DNA and RNA Alterations Associated with Colorectal Peritoneal Metastases: A Systematic Review
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Study Protocol and Registration
2.2. Search and Information Sources
2.3. Selection Process
2.3.1. Inclusion and Exclusion Criteria
2.3.2. Study Selection
2.3.3. Data Items and Collection Process
2.3.4. Study Risk of Bias Assessment
3. Results
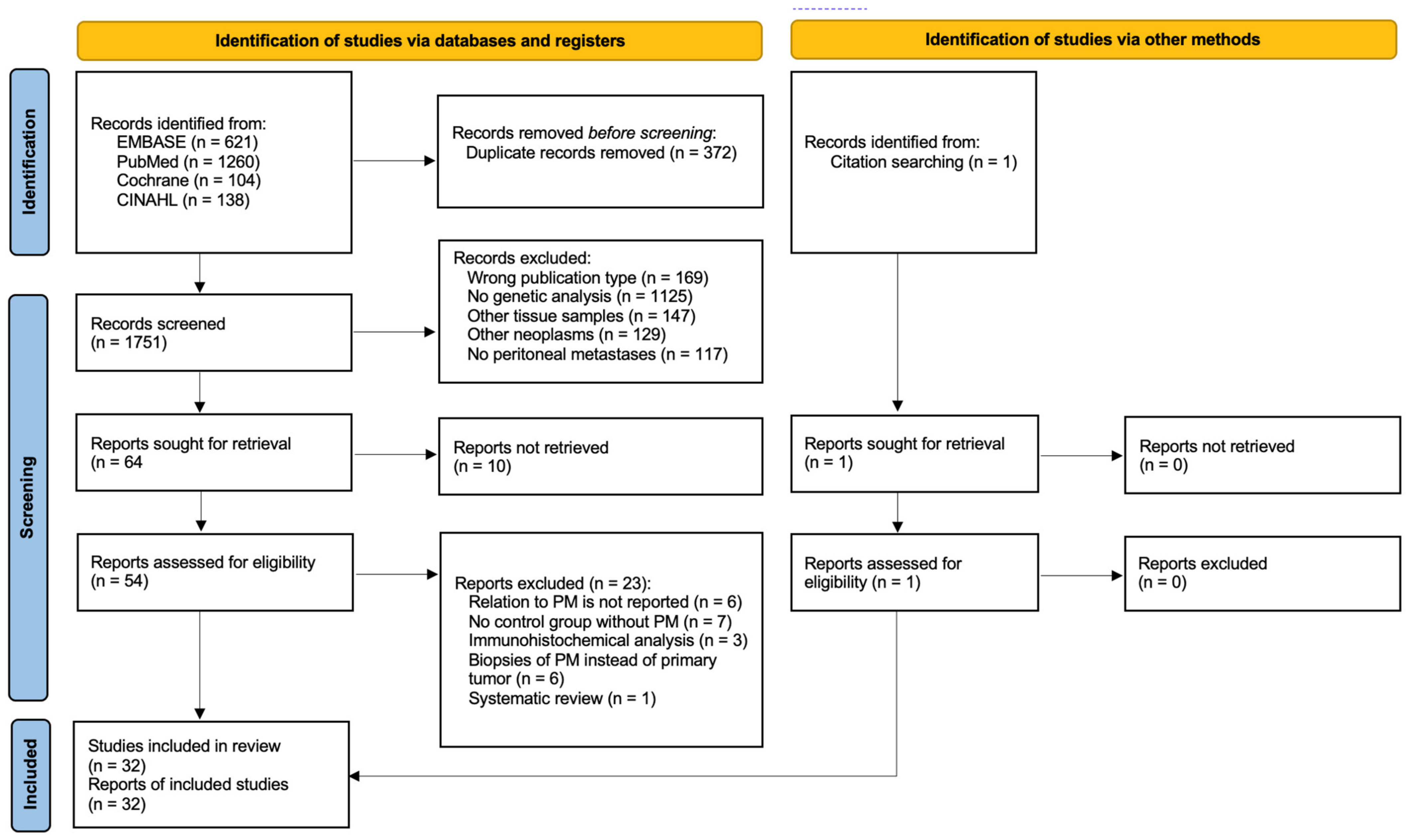
3.1. Study Selection
3.2. Study Characteristics
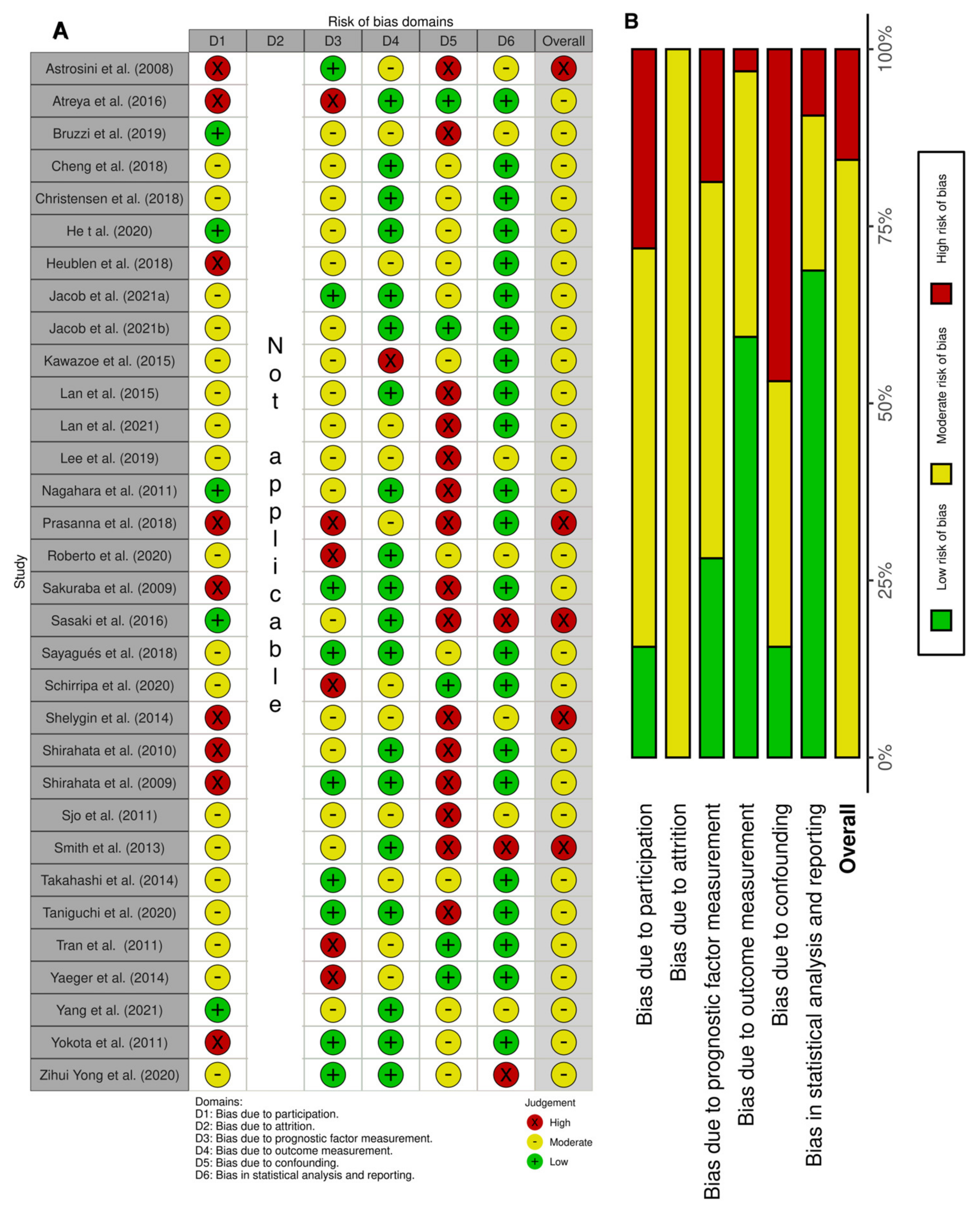
3.3. Risk of Bias in Studies
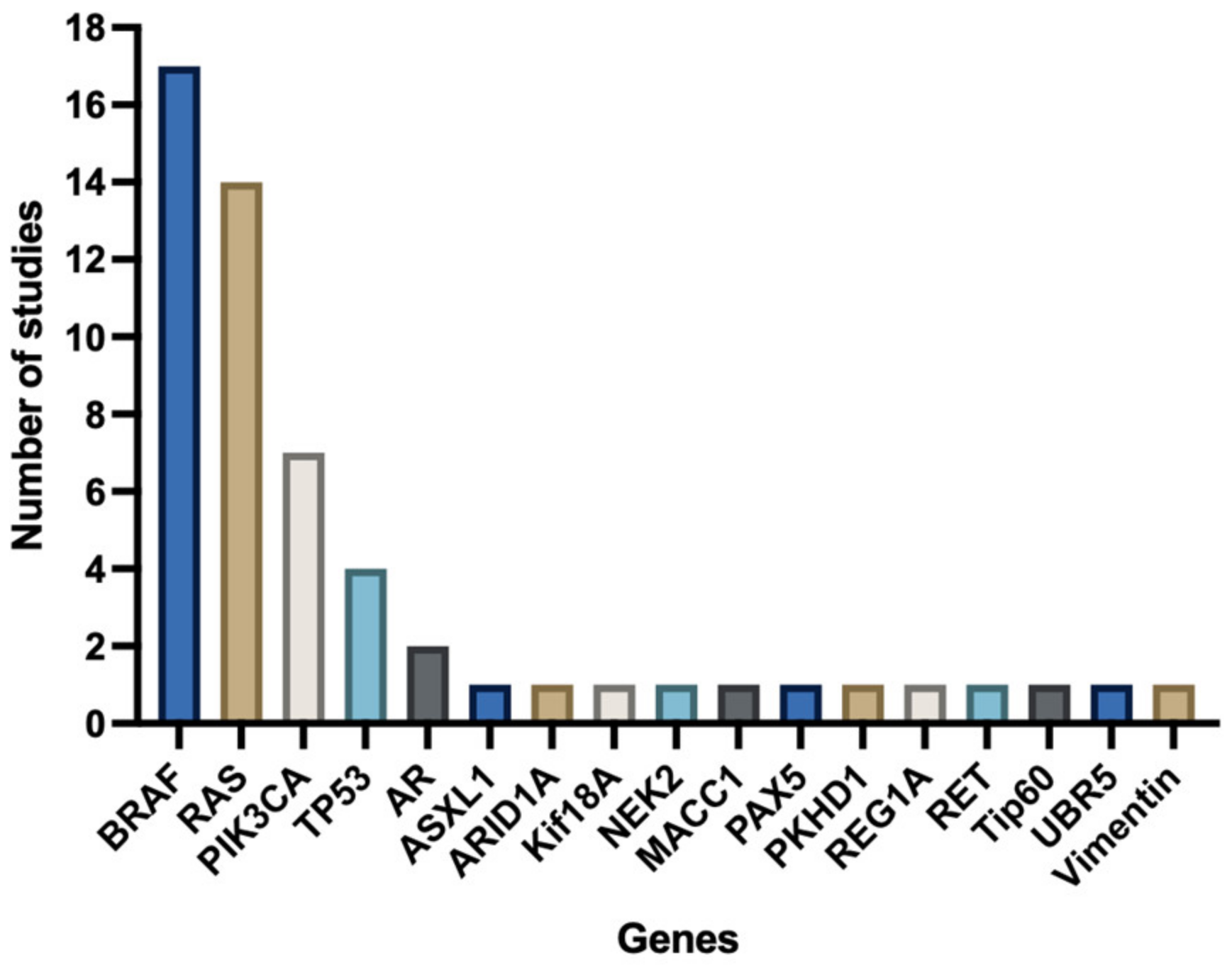
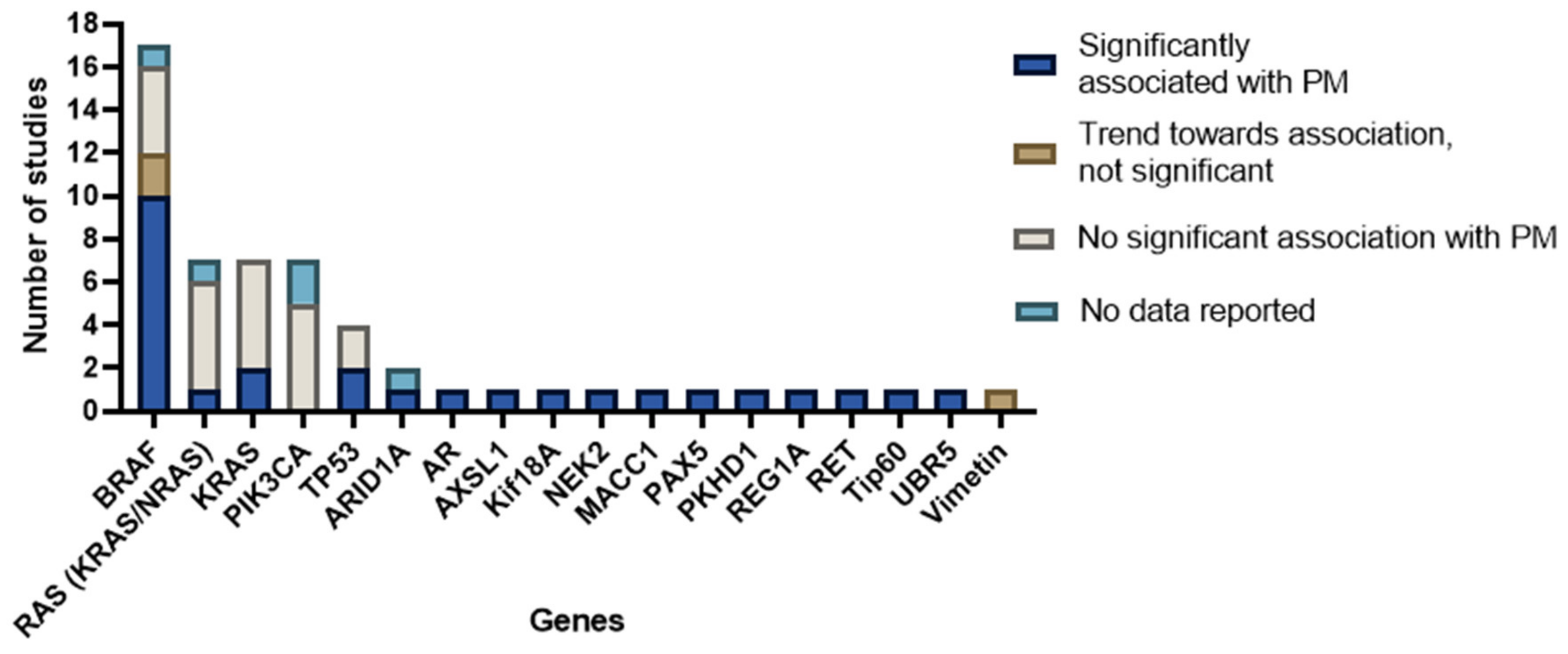
3.4. Reported Genes
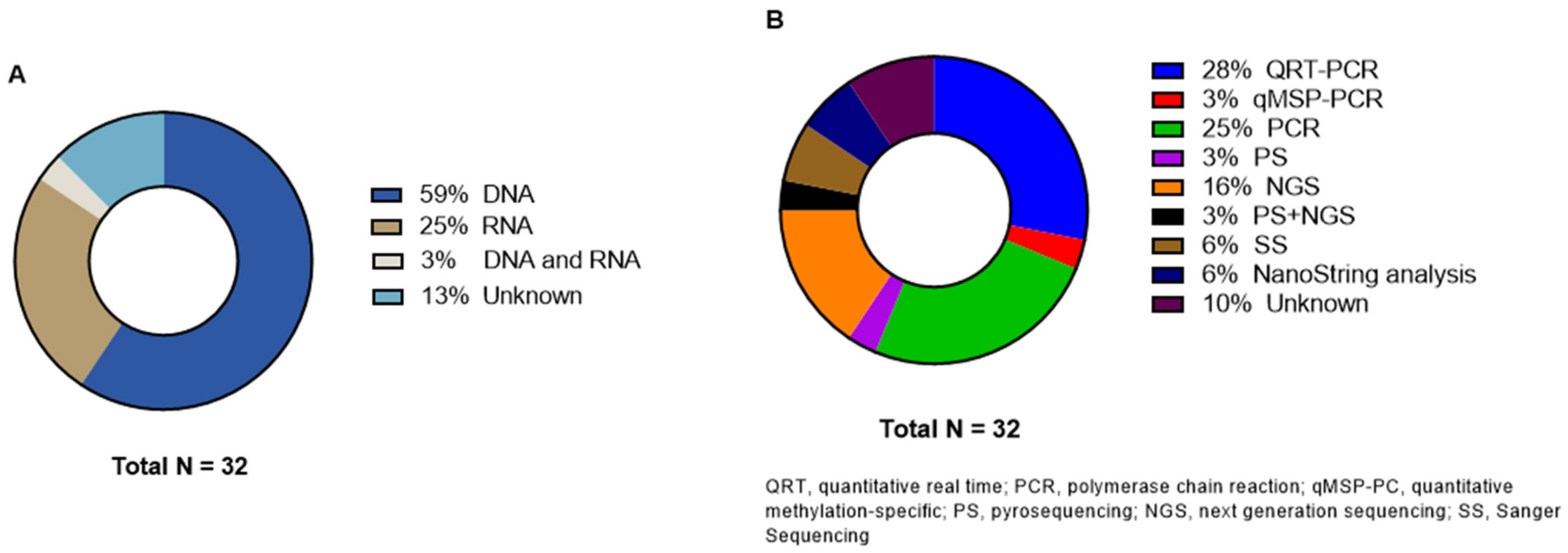
3.5. Genetic Analysis Methods
3.6. DNA/RNA Alterations Outcomes and Association with PM
3.6.1. Mitogen-Activated Protein Kinase (MAPK) Pathway Outcomes
3.6.2. PIK3CA Outcomes
3.6.3. TP53 Outcomes
3.6.4. Other DNA Outcomes
3.6.5. RNA Outcomes
3.6.6. Results of Broader Panel Analyses
3.7. MSI Status
4. Discussion
4.1. BRAF Mutations
4.2. Other Mutations
4.3. MSI Status
4.4. Clinical Relevancy
4.5. Techniques
4.6. Limitations
4.7. Future Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Database | Search Syntax |
|---|---|
| Pubmed | (((“Colorectal Neoplasms”[Mesh]) OR (((((((((“Neoplasms”[Mesh]) OR (carcinoma*[tw])) OR (adenocarcinoma*[tw])) OR (neoplas*[tw])) OR (tumour*[tw])) OR (tumor*[tw])) OR (oncolog*[tw])) OR (malignan*[tw])) AND ((((colorectal[tiab]) OR (colon[tw])) OR (rectal[tw])) OR (rectum[tw])))) AND ((“Peritoneal Neoplasms”[Mesh]) OR ((((“Peritoneum”[Mesh]) OR (peritoneal[tiab])) OR (peritoneum[tiab])) AND (((((((cancer[tiab]) OR (carcinomatos*[tiab])) OR (metastas*[tiab])) OR (neoplas*[tiab])) OR (tumor*[tiab])) OR (tumour*[tiab])) OR (malignan*[tiab]))))) AND ((((((((“Mutation”[Mesh]) OR (“Genetic Testing”[Mesh])) OR (“Genetic Association Studies”[Mesh])) OR (“Gene Expression Profiling”[Mesh])) OR (“Biomarkers, Tumor”[Mesh])) OR (((((tumor*[tw]) OR (tumour*[tw])) OR (cancer[tw])) OR (predictive[tw])) AND ((biomarker*[tw]) OR (marker*[tw])))) OR (((((mutation*[tiab]) OR (next generation sequencing[tiab])) OR (Gene*[tiab])) OR (RNA[tw])) OR (DNA[tw])))) |
| Embase | (exp colorectal tumor/ or ((neoplasm/ or (carcinoma* or adenocarcinoma* or neoplas* or tumour* or tumor* or oncolog* or malignan*).ti,ab,kw.) ADJ3 (colorectal or colon or rectal or rectum).ti,ab,kw.)) and (peritoneum tumor/ or ((exp peritoneum/ or (peritoneal or peritoneum).ti,ab,kw.) ADJ3 (cancer or carcinomatos* or metastas* or neoplas* or tumor or tumour or malignan*).ti,ab,kw.)) and ((exp mutation/ or exp sequence analysis/ or exp genetic association study/ or exp gene expression profiling/ or exp tumor marker/) or ((((tumour or tumor or cancer or predictive) ADJ3 (biomarker* or marker*)) or (mutation* or next generation sequencing or Gene* or RNA or DNA)).ti,ab,kw.)) |
| Cochrane | ([mh “Colorectal Neoplasms”] or (([mh Neoplasms] or (carcinoma* or adenocarcinoma* or neoplas* or tumour* or tumor* or oncolog* or malignan*):ti,ab,kw) and (colorectal OR colon OR rectal OR rectum):ti,ab,kw)) and ([mh “Peritoneal Neoplasms”] or ((peritoneal or peritoneum):ti,ab,kw and (cancer OR carcinomatos* OR metastas* OR neoplas* OR tumor* OR tumour* OR malignan*):ti,ab,kw)) and (([mh Mutation] or [mh “Genetic Testing”] or [mh “Genetic Association Studies”] or [mh “Gene Expression Profiling”] or [mh “Biomarkers, Tumor”]) or (((tumor* OR tumour* OR cancer* OR predictive):ti,ab,kw and (biomarker* OR marker*):ti,ab,kw) or (mutation* or Gene* or RNA or DNA):ti,ab,kw)) |
| CINAHL | (MH Colorectal Neoplasms OR (MH neoplasms OR carcinoma OR adenocarcinoma* OR neoplas* OR tumour* OR tumor* OR oncolog* OR malignan*) AND (colorectal OR colon OR rectal OR rectum)) AND (MH Peritoneal Neoplasms OR ((peritoneum OR peritoneal) AND (cancer OR carcinomatos* OR metastas* OR neoplas* OR tumor* OR tumour* OR malignan*))) AND ((MH mutation OR MH genetic testing OR MH Genetic Association Studied OR MH Gene Expression Profiling OR MH biomarkers) OR ((tumor* OR tumour* OR cancer OR predictive) AND (biomarker* OR marker*)) OR (mutation OR next-generation sequencing OR Gene* OR DNA OR RNA)) |
References
- Kamiyama, H.; Noda, H.; Konishi, F.; Rikiyama, T. Molecular biomarkers for the detection of metastatic colorectal cancer cells. World J. Gastroenterol. 2014, 20, 8928–8938. [Google Scholar] [CrossRef] [PubMed]
- Simkens, G.A.; Wintjens, A.; Rovers, K.P.; Nienhuijs, S.W.; de Hingh, I.H. Effective Strategies to Predict Survival of Colorectal Peritoneal Metastases Patients Eligible for Cytoreductive Surgery and HIPEC. Cancer Manag. Res. 2021, 13, 5239–5249. [Google Scholar] [CrossRef] [PubMed]
- Van Gestel, Y.R.; Thomassen, I.; Lemmens, V.E.; Pruijt, J.F.; van Herk-Sukel, M.P.; Rutten, H.J.; Creemers, G.J.; de Hingh, I.H. Metachronous peritoneal carcinomatosis after curative treatment of colorectal cancer. Eur. J. Surg. Oncol. 2014, 40, 963–969. [Google Scholar] [CrossRef]
- Lurvink, R.J.; Bakkers, C.; Rijken, A.; van Erning, F.N.; Nienhuijs, S.W.; Burger, J.W.; Creemers, G.J.; Verhoef, C.; Lemmens, V.E.; De Hingh, I.H. Increase in the incidence of synchronous and metachronous peritoneal metastases in patients with colorectal cancer: A nationwide study. Eur. J. Surg. Oncol. 2021, 47, 1026–1033. [Google Scholar] [CrossRef]
- Koppe, M.J.; Boerman, O.C.; Oyen, W.J.; Bleichrodt, R.P. Peritoneal carcinomatosis of colorectal origin: Incidence and current treatment strategies. Ann. Surg. 2006, 243, 212–222. [Google Scholar] [CrossRef]
- Klaver, Y.L.; Lemmens, V.E.; Nienhuijs, S.W.; Luyer, M.D.; de Hingh, I.H. Peritoneal carcinomatosis of colorectal origin: Incidence, prognosis and treatment options. World J. Gastroenterol. 2012, 18, 5489–5494. [Google Scholar] [CrossRef] [PubMed]
- Kranenburg, O.; van der Speeten, K.; de Hingh, I. Peritoneal Metastases From Colorectal Cancer: Defining and Addressing the Challenges. Front. Oncol. 2021, 11, 650098. [Google Scholar] [CrossRef] [PubMed]
- Jayne, D.G.; Fook, S.; Loi, C.; Seow-Choen, F. Peritoneal carcinomatosis from colorectal cancer. Br. J. Surg. 2002, 89, 1545–1550. [Google Scholar] [CrossRef] [PubMed]
- Bakkers, C.; Lurvink, R.J.; Rijken, A.; Nienhuijs, S.W.; Kok, N.F.; Creemers, G.J.; Verhoef, C.; Lemmens, V.E.; van Erning, F.N.; De Hingh, I.H. Treatment Strategies and Prognosis of Patients With Synchronous or Metachronous Colorectal Peritoneal Metastases: A Population-Based Study. Ann. Surg. Oncol. 2021, 28, 9073–9083. [Google Scholar] [CrossRef]
- Maggiori, L.; Elias, D. Curative treatment of colorectal peritoneal carcinomatosis: Current status and future trends. Eur. J. Surg. Oncol. 2010, 36, 599–603. [Google Scholar] [CrossRef]
- Klaver, C.E.L.; Wisselink, D.D.; Punt, C.J.A.; Snaebjornsson, P.; Crezee, J.; Aalbers, A.G.J.; Brandt, A.; Bremers, A.J.A.; Burger, J.W.A.; Fabry, H.F.J.; et al. Adjuvant hyperthermic intraperitoneal chemotherapy in patients with locally advanced colon cancer (COLOPEC): A multicentre, open-label, randomised trial. Lancet Gastroenterol. Hepatol. 2019, 4, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Neumann, J.; Löhrs, L.; Albertsmeier, M.; Reu, S.; Guba, M.; Werner, J.; Kirchner, T.; Angele, M. Cancer Stem Cell Markers Are Associated With Distant Hematogenous Liver Metastases But Not With Peritoneal Carcinomatosis in Colorectal Cancer. Cancer Investig. 2015, 33, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Karunasena, E.; Sham, J.; McMahon, K.W.; Ahuja, N. Genomics of Peritoneal Malignancies. Surg. Oncol. Clin. 2018, 27, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Roth, L.; Russo, L.; Ulugoel, S.; Freire Dos Santos, R.; Breuer, E.; Gupta, A.; Lehmann, K. Peritoneal Metastasis: Current Status and Treatment Options. Cancers 2021, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Pino, M.S.; Chung, D.C. The chromosomal instability pathway in colon cancer. Gastroenterology 2010, 138, 2059–2072. [Google Scholar] [CrossRef]
- Lipsyc, M.; Yaeger, R. Impact of somatic mutations on patterns of metastasis in colorectal cancer. J. Gastrointest. Oncol. 2015, 6, 645–649. [Google Scholar] [CrossRef]
- Venkatachalam, R.; Ligtenberg, M.J.; Hoogerbrugge, N.; Geurts van Kessel, A.; Kuiper, R.P. Predisposition to colorectal cancer: Exploiting copy number variation to identify novel predisposing genes and mechanisms. Cytogenet. Genome Res. 2008, 123, 188–194. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Syst. Rev. 2021, 10, 89. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Astrosini, C.; Roeefzaad, C.; Dai, Y.Y.; Dieckgraefe, B.K.; Jöns, T.; Kemmner, W. REG1A expression is a prognostic marker in colorectal cancer and associated with peritoneal carcinomatosis. Int. J. Cancer 2008, 123, 409–413. [Google Scholar] [CrossRef]
- Bruzzi, M.; Auclin, E.; Lo Dico, R.; Voron, T.; Karoui, M.; Espin, E.; Cianchi, F.; Weitz, J.; Buggenhout, A.; Malafosse, R.; et al. Influence of Molecular Status on Recurrence Site in Patients Treated for a Stage III Colon Cancer: A Post Hoc Analysis of the PETACC-8 Trial. Ann. Surg. Oncol. 2019, 26, 3561–3567. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.H.; Lin, J.K.; Chen, W.S.; Jiang, J.K.; Yang, S.H.; Chang, S.C. Clinical significance of the BRAFV600E mutation in Asian patients with colorectal cancer. Int. J. Color. Dis. 2018, 33, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Wang, Y.; Zhong, Y.; Pan, X.; Si, L.; Lu, J. KRAS Codon 12 Mutation is Associated with More Aggressive Invasiveness in Synchronous Metastatic Colorectal Cancer (mCRC): Retrospective Research. Onco Targets Ther. 2020, 13, 12601–12613. [Google Scholar] [CrossRef] [PubMed]
- Heublein, S.; Albertsmeier, M.; Pfeifer, D.; Loehrs, L.; Bazhin, A.V.; Kirchner, T.; Werner, J.; Neumann, J.; Angele, M.K. Association of differential miRNA expression with hepatic vs. peritoneal metastatic spread in colorectal cancer. BMC Cancer 2018, 18, 201. [Google Scholar] [CrossRef]
- Jacob, S.; Bösch, F.; Schoenberg, M.B.; Pretzsch, E.; Lampert, C.; Haoyu, R.; Renz, B.W.; Michl, M.; Kumbrink, J.; Kirchner, T.; et al. Expression of CIB1 correlates with colorectal liver metastases but not with peritoneal carcinomatosis. BMC Cancer 2021, 21, 1243. [Google Scholar] [CrossRef]
- Jacob, S.; Jurinovic, V.; Lampert, C.; Pretzsch, E.; Kumbrink, J.; Neumann, J.; Haoyu, R.; Renz, B.W.; Kirchner, T.; Guba, M.O.; et al. The association of immunosurveillance and distant metastases in colorectal cancer. J. Cancer Res. Clin. Oncol. 2021, 147, 3333–3341. [Google Scholar] [CrossRef]
- Kawazoe, A.; Shitara, K.; Fukuoka, S.; Kuboki, Y.; Bando, H.; Okamoto, W.; Kojima, T.; Fuse, N.; Yamanaka, T.; Doi, T.; et al. A retrospective observational study of clinicopathological features of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with metastatic colorectal cancer. BMC Cancer 2015, 15, 258. [Google Scholar] [CrossRef]
- Lan, Y.T.; Jen-Kou, L.; Lin, C.H.; Yang, S.H.; Lin, C.C.; Wang, H.S.; Chen, W.S.; Lin, T.C.; Jiang, J.K.; Chang, S.C. Mutations in the RAS and PI3K pathways are associated with metastatic location in colorectal cancers. J. Surg. Oncol. 2015, 111, 905–910. [Google Scholar] [CrossRef]
- Lan, Y.T.; Chang, S.C.; Lin, P.C.; Lin, C.H.; Liang, W.Y.; Chen, W.S.; Jiang, J.K.; Yang, S.H.; Lin, J.K. High concordance of mutation patterns in 10 common mutated genes between tumor tissue and cell-free DNA in metastatic colorectal cancer. Am. J. Cancer Res. 2021, 11, 2228–2237. [Google Scholar]
- Lee, J.H.; Ahn, B.K.; Baik, S.S.; Lee, K.H. Comprehensive Analysis of Somatic Mutations in Colorectal Cancer With Peritoneal Metastasis. Vivo 2019, 33, 447–452. [Google Scholar] [CrossRef]
- Nagahara, M.; Nishida, N.; Iwatsuki, M.; Ishimaru, S.; Mimori, K.; Tanaka, F.; Nakagawa, T.; Sato, T.; Sugihara, K.; Hoon, D.S.; et al. Kinesin 18A expression: Clinical relevance to colorectal cancer progression. Int. J. Cancer 2011, 129, 2543–2552. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, K.; Yasuda, T.; Sakata, M.; Kitamura, Y.H.; Shirahata, A.; Goto, T.; Mizukami, H.; Saito, M.; Ishibashi, K.; Kigawa, G.; et al. Down-regulation of Tip60 gene as a potential marker for the malignancy of colorectal cancer. Anticancer Res. 2009, 29, 3953–3955. [Google Scholar] [PubMed]
- Sasaki, Y.; Hamaguchi, T.; Yamada, Y.; Takahashi, N.; Shoji, H.; Honma, Y.; Iwasa, S.; Okita, N.; Takashima, A.; Kato, K.; et al. Value of KRAS, BRAF, and PIK3CA Mutations and Survival Benefit from Systemic Chemotherapy in Colorectal Peritoneal Carcinomatosis. Asian Pac. J. Cancer Prev. 2016, 17, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Sayagués, J.M.; Del Carmen, S.; Del Mar Abad, M.; Corchete, L.A.; Bengoechea, O.; Anduaga, M.F.; Baldeón, M.J.; Cruz, J.J.; Alcazar, J.A.; Angoso, M.; et al. Combined assessment of the TNM stage and BRAF mutational status at diagnosis in sporadic colorectal cancer patients. Oncotarget 2018, 9, 24081–24096. [Google Scholar] [CrossRef] [PubMed]
- Shelygin, Y.A.; Pospekhova, N.I.; Shubin, V.P.; Kashnikov, V.N.; Frolov, S.A.; Sushkov, O.I.; Achkasov, S.I.; Tsukanov, A.S. Epithelial-mesenchymal transition and somatic alteration in colorectal cancer with and without peritoneal carcinomatosis. Biomed. Res. Int. 2014, 2014, 629496. [Google Scholar] [CrossRef] [PubMed]
- Sjo, O.H.; Berg, M.; Merok, M.A.; Kolberg, M.; Svindland, A.; Lothe, R.A.; Nesbakken, A. Peritoneal carcinomatosis of colon cancer origin: Highest incidence in women and in patients with right-sided tumors. J. Surg. Oncol. 2011, 104, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.G.; Fisher, D.; Claes, B.; Maughan, T.S.; Idziaszczyk, S.; Peuteman, G.; Harris, R.; James, M.D.; Meade, A.; Jasani, B.; et al. Somatic profiling of the epidermal growth factor receptor pathway in tumors from patients with advanced colorectal cancer treated with chemotherapy ± cetuximab. Clin. Cancer Res. 2013, 19, 4104–4113. [Google Scholar] [CrossRef]
- Takahashi, Y.; Iwaya, T.; Sawada, G.; Kurashige, J.; Matsumura, T.; Uchi, R.; Ueo, H.; Takano, Y.; Eguchi, H.; Sudo, T.; et al. Up-regulation of NEK2 by microRNA-128 methylation is associated with poor prognosis in colorectal cancer. Ann. Surg. Oncol. 2014, 21, 205–212. [Google Scholar] [CrossRef]
- Taniguchi, H.; Uehara, K.; Nakayama, G.; Nakayama, H.; Aiba, T.; Hattori, N.; Kataoka, M.; Nakano, Y.; Kawase, Y.; Okochi, O.; et al. Tumor Location Is Associated With the Prevalence of Braf And PIK3CA Mutations in Patients with Wild-Type Ras Colorectal Cancer: A Prospective Multi-Center Cohort Study in Japan. Transl. Oncol. 2020, 13, 100786. [Google Scholar] [CrossRef]
- Yokota, T.; Ura, T.; Shibata, N.; Takahari, D.; Shitara, K.; Nomura, M.; Kondo, C.; Mizota, A.; Utsunomiya, S.; Muro, K.; et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br. J. Cancer 2011, 104, 856–862. [Google Scholar] [CrossRef]
- Shirahata, A.; Sakata, M.; Sakuraba, K.; Goto, T.; Mizukami, H.; Saito, M.; Ishibashi, K.; Kigawa, G.; Nemoto, H.; Sanada, Y.; et al. Vimentin methylation as a marker for advanced colorectal carcinoma. Anticancer Res. 2009, 29, 279–281. [Google Scholar] [PubMed]
- Shirahata, A.; Shinmura, K.; Kitamura, Y.; Sakuraba, K.; Yokomizo, K.; Goto, T.; Mizukami, H.; Saito, M.; Ishibashi, K.; Kigawa, G.; et al. MACC1 as a marker for advanced colorectal carcinoma. Anticancer Res. 2010, 30, 2689–2692. [Google Scholar] [PubMed]
- Atreya, C.E.; Greene, C.; McWhirter, R.M.; Ikram, N.S.; Allen, I.E.; Van Loon, K.; Venook, A.P.; Yeh, B.M.; Behr, S.C. Differential Radiographic Appearance of BRAF V600E-Mutant Metastatic Colorectal Cancer in Patients Matched by Primary Tumor Location. J. Natl. Compr. Cancer Netw. 2016, 14, 1536–1543. [Google Scholar] [CrossRef] [PubMed]
- Christensen, T.D.; Palshof, J.A.; Larsen, F.O.; Poulsen, T.S.; Høgdall, E.; Pfeiffer, P.; Jensen, B.V.; Yilmaz, M.K.; Nielsen, D. Associations between primary tumor RAS, BRAF and PIK3CA mutation status and metastatic site in patients with chemo-resistant metastatic colorectal cancer. Acta Oncol. 2018, 57, 1057–1062. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, T.; Karapetis, C.S.; Roder, D.; Tie, J.; Padbury, R.; Price, T.; Wong, R.; Shapiro, J.; Nott, L.; Lee, M.; et al. The survival outcome of patients with metastatic colorectal cancer based on the site of metastases and the impact of molecular markers and site of primary cancer on metastatic pattern. Acta Oncol. 2018, 57, 1438–1444. [Google Scholar] [CrossRef]
- Roberto, M.; Marchetti, P.; Arrivi, G.; Di Pietro, F.R.; Cascinu, S.; Gelsomino, F.; Caputo, F.; Cerma, K.; Ghidini, M.; Ratti, M.; et al. The treatment paradigm of right-sided metastatic colon cancer: Harboring BRAF mutation makes the difference. Int. J. Color. Dis. 2020, 35, 1513–1527. [Google Scholar] [CrossRef]
- Schirripa, M.; Nappo, F.; Cremolini, C.; Salvatore, L.; Rossini, D.; Bensi, M.; Businello, G.; Pietrantonio, F.; Randon, G.; Fucà, G.; et al. KRAS G12C Metastatic Colorectal Cancer: Specific Features of a New Emerging Target Population. Clin. Color. Cancer 2020, 19, 219–225. [Google Scholar] [CrossRef]
- Tran, B.; Kopetz, S.; Tie, J.; Gibbs, P.; Jiang, Z.Q.; Lieu, C.H.; Agarwal, A.; Maru, D.M.; Sieber, O.; Desai, J. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer 2011, 117, 4623–4632. [Google Scholar] [CrossRef]
- Yaeger, R.; Cercek, A.; Chou, J.F.; Sylvester, B.E.; Kemeny, N.E.; Hechtman, J.F.; Ladanyi, M.; Rosen, N.; Weiser, M.R.; Capanu, M.; et al. BRAF mutation predicts for poor outcomes after metastasectomy in patients with metastatic colorectal cancer. Cancer 2014, 120, 2316–2324. [Google Scholar] [CrossRef]
- Yang, Y.Z.; Hu, W.M.; Xia, L.P.; He, W.Z. Association between somatic RET mutations and clinical and genetic characteristics in patients with metastatic colorectal cancer. Cancer Med. 2021, 10, 8876–8882. [Google Scholar] [CrossRef]
- Zihui Yong, Z.; Ching, G.T.H.; Ching, M.T.C. Metastatic Profile of Colorectal Cancer: Interplay Between Primary Tumor Location and KRAS Status. J. Surg. Res. 2020, 246, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, T.; Wong, R.; Price, T.; Shapiro, J.; Tie, J.; Wong, H.L.; Nott, L.; Roder, D.; Lee, M.; Kosmider, S.; et al. Metastasectomy and BRAF mutation; an analysis of survival outcome in metastatic colorectal cancer. Curr. Probl. Cancer 2021, 45, 100637. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.H.; Joo, Y.E. Novel biomarkers for the diagnosis and prognosis of colorectal cancer. Intest. Res. 2020, 18, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Caputo, F.; Santini, C.; Bardasi, C.; Cerma, K.; Casadei-Gardini, A.; Spallanzani, A.; Andrikou, K.; Cascinu, S.; Gelsomino, F. BRAF-Mutated Colorectal Cancer: Clinical and Molecular Insights. Int. J. Mol. Sci. 2019, 20, 5369. [Google Scholar] [CrossRef]
- Sinicrope, F.A.; Shi, Q.; Smyrk, T.C.; Thibodeau, S.N.; Dienstmann, R.; Guinney, J.; Bot, B.M.; Tejpar, S.; Delorenzi, M.; Goldberg, R.M.; et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology 2015, 148, 88–99. [Google Scholar] [CrossRef]
- Argilés, G.; Tabernero, J.; Labianca, R.; Hochhauser, D.; Salazar, R.; Iveson, T.; Laurent-Puig, P.; Quirke, P.; Yoshino, T.; Taieb, J.; et al. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2020, 31, 1291–1305. [Google Scholar] [CrossRef]
- Janakiraman, M.; Vakiani, E.; Zeng, Z.; Pratilas, C.A.; Taylor, B.S.; Chitale, D.; Halilovic, E.; Wilson, M.; Huberman, K.; Ricarte Filho, J.C.; et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010, 70, 5901–5911. [Google Scholar] [CrossRef]
- De Roock, W.; Claes, B.; Bernasconi, D.; De Schutter, J.; Biesmans, B.; Fountzilas, G.; Kalogeras, K.T.; Kotoula, V.; Papamichael, D.; Laurent-Puig, P.; et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010, 11, 753–762. [Google Scholar] [CrossRef]
- Popat, S.; Hubner, R.; Houlston, R.S. Systematic review of microsatellite instability and colorectal cancer prognosis. J. Clin. Oncol. 2005, 23, 609–618. [Google Scholar] [CrossRef]
- Segelman, J.; Granath, F.; Holm, T.; Machado, M.; Mahteme, H.; Martling, A. Incidence, prevalence and risk factors for peritoneal carcinomatosis from colorectal cancer. Br. J. Surg. 2012, 99, 699–705. [Google Scholar] [CrossRef]
- Arjona-Sánchez, A.; Barrios, P.; Boldo-Roda, E.; Camps, B.; Carrasco-Campos, J.; Concepción Martín, V.; García-Fadrique, A.; Gutiérrez-Calvo, A.; Morales, R.; Ortega-Pérez, G.; et al. HIPECT4: Multicentre, randomized clinical trial to evaluate safety and efficacy of Hyperthermic intra-peritoneal chemotherapy (HIPEC) with Mitomycin C used during surgery for treatment of locally advanced colorectal carcinoma. BMC Cancer 2018, 18, 183. [Google Scholar] [CrossRef] [PubMed]
- Sorich, M.J.; Wiese, M.D.; Rowland, A.; Kichenadasse, G.; McKinnon, R.A.; Karapetis, C.S. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: A meta-analysis of randomized, controlled trials. Ann. Oncol. 2015, 26, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Graf, W.; Cashin, P.H.; Ghanipour, L.; Enblad, M.; Botling, J.; Terman, A.; Birgisson, H. Prognostic Impact of BRAF and KRAS Mutation in Patients with Colorectal and Appendiceal Peritoneal Metastases Scheduled for CRS and HIPEC. Ann. Surg. Oncol. 2020, 27, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef] [PubMed]
- Damodaran, S.; Berger, M.F.; Roychowdhury, S. Clinical tumor sequencing: Opportunities and challenges for precision cancer medicine. Am. Soc. Clin. Oncol. Educ. Book 2015, 35, e175–e182. [Google Scholar] [CrossRef] [PubMed]
- Alorda-Clara, M.; Torrens-Mas, M.; Morla-Barcelo, P.M.; Martinez-Bernabe, T.; Sastre-Serra, J.; Roca, P.; Pons, D.G.; Oliver, J.; Reyes, J. Use of Omics Technologies for the Detection of Colorectal Cancer Biomarkers. Cancers 2022, 14, 817. [Google Scholar] [CrossRef]
- Lenos, K.J.; Bach, S.; Ferreira Moreno, L.; ten Hoorn, S.; Sluiter, N.R.; Bootsma, S.; Vieira Braga, F.A.; Nijman, L.E.; van den Bosch, T.; Miedema, D.M.; et al. Molecular characterization of colorectal cancer related peritoneal metastatic disease. Nat. Commun. 2022, 13, 4443. [Google Scholar] [CrossRef]

| Reference | Year | Country | Study Design | Patient Inclusion Period | Reasons for Patient Exclusion | No. of Subjects | No. of Genes Investigated | Aim of the Study | |
|---|---|---|---|---|---|---|---|---|---|
| Start | End | ||||||||
| Astrosini et al. [20] | 2008 | Germany | CH | - | - | - | 63 | 1 | To investigate if REG1A is upregulated in CRC patients with unfavorable clinical outcome. |
| Atreya et al. [43] | 2016 | USA | MCH | 01/2013 | 09/2015 | - | 120 | 1 | To investigate if BRAF V600E mutation is associated with sites and radiographic appearance of metastatic disease in patients matched by primary tumor location. |
| Bruzzi et al. [21] | 2019 | France | CH | 12/2005 | 11/2009 | - | 1650 | 2 | To assess recurrence patterns according to microsatellite instability, RAS and BRAFV600E status in stage III. |
| Cheng et al. [22] | 2018 | Taiwan | CH | 2000 | 2013 | History of other malignancies, inflammatory bowel disease or death within 30 days after surgery. | 1969 | 1 | To evaluate clinicopathological features, metastatic patterns, and prognostic value of CRC with the BRAFV600E mutation. |
| Christensen et al. [44] | 2018 | Denmark | CH | 01/2005 | 08/2008 | Presence of other active tumors and no tissue sample or medical charts available. | 448 | 3 | To investigate associations between mutations and pattern of metastases. |
| He et al. [23] | 2020 | China | CH | 12/2015 | 02/2020 | Non-metastatic synchronous CRC; neo-adjuvant therapy; location cecum, appendix or ileocecal junction; neuroendocrine components. | 194 | 3 | To investigate the connection between mutant KRAS, NRAS, and BRAF and clinicopathological characteristics in therapy-naïve synchronous mCRC in Chinese populations. |
| Heublein et al. [24] | 2018 | Germany | CH | 1988 | 2012 | - | 23 | 754 miRNAs | miRNA profiling of primary CRC tissue to identify miRNAs potentially associated with defining the site of metastatic spread in CRC. |
| Jacob et al. [25] | 2021 | Germany | CC | 01/2005 | 12/2014 | Lack of any of the baseline variables or specimens, HNPCC or FAP co-malignancies. | 18 | 770 | To identify genes associated with the metastatic route in CRC. |
| Jacob et al. [26] | 2021 | Germany | CH | - | - | Missing FFPE tissue of the primary tumor, co-malignancies, Lynch-syndrome or other hereditary diseases. | 18 | 770 | To elucidate the link between immunosurveillance and organotropism of metastases in CRC by evaluating different gene signatures and pathways. |
| Kawazoe et al. [27] | 2015 | Japan | CH | 01/2013 | 06/2014 | - | 264 | 4 | To evaluate mutations in Japanese mCRC patients and assessing their corresponding effects on the efficacy of anti-EGFR therapy. |
| Lan et al. [28] | 2015 | Taiwan | CH | 03/2000 | 01/2010 | No tissue sample available in the biobank. | 1492 | 7 | To analyze mutation spectra of the PI3K and RAS pathways in CRC and the associations with sites of metastases of recurrence. |
| Lan et al. [29] | 2021 | Taiwan | CH | - | - | Patients who had stage I–III CRC, received emergent surgery, or who did not have available tumor or preoperative serum samples in the biobank. | 95 | 10 | To evaluate the concordance of mutation patterns between tumor tissue DNA and circulating cell-free DNA in stage IV CRC patients and to analyze relationship between the mutational patterns and site of metastases. |
| Lee et al. [30] | 2019 | Korea | CH | 2004 | 2008 | - | 15 | 409 | Analyzing genetic mutations which may be presage PM. |
| Nagahara et al. [31] | 2011 | Japan | CH | 1993 | 2000 | Chemotherapy or radiotherapy before surgery. | 113 | 1 | To investigate if Kif18A has a role in the progression of CRC. |
| Prasanna et al. [45] | 2018 | Australia | CH | 01/2005 | 12/2015 | - | 5967 | 2 | To explore the outcome of patients with mCRC based on their site of metastases at diagnosis and to explore the association between tumor characteristics and site of metastases. |
| Roberto et al. [46] | 2020 | Italy | CH | 2008 | 2019 | - | 207 | 1 | To evaluate the outcome of right CRC patients according to BRAF status and the treatment performed. |
| Sakuraba et al. [32] | 2009 | Japan | CH | - | - | - | 38 | 1 | To evaluate the correlation between Tip60 expression and the clinicopathological findings. |
| Sasaki et al. [33] | 2016 | Japan | CH | 02/2006 | 10/2011 | Previous chemotherapy for advanced disease. | 526 | 3 | To compare the prognostic impact of modern chemotherapy or anti-EGFR monoclonal antibody between CRC patients with and without PM. |
| Sayagués et al. [34] | 2018 | Spain | CH | - | - | - | 87 | 4 | To investigate the frequency of mutations in primary sCRC tumors and their impact on patient progression-free survival and overall survival. |
| Schirripa et al. [47] | 2020 | Italy | CH | 01/2010 | 12/2018 | - | 499 | 1 | Description of the clinicopathologic features and prognosis of KRAS G12C-mutated metastatic CRC. |
| Shelygin et al. [35] | 2014 | Russia | CH | 11/2012 | 02/2014 | - | 58 | 7 | To describe the epithelial–mesenchymal transition in terms of gene expression profile and somatic changes in CRC patients with or without PM. |
| Shirahata et al. [42] | 2010 | Japan | CH | - | - | - | 52 | 1 | To examine the expression of the MACC1 gene in primary tumors and to evaluate the correlation between the MACC1 expression and the clinicopathological findings. |
| Shirahata et al. [41] | 2009 | Japan | CH | - | - | - | 48 | 1 | To examine the methylation status of the Vimentin gene in CRC patients and to evaluate the correlation between this status and the clinicopathological findings. |
| Sjo et al. [36] | 2011 | Norway | CH | 1987 | 2006 | Stage I–III or stage unknown, distant metastases without PM, no surgery or diverting procedures. | 57 | 1 | To evaluate the incidence of PM in CRC patients and to compare clinicopathological characteristics, survival and TP53 mutation status in primary tumors. |
| Smith et al. [37] | 2013 | UK | CH | - | - | Patients with resectable disease. | 2161 | 4 | To study the somatic molecular profile of the epidermal growth factor receptor pathway in advanced CRC, its relationship to prognosis, the site of the primary and metastases, and response to cetuximab. |
| Takahashi et al. [38] | 2013 | Japan | CH | 1992 | 2002 | - | 180 | 1 | To investigate the clinical significance of NEK2/miR128 expression in CRC. |
| Taniguchi et al. [39] | 2020 | Japan | CH | 08/2014 | 04/2016 | RAS-mutant tumors or unknown RAS status. | 331 | 2 | To present institutional experience with patients with CRC who underwent clinical mutation profiling and to evaluate the differences in patient characteristics with BRAF mutations. |
| Tran et al. [48] | 2011 | Australia | CH | - | - | - | 524 | 1 | To investigate whether BRAF mutant CRC is further defined by metastatic spread and evaluation of the impact of this mutation on prognosis. |
| Yaeger et al. [49] | 2014 | USA | CC | 2009 | 2012 | - | 515 | 1 | To determine the clinicopathologic characteristics, PIK3CA mutation frequency, and outcomes after metastasectomy in patients with BRAF-mutant mCRC. |
| Yang et al. [50] | 2021 | China | CH | 01/2015 | 03/2020 | Chemo- or radiotherapy before NGS and no follow-up information. | 582 | 1 | To evaluate the frequency and phenotypic characteristics of mCRC with somatic RET mutation. |
| Yokota et al. [40] | 2011 | Japan | CH | 2002 | 2010 | - | 229 | 2 | To investigate the clinicopathological features and prognostic impact of KRAS/BRAF mutation in advanced and recurrent CRC patients. |
| Zihui Yong et al. [51] | 2018 | Singapore | CH | 01/2010 | 12/2014 | Appendiceal tumors, other stages than stage 4 and patients without metastases. | 363 | 1 | To describe the metastatic pattern of advanced CRC by assessing the interaction between the KRAS mutational status and the location of primary tumors. |
| Reference | Patients’ Specification | No. of Patients for Analysis | Tissue Samples | Tissue Collection | |
|---|---|---|---|---|---|
| Without PM | With PM | ||||
| Astrosini et al. [20] | Nonpretreated CRC patients. | 54 | 9 | Primary tumor tissue | Retrospect, archives of pathology department |
| Atreya et al. [43] | Cohort of patients with BRAF mutant mCRC was matched 1:2 to patients with BRAF WT mCRC. | 75 | 45 | Not specified | Not performed, BRAF status was already known |
| Bruzzi et al. [21] | Patients with signed informed consent for biological sample collection from the PETACC-8 trial (stage III CRC patients) were included. | 1446 | 38 | FFPE tumor tissues | Retrospect, archives of pathology department |
| Cheng et al. [22] | Patients who underwent surgery for CRC. Stage IV patients are included for genetic analysis. | 260 | 76 | Frozen tumor tissue in liquid nitrogen | Retrospect, hospital biobank |
| Christensen et al. [44] | Patients with mCRC who had received multiple treatment line (irinotecaban and cetiuximab). | 405 | 43 | FFPE tumor tissue blocks from primary tumors | Not performed, BRAF, RAS and PIK3CA status was already known |
| He et al. [23] | Therapy-naïve synchronous mCRC patients at first diagnosis. | 168 | 26 | FFPE tumor tissue blocks | Retrospect, archives of pathology department |
| Heublein et al. [24] | Patients underwent surgical resection and were divided into three groups according to metastases location. | 10 | 13 | FFPE tissue from primary tumors | Retrospect, archives of pathology department |
| Jacob et al. [25] | Patients undergoing surgery. Four groups; patients without metastases, with LM, with PM and with LM and PM. | 12 | 6 | FFPE tissue from primary tumors | Retrospect, national database and biobank |
| Jacob et al. [26] | CRC patients surgically treated and divided in three groups: patients without metastases, with LM and with PM. | 12 | 6 | FFPE tissue from primary tumors | Retrospect, archives of pathology department |
| Kawazoe et al. [27] | CRC patients with histologically confirmed adenocarcinoma and presence of unresectable metastatic disease. | 212 | 52 | FFPE cancer specimens (239 primary tumors and 25 metastases) | Retrospect, archives of pathology department |
| Lan et al. [28] | Patients with stages I–IV CRC who underwent surgery. | 1388 | 104 | Surgery tissue samples | Retrospect, hospital biobank |
| Lan et al. [29] | Patients with stage IV CRC who underwent surgery. | 63 | 32 | Surgery tissue samples | Retrospect, hospital biobank |
| Lee et al. [30] | Small obstructive colorectal cancer group compared to large non-obstructing colorectal cancer group (contrast group). | 5 | 10 | FFPE surgical specimens | Retrospect, archives of pathology department |
| Nagahara et al. [31] | Patients identified as having primary CRC based on the clinicopathologic criteria described by the Japanese Society for Cancer of the Colon and Rectum. | 107 | 6 | Frozen tissue specimens in liquid nitrogen from colorectal tumor tissue and paired healthy tissue at least 10 cm distal from primary tumor | Retrospect, archives of pathology department |
| Prasanna et al. [45] | Patients with proven mCRC who were registered in the included databases. | 4755 | 1212 | Unknown | Not performed, BRAF and KRAS/RAS status was already known |
| Roberto et al. [46] | Patients with right mCRC with known BRAF mutation status. | 154 | 53 | Unknown | Not performed, BRAF status was already known |
| Sakuraba et al. [32] | Patients undergoing surgery for CRC. | 33 | 5 | Frozen tumor specimens and corresponding normal tissues | Retrospect, archives of pathology department |
| Sasaki et al. [33] | Patients with mCRC treated with systemic chemotherapy, combined with or without bevacizumab, cetuximab or panitumumab. | 409 | 117 | FFPE tumor samples | Retrospect, archives of pathology department |
| Sayagués et al. [34] | Caucasian patients diagnosed with CRC who underwent surgical resection of primary tumor tissues. | 80 | 7 | Freshly frozen primary tumor tissues | Retrospect, archives of pathology department |
| Schirripa et al. [47] | Patients with presence of a KRAS mutation with the focus of the specific variant. | 391 | 108 | FFPE tissue from primary tumors and/or paired metastases | Not performed, KRAS status was already known |
| Shelygin et al. [35] | Patients undergoing surgery for colorectal cancer. | 38 | 20 | Tissue samples from primary tumor, peritoneal metastases, and healthy tissue | Retrospect, archives of pathology department |
| Shirahata et al. [42] | CRC patients who underwent surgery. | 47 | 5 | Frozen colorectal cancer tissue and corresponding normal tissues | Were collected at surgical resection and stored at the pathology department |
| Shirahata et al. [41] | CRC patients who underwent surgery. | 43 | 5 | Frozen primary tumor specimens and corresponding normal tissue | Were collected at surgical resection and stored at the pathology department |
| Sjo et al. [36] | CRC patients with PM. | 57 | 148 * | FFPE tumor samples | Retrospect, archives of pathology department |
| Smith et al. [37] | Patients with measurable metastatic of locally advanced colorectal adenocarcinoma and unresectable disease. | 1667 | 283 | FFPE tumor samples (adenocarcinomas) | Retrospect, archives of pathology department |
| Takahashi et al. [38] | Patients with CRC who underwent surgical treatment. | 174 | 6 | Frozen resected tumor samples in liquid nitrogen | Retrospect, archives of pathology department |
| Taniguchi et al. [39] | Patients with CRC who received any treatment at one of the 15 study hospitals that participated, and who had RAS WT tumors. | 62 | 281 | FFPE tumor samples | Retrospect, archives of pathology department |
| Tran et al. [48] | Patients with mCRC with known BRAF mutation status from two institutional databases. | 385 | 139 | Tumor tissue | Not performed, BRAF status was already known |
| Yaeger et al. [49] | Patients with mCRC with available tumor sequencing. | 431 | 84 | FFPE primary tumor samples or metastatic tissue | Not performed, BRAF status was already known |
| Yang et al. [50] | Cohort of patients with mCRC. | 412 | 170 | Unknown | Not performed, RET status was already known |
| Yokota et al. [40] | Cohort of patients with CRC. | 175 | 54 | Frozen or FFPE tissues | Retrospect, archives of pathology department |
| Zihui Yong et al. [51] | Stage 4 CRC patients with metastases to the liver, lung, and/or peritoneum. | 266 | 89 | FFPE surgical specimens | Not performed, KRAS status was already known |
| Reference | Level of Testing | Name Genes, Molecules or Panel Investigated | Type of Analysis Performed | Gene or Molecule Name and Mutation or Expression Status (n) | No. of Patients with PM (N) and Outcomes (n) | No. of Patients without PM (N) and Outcomes (n) | MMR Status (MSI/MSS) | Findings as Reported by Authors in Studies | |
|---|---|---|---|---|---|---|---|---|---|
| Astrosini et al. [20] | RNA | REG1A | RT-PCR | N = 9 | N = 54 | N/A | REG1A expression levels highly correlated with formation of PM (median relative amount of 10.36 vs. 0.94, p = 0.0039 a). | ||
| REG1A expression | - | - | |||||||
| Atreya et al. [43] | DNA | BRAF | - | N = 45 | N = 75 | Total: 10/68 PM: 2/23 | No significant differences in metastatic sites were observed, although PM were more common in BRAF mutant patients (p = 0.045 †,b). | ||
| BRAF mutant (40) | 20 | 20 | |||||||
| BRAF wild-type (80) | 25 | 55 | |||||||
| Bruzzi et al. [21] | DNA | BRAF, RAS (KRAS and NRAS) | RT-PCR | N = 38 | N = 1446 | Only MSS included. | There is a trend for a higher rate of PM in BRAFV600E mutant compared to RAS mutant and wild-type patients (12.2% vs. 7.44% vs. 9.96% respectively, p > 0.05 c,d). | ||
| BRAF V600E mutant (127) | 15 | 112 | |||||||
| RAS mutant (748) | 56 | 692 | |||||||
| Double wild-type (609) | 61 | 548 | |||||||
| Cheng et al. [22] | DNA | BRAF | PCR or SNP genotyping assay | N = 76 | N = 260 | N/A | Stage IV CRC patients with a BRAFV600E mutation had a higher frequency of PM (41.7% vs. 21.2%, p = 0.04 d). | ||
| BRAF V600E mutant (312) | 66 | 246 | |||||||
| BRAF wild-type (24) | 10 | 14 | |||||||
| Heublein et al. [24] | miRNA | miRNAs | RT-PCR | N = 10 | N = 13 | N/A | A set of 31 miRNAs was significantly upregulated in the PM group, while 10 miRNAs were repressed as compared to LM. A set of 2 miRNAs was significantly upregulated in the PM group, while 25 were repressed as compared to M0. hsa-miR-31-5p was significantly overexpressed in PM patients (p = 0.002 b,d). | ||
| hsa-miR-215-5p | Induced 17-fold | Compared to LM | |||||||
| hsa-miR-31-3p | Induced 8.9-fold | Compared to LM | |||||||
| hsa- miR-31-5p | Induced 5.4-fold | Compared to LM | |||||||
| hsa-miR-483-5p | Repressed 0.04-fold | Compared to LM | |||||||
| hsa-miR-1226-5p | Repressed 0.29-fold | Compared to LM | |||||||
| hsa-miR- 296-5p | Repressed 0.32-fold | Compared to LM | |||||||
| hsa-miR-215-5p | Induced 3.6-fold | Compared to M0 | |||||||
| hsa-miR-148a-3p | Induced 2.8-fold | Compared to M0 | |||||||
| Jacob et al. [25] | mRNA | PanCancer Progression Panel | NanoString analysis | N = 6 | N = 12 | N/A | In PM patients, 18 genes demonstrated significant different expression rates (p < 0.05 b,d). | ||
| Not specified | - | - | |||||||
| Jacob et al. [26] | RNA | PanCancer Progression Panel | NanoString analysis | N = 6 | N = 12 | N/A | The analysis between patients with PM and M0 did not show a significant down- or upregulation of distinct gene sets. | ||
| Not described | - | - | |||||||
| Kawazoe et al. [27] | DNA | KRAS, NRAS, BRAF and PIK3CA | PCR | N = 52 | N = 212 | N/A | BRAF mutant tumors were more likely to have PM in comparison with BRAF wild-type tumors (50.0% vs. 18.0%, p = 0.009 c). No significant differences for PM according to RAS mutation (p = 0.64 d). | ||
| BRAF mutant (14) | 7 | 7 | |||||||
| RAS pathway mutant (21) | 4 | 17 | |||||||
| KRAS exon 2 mutant (90) | 15 | 75 | |||||||
| PIK3CA | N/A | N/A | |||||||
| Lan et al. [28] | DNA | RAS pathway (KRAS, NRAS, HRAS, BRAF) PI3K pathway | PCR | N = 104 | N = 1388 | Total: 154/1492 | PM was significantly higher in RAS pathway mutated patients compared to wild-type tumors (p = 0.009 d). Tumors with KRAS mutation had a trend toward a higher proportion of PM (p = 0.061 d). There was no association between PM and the presence of a PI3K pathway mutation (p = 0.408 d). | ||
| PI3K pathway mutant (213) | 12 | 201 | 36/177 | ||||||
| RAS pathway mutant (706) | 62 | 644 | 91/615 | ||||||
| BRAF mutant (70) | 8 | 62 | N/A | ||||||
| KRAS mutant (602) | 51 | 551 | |||||||
| NRAS mutant (49) | 5 | 44 | |||||||
| HRAS mutant (21) | 4 | 17 | |||||||
| Lan et al. [29] | DNA | TP53, APC, KRAS, FAT4, ARID1A, FBXW7, SMAD4, PIK3CA, NRAS and BRAF | NGS | N = 32 | N = 63 | N/A | For patients with PM, the frequency of genetic mutations was the highest in TP53. The authors conducted analysis to compare left- and right-sided CRC with mutation status but not between the non-PM and PM group. | ||
| TP53 mutant (59) | 19 | 40 | |||||||
| KRAS mutant (35) | 7 | 28 | |||||||
| APC mutant (45) | 17 | 28 | |||||||
| Lee et al. [30] | DNA | Life Technologies Ion AmpliSeq Comprehensive Cancer Panel | NGS | N = 10 | N = 5 | N/A | ARID1A, PKHD1, UBR5, PAX5, TP53, ASXL1 and AR were detected more frequently in the SOC group with PM (p values of 0.002, 0.019, 0.002, <0.001, 0.007, 0.047, 0.019 respectively d). TNFRSF14, VHL, MTRR, MLLT10, BIRC2, EP400, IRS2, PER1, TCF3 and CYP2D6 were detected more frequently in the LNOC group without PM (p values of 0.019, 0.004, 0.047, 0.022, <0.001, 0.022, <0.001, 0.022, 0.026, 0.004 respectively d). | ||
| ARID1A mutant | 9 | 0 | |||||||
| PKHD1 mutant | 7 | 0 | |||||||
| UBR5 mutant | 9 | 0 | |||||||
| PAX5 mutant | 10 | 0 | |||||||
| TP53 mutant | 8 | 0 | |||||||
| ASXL1 mutant | 8 | 1 | |||||||
| AR mutant | 7 | 0 | |||||||
| TNFRSF14 mutant | 5 | 3 | |||||||
| VHL mutant | 4 | 0 | |||||||
| MTRR mutant | 4 | 2 | |||||||
| MLLT10 mutant | 3 | 0 | |||||||
| BIRC2 mutant | 5 | 0 | |||||||
| EP400 mutant | 3 | 0 | |||||||
| IRS2 mutant | 5 | 0 | |||||||
| PER1 mutant | 3 | 0 | |||||||
| TCF3 mutant | 5 | 3 | |||||||
| CYP2D6 mutant | 4 | 0 | |||||||
| Nagahara et al. [31] | RNA | Kif18A | RT-PCR | N = 6 | N = 107 | N/A | Kif18A overexpression in CRC significantly correlated with PM (p = 0.02 d). | ||
| Kif18A low expression | 0 | 38 | |||||||
| Kif18A high expression | 6 | 69 | |||||||
| Prasanna et al. [45] | - | BRAF, RAS | - | N = unknown | N = unknown | PM: 29/239 M0: 77/940 | BRAF-mutated colorectal cancer showed higher incidence of PM with a relative risk of 1.8 (p < 0.001 e). KRAS-mutated patients showed no higher incidence of PM with a relative risk of 0.95 (p = 0.63 e). | ||
| RAS mutant (965) | 199 | 766 | |||||||
| RAS wild-type (1271) | 274 | 997 | |||||||
| BRAF mutant (143) | 51 | 92 | |||||||
| BRAF wild-type (1058) | 208 | 850 | |||||||
| Roberto et al. [46] | - | BRAF | - | N = 53 | N = 154 | Total: 19/66 | BRAF mutant right colorectal cancer was significantly more likely to occur with peritoneal metastases (38.1% vs. 22.4%, p = 0.003 d). | ||
| BRAF mutant (42) | 16 | 26 | |||||||
| Sakuraba et al. [32] | RNA | Tip60 | RT-PCR | N = 5 | N = 33 | N/A | The authors found that Tip60 downregulation (compared to healthy tissue expression) showed significant correlation with PM (p = 0.0053 d). | ||
| Downregulation of Tip60 expression | 3 | 2 | |||||||
| Sasaki et al. [33] | DNA | BRAF, KRAS, PIK3CA | PCR | N = 117 | N = 409 | N/A | The PM group had a significantly higher incidence of the BRAF V600E mutation than the non-PM group (27.7% vs. 7.3%, p < 0.01 d). In contrast, no differences were observed between the two groups in KRAS and PIK3CA mutations (p 0.42 d and 0.76, d respectively). | ||
| KRAS wild-type | 54 | 163 | |||||||
| KRAS mutation | 46 | 115 | |||||||
| BRAF wild-type | 34 | 115 | |||||||
| BRAF mutation | 13 | 9 | |||||||
| PIK3CA wild-type | 53 | 181 | |||||||
| PIK3CA mutation | 5 | 20 | |||||||
| Sayagués et al. [34] | DNA | KRAS/NRAS, BRAF and TP53 | NGS | N = 7 | N = 80 | Total: 6/48 | BRAF-mutated CRC tumors were significantly associated with PM (p = 0.006 d). | ||
| KRAS mutant (24) | 1 | 23 | 0/16 | ||||||
| NRAS mutant (1) | 0 | 1 | 0/0 | ||||||
| BRAF mutant (6) | 3 | 3 | 3/2 | ||||||
| TP53 mutant (29) | 2 | 27 | 1/21 | ||||||
| Schirripa et al. [47] | - | KRAS | Sanger sequencing, Sequenom MassArray | N = 108 | N = 391 | N/A | Compared to other KRAS-mutated cases, KRAS G12C mutations had a lower frequency in PM patients (13.5% vs. 25%, p = 0.008 d). | ||
| KRAS mutant (694) | 90 | 276 | |||||||
| KRAS G12C mutant (145) | 18 | 115 | |||||||
| Shelygin et al. [35] | DNA and RNA | KRAS, BRAF and EMT * | RT-PCR | N = 20 | N = 38 | PM: 2/18 M0: 6/32 | Mutations in KRAS and BRAF with PM was 70%, compared to 42.1% in M0 patients (p = 0.04 d). The frequency of wild types in both genes was 57.9% in CRC without PM compared to 30% with PM (p = 0.04 d). No differences were observed between the two groups in KRAS and BRAF mutations solely. | ||
| KRAS mutant | 11 | 15 | |||||||
| BRAF V600E | 3 | 1 | |||||||
| KRAS/BRAF wild-type | 6 | 22 | |||||||
| KRAS/BRAF mutant | 14 | 16 | |||||||
| EMT + (13) | 7 | 6 | |||||||
| EMT − (45) | 13 | 32 | |||||||
| Shirahata et al. [42] | RNA | MACC1 expression | RT-PCR | N = 5 | N = 47 | N/A | MACC1 expression showed significant correlation with PM compared to the group without PM (5.75 ± 4.58 vs. 2.57 ± 3.09, p = 0.042 b). | ||
| MACC1 expression | - | - | |||||||
| Shirahata et al. [41] | DNA | Vimentin methylation | qMSP | N = 5 | N = 43 | N/A | A trend was shown toward preferentially developing PM with Vimentin methylation (p = 0.080 d). | ||
| Vimentin + | 5 | 26 | |||||||
| Vimentin - | 0 | 17 | |||||||
| Sjo et al. [36] | DNA | TP53 | PCR | N = 49 | N = 148 | N/A | Univariate analyses demonstrated that PM was significantly associated with mutations in the TP53 gene (p = 0.05 f). Multivariate analyses confirmed the previous finding (OR = 2.4; 95%CI = 1.2–4.8, p = 0.013 g). | ||
| TP53 mutant | - | - | |||||||
| TP53 wild-type | - | - | |||||||
| Smith et al. [37] | DNA | KRAS, NRAS, BRAF, and PIK3CA | PS and Sequenom | N = 283 | N = 1667 | Co-occurred with BRAF wild-type tumors | BRAF mutations were more common in patients with PM-only compared to LM-only (22.2% vs. 6.7%, p = 0.00092 d), although this association with PM did not withstand correction for multiple testing. BRAF mutations were significantly associated with PM (p = 0.018), which did not remain significant after Bonferroni correction (p = 0.36). For KRAS, NRAS, and PIK3CA, there was no association found for PM. | ||
| KRAS mutant | 131 (/282) | 693 (/1667) | |||||||
| BRAF mutant | 36 (/283) | 193 (/1663) | |||||||
| NRAS mutant | 7 (/283) | 62 (/1656) | |||||||
| PIK3CA mutant | 40 (/280) | 293 (/1627) | |||||||
| Takahashi et al. [38] | RNA | NEK2 | RT-PCR | N = 6 | N = 174 | N/A | The high NEK2 expression group showed greater PM then the low NEK2 mRNA expression group (p = 0.004 b,d). | ||
| NEK2 low (90) | 0 | 90 | |||||||
| NEK2 high (90) | 6 | 84 | |||||||
| Taniguchi et al. [39] | DNA | RAS, NRAS, BRAF and PIK3CA | PCR | N = 62 | N = 281 | N/A | The frequencies of RAS/BRAFV600E wild-type over either BRAF or PIK3CA mutations were higher for PM (35% vs. 15%, p = 0.003 d). | ||
| RAS and BRAF wild-type (291) | 44 | 247 | |||||||
| RAS wild-type + BRAF or PIK3CA mutation (52) | 18 | 34 | |||||||
| Tran et al. [48] | DNA | BRAF | RT-PCR | N = 139 | N = 385 | Total: 40/310 | BRAF mutant tumors had significantly higher rates of PM (46% vs. 24%, p = 0.001 d). | ||
| BRAF mutant (57) | 26 | 31 | 12/30 | ||||||
| Yaeger et al. [49] | DNA | BRAF | Sanger sequencing, Sequenom MassArray | N = 84 | N = 431 | N/A | PM was significantly more common at the time of diagnosis of metastatic disease in the BRAF-mutant cases (26% vs. 14%, p < 0.01 d). | ||
| BRAF mutant (92) | 24 | 68 | |||||||
| Yang et al. [50] | DNA | RET | NGS | N = 170 | N = 412 | Total: 24/558 | The presence of RET mutations was associated with PM compared to wild-type tumors (56.2% vs. 28.4%, p = 0.024 d). | ||
| RET mutant (16) | 9 | 7 | 6/10 | ||||||
| Yokota et al. [40] | DNA | KRAS and BRAF | PCR | N = 54 | N = 175 | N/A | 60.0% of CRCs with BRAF mutation develops PM compared with 15% of CRCs with other subtypes (p = 0.0062 b,d). | ||
| KRAS/BRAF wild-type (135) | 30 | 105 | |||||||
| KRASG12X mutant (53) | 11 | 42 | |||||||
| KRASG13X mutant (26) | 4 | 22 | |||||||
| BRAFV600E mutant (15) | 9 | 6 | |||||||
| Zihui Yong et al. [51] | DNA | KRAS | PCR | N = 89 | N = 266 | N/A | After stratification, PM was associated with mutant KRAS tumors (26.6% vs. 15.1%, p = 0.02 d). | ||
| KRAS mutant (126) | 37 | 89 | |||||||
| Studies with synchronous and metachronous peritoneal metastases population | |||||||||
| Reference | Level of Testing | Genes or Panel Investigated | Type of Analysis | Gene or Molecule Name and Mutation or Expression Status (n) | No. of Patients with PM (N) with Outcomes (n) | No. of Patients without PM (N) with Corresponding Outcomes (n) | MMR Status (MSI/MSS) | Conclusions Findings as Reported by Authors in Studies | |
| Synch. | Metach. | ||||||||
| Christensen et al. [44] | DNA | RAS (KRAS and NRAS), BRAF and PIK3CA | NGS or Mutation kit and PS | N = 43 | N = 33 | N = 372 | N/A | PIK3CA mutations were significantly associated with absence of PM (OR = 0.10; 95%CI = 0.01–0.79, p = 0.028 g) and with a decreased hazard of developing PM (HR = 0.31; 95%CI = 0.11–0.86, p = 0.024 g). The hazard ratio of developing PM and having BRAF mutations were not associated with PM (OR = 2.07; 95%CI = 0.60-6.19, p = 0.192 g and (HR = 1.82; 95%CI = 0.81–4.08, p = 0.146 g).). | |
| RAS mutant (206) | 21 | 16 | 169 | ||||||
| BRAF V600E mutant (30) | 7 | 3 | 20 | ||||||
| PIK3CA mutant (61) | 1 | 3 | 57 | ||||||
| He et al. [23] | DNA | KRAS, BRAF, NRAS | NGS | N = 26 | N = 0 | N = 174 | N/A | Mutant KRAS tumors had a significant relevance with PM (p = 0.017 d). KRAS codon 12 mutation was more likely to present with PM (p = 0.014 d). Patients with PM had the tendency to carry mutant KRAS G12D (p = 0.052 d). Tumors with mutated BRAF were more likely to develop PM (p = 0.052 d). | |
| Any mutation (108) | 20 | - | 88 | ||||||
| KRAS mutant (77) | 15 | - | 62 | ||||||
| NRAS mutant (8) | 0 | - | 8 | ||||||
| BRAF mutant (23) | 5 | - | 18 | ||||||
| All wild-type (86) | 6 | - | 80 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heuvelings, D.J.I.; Wintjens, A.G.W.E.; Luyten, J.; Wilmink, G.E.W.A.; Moonen, L.; Speel, E.-J.M.; de Hingh, I.H.J.T.; Bouvy, N.D.; Peeters, A. DNA and RNA Alterations Associated with Colorectal Peritoneal Metastases: A Systematic Review. Cancers 2023, 15, 549. https://doi.org/10.3390/cancers15020549
Heuvelings DJI, Wintjens AGWE, Luyten J, Wilmink GEWA, Moonen L, Speel E-JM, de Hingh IHJT, Bouvy ND, Peeters A. DNA and RNA Alterations Associated with Colorectal Peritoneal Metastases: A Systematic Review. Cancers. 2023; 15(2):549. https://doi.org/10.3390/cancers15020549
Chicago/Turabian StyleHeuvelings, Danique J. I., Anne G. W. E. Wintjens, Julien Luyten, Guus E. W. A. Wilmink, Laura Moonen, Ernst-Jan M. Speel, Ignace H. J. T. de Hingh, Nicole D. Bouvy, and Andrea Peeters. 2023. "DNA and RNA Alterations Associated with Colorectal Peritoneal Metastases: A Systematic Review" Cancers 15, no. 2: 549. https://doi.org/10.3390/cancers15020549
APA StyleHeuvelings, D. J. I., Wintjens, A. G. W. E., Luyten, J., Wilmink, G. E. W. A., Moonen, L., Speel, E.-J. M., de Hingh, I. H. J. T., Bouvy, N. D., & Peeters, A. (2023). DNA and RNA Alterations Associated with Colorectal Peritoneal Metastases: A Systematic Review. Cancers, 15(2), 549. https://doi.org/10.3390/cancers15020549






