A Combinatorial Regulatory Platform Determines Expression of RNA Polymerase III Subunit RPC7α (POLR3G) in Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Reagents and Chemicals
2.3. Antibodies
2.4. Plasmids and Transfection
2.5. Western Blots (WB)
2.6. RNA Extraction and Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction (RT−qPCR)
2.7. Statistical Analysis (mRNA and Protein Quantification)
2.8. Data Acquisition
2.9. Pan-Cancer Co-Expression and Chromatin Accessibility Correlation Analyses
2.10. Overlap Enrichment Analyses (TFs, DNA-Methylation, and ATAC)
2.11. Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA)
2.12. Gene- and TF-Distance Analyses
3. Results
3.1. The Gene Encoding RPC7 Subunit RPC7α Is a Distinctively Negative Prognostic Factor in Multiple Cancer Subtypes
3.2. An Integrated Survey of POLR3G mRNA and Chromatin Correlates in Cancer Identifies Candidate Regulatory Factors and Sequence Elements

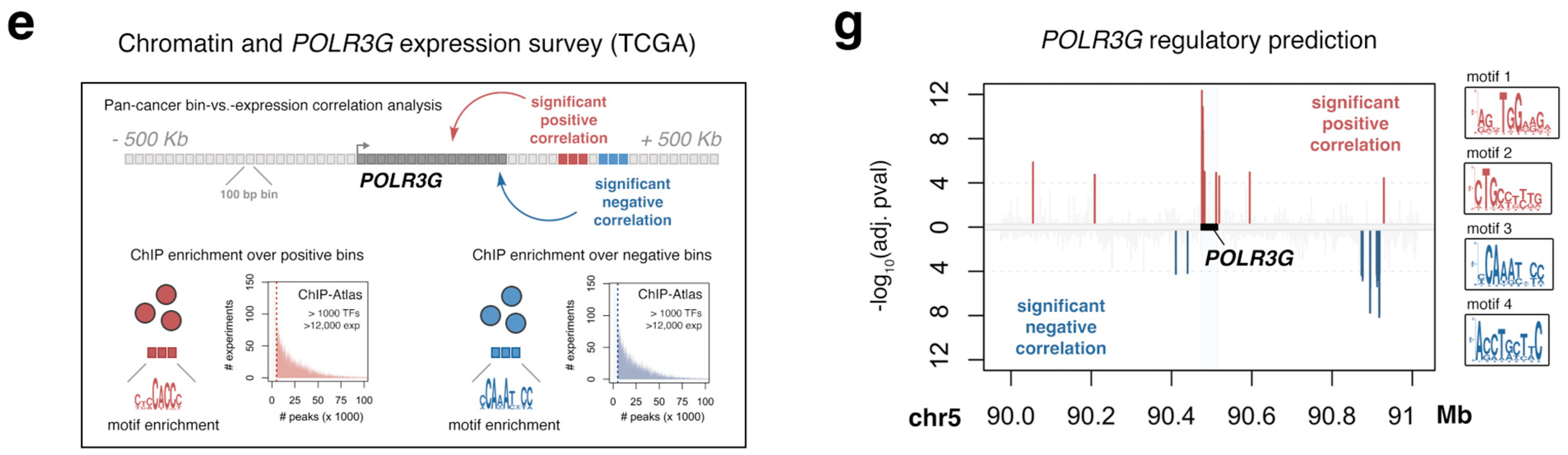
3.3. A Gene-Internal Element, Corresponding to an Early Developmental Super-Enhancer, Is the Strongest Chromatin Correlate with POLR3G Expression in Cancer
3.4. MYC Promotes POLR3G Expression Independently of the Gene-Internal Regulatory Element
3.5. Identification of Zinc Finger Proteins ZNF131 (ZBTB35) and ZNF207 (BuGZ) as Additional Regulatory Factors That Promote POLR3G Expression
3.6. Gain of DNA Methylation over the Gene-Internal Regulatory Element Coincides with Developmental Loss of POLR3G Expression
3.7. POLR3G mRNA Levels Decrease Early in Response to Retinoic Acid, but Subsequent to MYC Downregulation and Concomitant with Markers of Differentiation
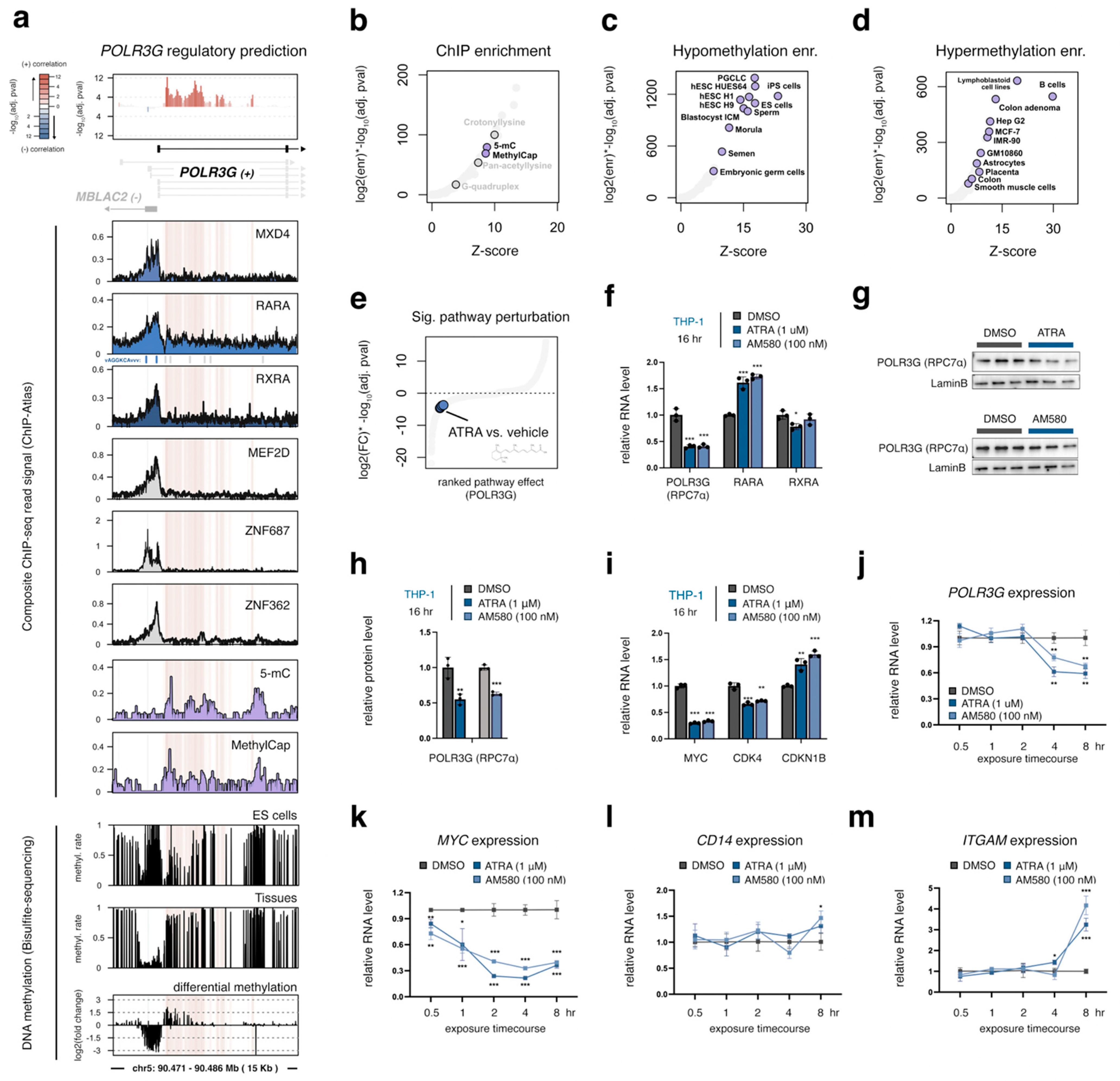
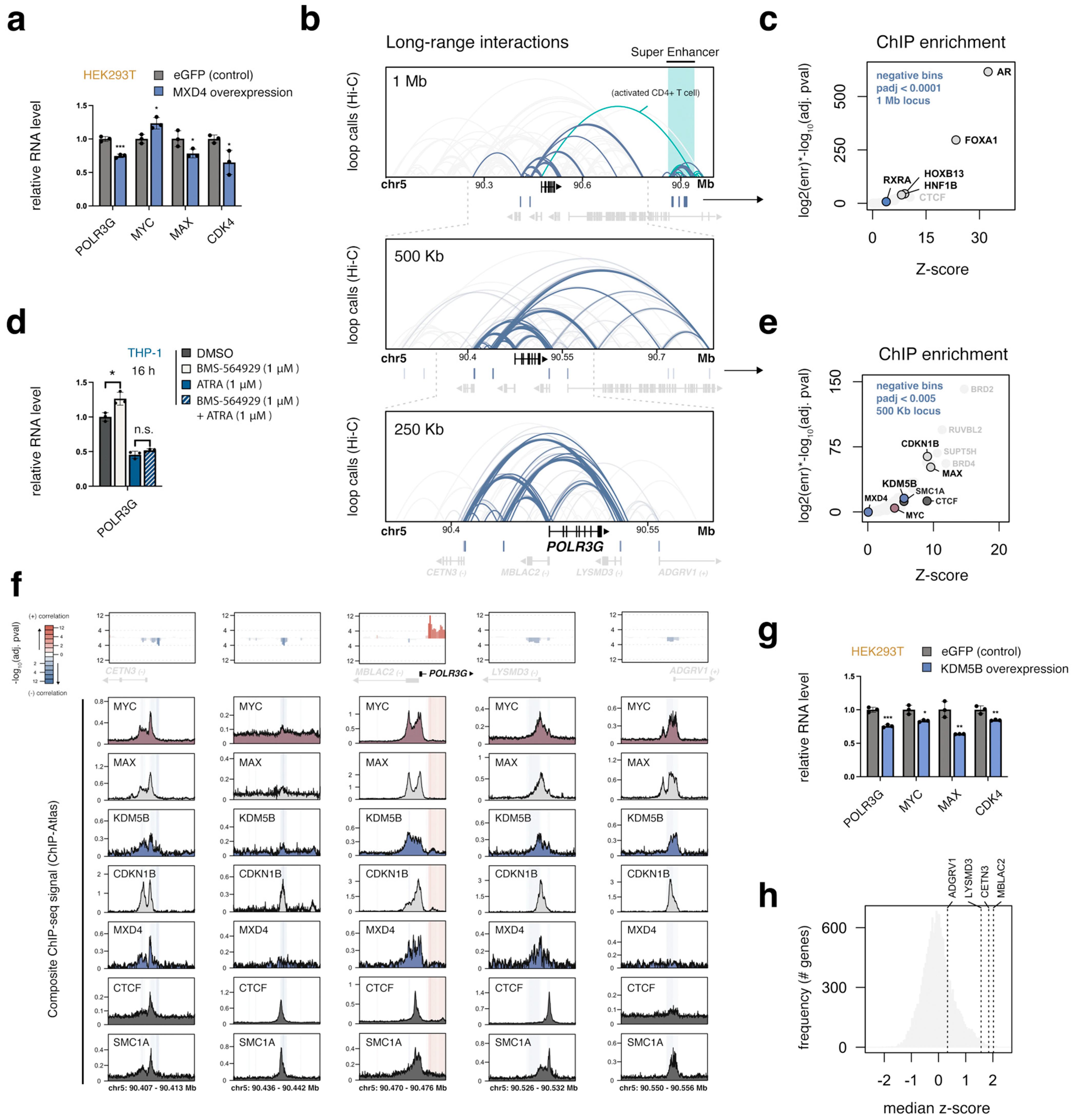
3.8. MXD4, the Strongest Negative POLR3G Correlate, Limits POLR3G Expression
3.9. A Local Multi-Promoter Hub Enriched for MAX, CDKN1B, and KDM5B Is Negatively Linked to POLR3G Expression
3.10. MYC, MAX, MXD4, and KDM5B Target an Overlapping Set of Genes Important for Cell Growth, including Most RNA Polymerase Subunits
3.11. Integrated Regulatory Signatures Implicate Additional Factors as Putative Determinants of POLR3G Expression in Cancer

4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yeganeh, M.; Hernandez, N. RNA polymerase III transcription as a disease factor. Genes Dev. 2020, 34, 865–882. [Google Scholar] [CrossRef]
- Willis, I.M.; Moir, R.D. Signaling to and from the RNA Polymerase III Transcription and Processing Machinery. Annu. Rev. Biochem. 2018, 87, 75–100. [Google Scholar] [CrossRef]
- Vannini, A.; Cramer, P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell 2012, 45, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Roeder, R.G. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997, 11, 1315–1326. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kenneth, N.S.; Marshall, L.; White, R.J. Recruitment of RNA polymerase III in vivo. Nucleic Acids Res. 2008, 36, 3757–3764. [Google Scholar] [CrossRef][Green Version]
- Wang, Q.; Li, S.; Wan, F.; Xu, Y.; Wu, Z.; Cao, M.; Lan, P.; Lei, M.; Wu, J. Structural insights into transcriptional regulation of human RNA polymerase III. Nat. Struct. Mol. Biol. 2021, 28, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Renaud, M.; Praz, V.; Vieu, E.; Florens, L.; Washburn, M.P.; l’Hôte, P.; Hernandez, N. Gene duplication and neofunctionalization: POLR3G and POLR3GL. Genome Res. 2014, 24, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, L.B.; Sklar, V.E.; Jaehning, J.A.; Weinmann, R.; Roeder, R.G. Isolation and partial characterization of the multiple forms of deoxyribonucleic acid-dependent ribonucleic acid polymerase in the mouse myeloma, MOPC 315. J. Biol. Chem. 1974, 249, 5889–5897. [Google Scholar] [CrossRef] [PubMed]
- Haurie, V.; Durrieu-Gaillard, S.; Dumay-Odelot, H.; Da Silva, D.; Rey, C.; Prochazkova, M.; Roeder, R.G.; Besser, D.; Teichmann, M. Two isoforms of human RNA polymerase III with specific functions in cell growth and transformation. Proc. Natl. Acad. Sci. USA 2010, 107, 4176–4181. [Google Scholar] [CrossRef]
- Petrie, J.L.; Swan, C.; Ingram, R.M.; Frame, F.M.; Collins, A.T.; Dumay-Odelot, H.; Teichmann, M.; Maitland, N.J.; White, R.J. Effects on prostate cancer cells of targeting RNA polymerase III. Nucleic Acids Res. 2019, 47, 3937–3956. [Google Scholar] [CrossRef]
- McQueen, C.; Hughes, G.L.; Pownall, M.E. Skeletal muscle differentiation drives a dramatic downregulation of RNA polymerase III activity and differential expression of Polr3g isoforms. Dev. Biol. 2019, 454, 74–84. [Google Scholar] [CrossRef]
- Van Bortle, K.; Marciano, D.P.; Liu, Q.; Chou, T.; Lipchik, A.M.; Gollapudi, S.; Geller, B.S.; Monte, E.; Kamakaka, R.T.; Snyder, M.P. A cancer-associated RNA polymerase III identity drives robust transcription and expression of snaR-A noncoding RNA. Nat. Commun. 2022, 13, 3007. [Google Scholar] [CrossRef]
- Lautré, W.; Richard, E.; Feugeas, J.-P.; Dumay-Odelot, H.; Teichmann, M. The POLR3G Subunit of Human RNA Polymerase III Regulates Tumorigenesis and Metastasis in Triple-Negative Breast Cancer. Cancers 2022, 14, 5732. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, W.; Wang, H.; Lai, C.H.; Xu, K.; Hu, H. Increased expression of POLR3G predicts poor prognosis in transitional cell carcinoma. PeerJ 2020, 8, e10281. [Google Scholar] [CrossRef]
- Yang, J.; Wang, F.; Zhong, S.; Chen, B. Identification of hub genes with prognostic values in multiple myeloma by bioinformatics analysis. Hematology 2021, 26, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Jiang, W.; Ma, L.; Sun, J.; Yan, X.; Qian, J.; Wang, Y.; Shi, Y.; Ni, S.; Yao, N. A metabolism-related gene signature for predicting the prognosis and therapeutic responses in patients with hepatocellular carcinoma. Ann. Transl. Med. 2021, 9, 500. [Google Scholar] [CrossRef]
- Sun, N.; Chu, J.; Hu, W.; Chen, X.; Yi, N.; Shen, Y. A novel 14-gene signature for overall survival in lung adenocarcinoma based on the Bayesian hierarchical Cox proportional hazards model. Sci. Rep. 2022, 12, 27. [Google Scholar] [CrossRef]
- Wong, R.C.; Pollan, S.; Fong, H.; Ibrahim, A.; Smith, E.L.; Ho, M.; Laslett, A.L.; Donovan, P.J. A novel role for an RNA polymerase III subunit POLR3G in regulating pluripotency in human embryonic stem cells. Stem Cells 2011, 29, 1517–1527. [Google Scholar] [CrossRef]
- Smith, J.C.; Sheltzer, J.M. Genome-wide identification and analysis of prognostic features in human cancers. Cell Rep. 2022, 38, 110569. [Google Scholar] [CrossRef]
- Hutter, C.; Zenklusen, J.C. The Cancer Genome Atlas: Creating Lasting Value beyond Its Data. Cell 2018, 173, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Corces, M.R.; Granja, J.M.; Shams, S.; Louie, B.H.; Seoane, J.A.; Zhou, W.; Silva, T.C.; Groeneveld, C.; Wong, C.K.; Cho, S.W.; et al. The chromatin accessibility landscape of primary human cancers. Science 2018, 362, eaav1898. [Google Scholar] [CrossRef]
- Ochsner, S.A.; Abraham, D.; Martin, K.; Ding, W.; McOwiti, A.; Kankanamge, W.; Wang, Z.; Andreano, K.; Hamilton, R.A.; Chen, Y.; et al. The Signaling Pathways Project, an integrated ’omics knowledgebase for mammalian cellular signaling pathways. Sci. Data 2019, 6, 252. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Zou, Z.; Ohta, T.; Miura, F.; Oki, S. ChIP-Atlas 2021 update: A data-mining suite for exploring epigenomic landscapes by fully integrating ChIP-seq, ATAC-seq and Bisulfite-seq data. Nucleic Acids Res. 2022, 50, W175–W182. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Song, C.; Zhao, J.; Zhang, Y.; Zhao, X.; Feng, C.; Zhang, G.; Zhu, J.; Wang, F.; Qian, F.; et al. SEdb 2.0: A comprehensive super-enhancer database of human and mouse. Nucleic Acids Res. 2023, 51, D280–D290. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Girbig, M.; Misiaszek, A.D.; Vorländer, M.K.; Lafita, A.; Grötsch, H.; Baudin, F.; Bateman, A.; Müller, C.W. Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states. Nat. Struct. Mol. Biol. 2021, 28, 210–219. [Google Scholar] [CrossRef]
- Ramsay, E.P.; Abascal-Palacios, G.; Daiß, J.L.; King, H.; Gouge, J.; Pilsl, M.; Beuron, F.; Morris, E.; Gunkel, P.; Engel, C.; et al. Structure of human RNA polymerase III. Nat. Commun. 2020, 11, 6409. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yu, Z.; Zhao, D.; Ren, Y.; Hou, H.; Xu, Y. Structure of human RNA polymerase III elongation complex. Cell Res. 2021, 31, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.; Van Bortle, K. RNA polymerase III transcription and cancer: A tale of two RPC7 subunits. Front. Mol. Biosci. 2022, 9, 1073795. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L. STREME: Accurate and versatile sequence motif discovery. Bioinformatics 2021, 37, 2834–2840. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Xia, N.; Angulo, B.; Carey, J.; Cady, Z.; Durruthy-Durruthy, J.; Bennett, T.; Sebastiano, V.; Reijo Pera, R.A. A distinct isoform of ZNF207 controls self-renewal and pluripotency of human embryonic stem cells. Nat. Commun. 2018, 9, 4384. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, J.; Skinner, M.K. Basic helix-loop-helix proteins can act at the E-box within the serum response element of the c-fos promoter to influence hormone-induced promoter activation in Sertoli cells. Mol. Endocrinol. 1999, 13, 774–786. [Google Scholar] [CrossRef]
- Wang, R.; Xu, Q.; Wang, C.; Tian, K.; Wang, H.; Ji, X. Multiomic analysis of cohesin reveals that ZBTB transcription factors contribute to chromatin interactions. Nucleic Acids Res. 2023, 51, 6784–6805. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.J.; Cheng, Y.C.; Liu, C.R.; Lin, S.; Liu, H.E. A small-molecule c-Myc inhibitor, 10058-F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp. Hematol. 2006, 34, 1480–1489. [Google Scholar] [CrossRef]
- García-Gutiérrez, L.; Delgado, M.D.; León, J. MYC Oncogene Contributions to Release of Cell Cycle Brakes. Genes 2019, 10, 244. [Google Scholar] [CrossRef]
- Iguchi, T.; Aoki, K.; Ikawa, T.; Taoka, M.; Taya, C.; Yoshitani, H.; Toma-Hirano, M.; Koiwai, O.; Isobe, T.; Kawamoto, H.; et al. BTB-ZF Protein Znf131 Regulates Cell Growth of Developing and Mature T Cells. J. Immunol. 2015, 195, 982–993. [Google Scholar] [CrossRef]
- Bastien, J.; Rochette-Egly, C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 2004, 328, 1–16. [Google Scholar] [CrossRef]
- Chandra, V.; Wu, D.; Li, S.; Potluri, N.; Kim, Y.; Rastinejad, F. The quaternary architecture of RARβ-RXRα heterodimer facilitates domain-domain signal transmission. Nat. Commun. 2017, 8, 868. [Google Scholar] [CrossRef] [PubMed]
- Chatagnon, A.; Veber, P.; Morin, V.; Bedo, J.; Triqueneaux, G.; Sémon, M.; Laudet, V.; d’Alché-Buc, F.; Benoit, G. RAR/RXR binding dynamics distinguish pluripotency from differentiation associated cis-regulatory elements. Nucleic Acids Res. 2015, 43, 4833–4854. [Google Scholar] [CrossRef] [PubMed]
- Lund, R.J.; Rahkonen, N.; Malonzo, M.; Kauko, L.; Emani, M.R.; Kivinen, V.; Närvä, E.; Kemppainen, E.; Laiho, A.; Skottman, H.; et al. RNA Polymerase III Subunit POLR3G Regulates Specific Subsets of PolyA. Stem Cell Rep. 2017, 8, 1442–1454. [Google Scholar] [CrossRef]
- Heim, K.C.; White, K.A.; Deng, D.; Tomlinson, C.R.; Moore, J.H.; Freemantle, S.J.; Spinella, M.J. Selective repression of retinoic acid target genes by RIP140 during induced tumor cell differentiation of pluripotent human embryonal carcinoma cells. Mol. Cancer 2007, 6, 57. [Google Scholar] [CrossRef]
- Dimberg, A.; Bahram, F.; Karlberg, I.; Larsson, L.G.; Nilsson, K.; Oberg, F. Retinoic acid-induced cell cycle arrest of human myeloid cell lines is associated with sequential down-regulation of c-Myc and cyclin E and posttranscriptional up-regulation of p27(Kip1). Blood 2002, 99, 2199–2206. [Google Scholar] [CrossRef]
- Hurlin, P.J.; Quéva, C.; Koskinen, P.J.; Steingrímsson, E.; Ayer, D.E.; Copeland, N.G.; Jenkins, N.A.; Eisenman, R.N. Mad3 and Mad4: Novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995, 14, 5646–5659. [Google Scholar] [CrossRef]
- Ostrowski, J.; Kuhns, J.E.; Lupisella, J.A.; Manfredi, M.C.; Beehler, B.C.; Krystek, S.R.; Bi, Y.; Sun, C.; Seethala, R.; Golla, R.; et al. Pharmacological and x-ray structural characterization of a novel selective androgen receptor modulator: Potent hyperanabolic stimulation of skeletal muscle with hypostimulation of prostate in rats. Endocrinology 2007, 148, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, B.; Zuo, L.; Wang, B.; Yan, Y.; Tian, K.; Zhou, R.; Wang, C.; Chen, X.; Jiang, Y.; et al. The transcriptional coactivator RUVBL2 regulates Pol II clustering with diverse transcription factors. Nat. Commun. 2022, 13, 5703. [Google Scholar] [CrossRef]
- Xhabija, B.; Kidder, B.L. KDM5B is a master regulator of the H3K4-methylome in stem cells, development and cancer. Semin. Cancer Biol. 2019, 57, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Stine, Z.E.; Walton, Z.E.; Altman, B.J.; Hsieh, A.L.; Dang, C.V. MYC, Metabolism, and Cancer. Cancer Discov. 2015, 5, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Hu, G.; Wei, G.; Cui, K.; Yamane, A.; Resch, W.; Wang, R.; Green, D.R.; Tessarollo, L.; Casellas, R.; et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012, 151, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Wong, P.P.; Miranda, F.; Chan, K.V.; Berlato, C.; Hurst, H.C.; Scibetta, A.G. Histone demethylase KDM5B collaborates with TFAP2C and Myc to repress the cell cycle inhibitor p21(cip) (CDKN1A). Mol. Cell Biol. 2012, 32, 1633–1644. [Google Scholar] [CrossRef] [PubMed]
- Garipler, G.; Lu, C.; Morrissey, A.; Lopez-Zepeda, L.S.; Pei, Y.; Vidal, S.E.; Zen Petisco Fiore, A.P.; Aydin, B.; Stadtfeld, M.; Ohler, U.; et al. The BTB transcription factors ZBTB11 and ZFP131 maintain pluripotency by repressing pro-differentiation genes. Cell Rep. 2022, 38, 110524. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Guo, J.; Deng, W.; Zhang, C.; Du, P.; Shi, T.; Ma, D. High-throughput cell-based screening reveals a role for ZNF131 as a repressor of ERalpha signaling. BMC Genom. 2008, 9, 476. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions, and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions, or products referred to in the content. |

| Primer | Sequence |
|---|---|
| ZNF131-F | CGAGCTCGGATCCGCCACCATGGAGGCTGAAGAGACGATGG |
| ZNF131-R | CTTATCGTCGTCATCCTTGTAATCTTCTAAAACTGGCAGAGCTGTT |
| ZNF207-R | CTTATCGTCGTCATCCTTGTAATCGTAACGGCCACCTTGCGACATT |
| ZNF207-F | CGAGCTCGGATCCGCCACCATGGGTCGCAAGAAGAAGAAGCAG |
| pcDNA-BB-R | CATGGTGGCGGATCCGAGCT |
| pcDNA-BB-F | GATTACAAGGATGACGACGATAAGTGA |
| siRNA | Sequence | Concentration |
|---|---|---|
| ZNF207-siRNA1 | rGrArUrGrArArArGrArCrGrArCrGrArCrUrUrCTT rGrArArGrUrCrGrUrCrGrUrCrUrUrUrCrArUrCTT | 100 nM |
| ZNF207-siRNA2 | rCrUrUrArGrCrUrArUrUrCrArUrUrGrCrArUrGTT rCrArUrGrCrArArUrGrArArUrArGrCrUrArArGTT | 200 nM |
| ZNF131-siRNA1 | rArArGrGrUrArUrUrGrArArArUrUrGrUrGrGrArArCTT rGrUrUrCrCrArCrArArUrUrUrCrArArUrArCrCrUrUTT | 100 nM |
| ZNF131-siRNA2 | rArArGrGrUrArCrUrGrArArGrUrArCrArUrGrUrArGTT rCrUrArCrArUrGrUrArCrUrUrCrArGrUrArCrCrUrUTT | 100 nM |
| Scramble-siRNA | rUrUrCrUrCrCrGrArArCrGrUrGrUrCrArCrGrUTT rArCrGrUrGrArCrArCrGrUrUrCrGrGrArGrArATT | 100 nM/200 nM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, R.; Zhou, S.; K C, R.; Lizarazo, S.; Mouli, L.; Jayanth, A.; Liu, Q.; Van Bortle, K. A Combinatorial Regulatory Platform Determines Expression of RNA Polymerase III Subunit RPC7α (POLR3G) in Cancer. Cancers 2023, 15, 4995. https://doi.org/10.3390/cancers15204995
Cheng R, Zhou S, K C R, Lizarazo S, Mouli L, Jayanth A, Liu Q, Van Bortle K. A Combinatorial Regulatory Platform Determines Expression of RNA Polymerase III Subunit RPC7α (POLR3G) in Cancer. Cancers. 2023; 15(20):4995. https://doi.org/10.3390/cancers15204995
Chicago/Turabian StyleCheng, Ruiying, Sihang Zhou, Rajendra K C, Simon Lizarazo, Leela Mouli, Anshita Jayanth, Qing Liu, and Kevin Van Bortle. 2023. "A Combinatorial Regulatory Platform Determines Expression of RNA Polymerase III Subunit RPC7α (POLR3G) in Cancer" Cancers 15, no. 20: 4995. https://doi.org/10.3390/cancers15204995
APA StyleCheng, R., Zhou, S., K C, R., Lizarazo, S., Mouli, L., Jayanth, A., Liu, Q., & Van Bortle, K. (2023). A Combinatorial Regulatory Platform Determines Expression of RNA Polymerase III Subunit RPC7α (POLR3G) in Cancer. Cancers, 15(20), 4995. https://doi.org/10.3390/cancers15204995





