIM Domain-Encoding Genes: Their Role in Cancer Development" Cancers 15, no. 20: 5042. https://doi.org/10.3390/cancers15205042
APA StyleJiang, X., Xu, Z., Jiang, S., Wang, H., Xiao, M., Shi, Y., & Wang, K. (2023). PDZ and LIM Domain-Encoding Genes: Their Role in Cancer Development. Cancers, 15(20), 5042. https://doi.org/10.3390/cancers15205042






