G1 Dynamics at the Crossroads of Pluripotency and Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. G1 Dynamics in Stem Cells and Cancer
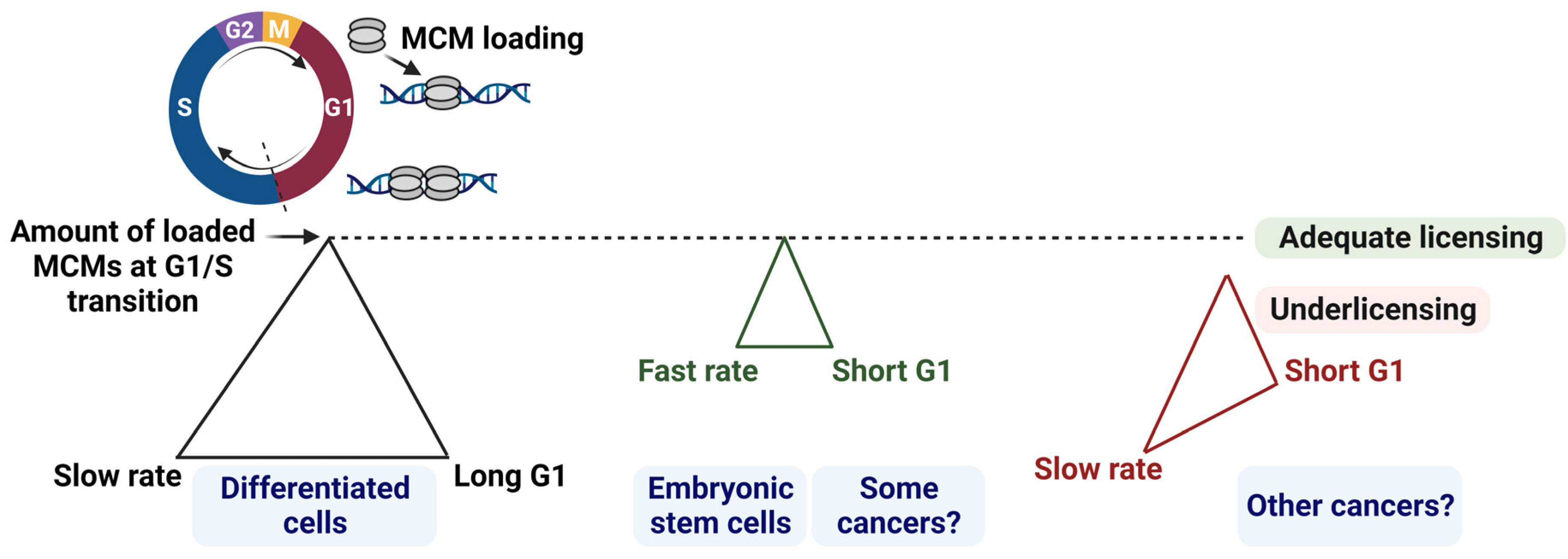
2.1. Balancing G1 Phase Length and Origin Licensing Dynamics
2.2. Consequences of a Short G1: A Common Feature in Stem Cells and Some Cancers
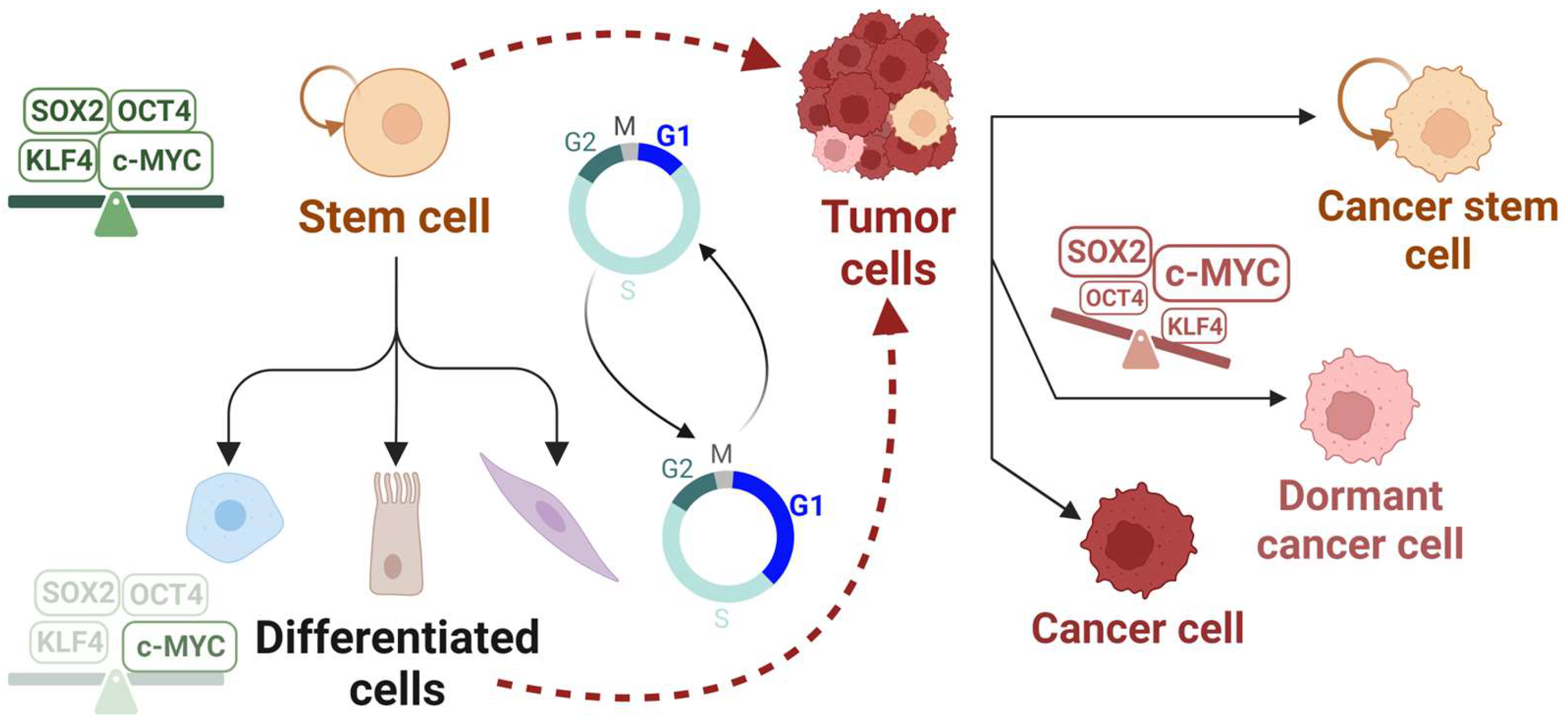
2.3. Cancer Resistance: Quiescence or a Cycling Stem Cell-like State?
3. Cell Cycle Regulators of G1 Progression in Stem Cells and Cancer
3.1. Cyclins and CDKs
3.2. CDK Inhibitors
4. G1 Origin Licensing Dynamics in Stems Cells and Cancer
4.1. Origin Licensing Factors
4.2. Dormant Origins in Cancer versus Stem Cells
5. Cell Cycle Regulation by the Reprogramming Factors in Stem Cells and Cancer
5.1. SOX2
5.2. OCT4
5.3. KLF4
5.4. c-MYC
6. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Becker, K.A.; Ghule, P.N.; Therrien, J.A.; Lian, J.B.; Stein, J.L.; van Wijnen, A.J.; Stein, G.S. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell. Physiol. 2006, 209, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, K.; Dakik, P.; Medkour, Y.; Mitrofanova, D.; Titorenko, V.I. Quiescence Entry, Maintenance, and Exit in Adult Stem Cells. Int. J. Mol. Sci. 2019, 20, 2158. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.G.; Croucher, P.I. The dormant cancer cell life cycle. Nat. Rev. Cancer 2020, 20, 398–411. [Google Scholar] [CrossRef]
- Wuebben, E.L.; Rizzino, A. The dark side of SOX2: Cancer—A comprehensive overview. Oncotarget 2017, 8, 44917–44943. [Google Scholar] [CrossRef]
- Wang, Y.-J.; Herlyn, M. The emerging roles of Oct4 in tumor-initiating cells. Am. J. Physiol.-Cell Physiol. 2015, 309, C709–C718. [Google Scholar] [CrossRef]
- Yoshida, G.J. Emerging roles of Myc in stem cell biology and novel tumor therapies. J. Exp. Clin. Cancer Res. 2018, 37, 1–20. [Google Scholar] [CrossRef]
- Tetreault, M.-P.; Yang, Y.; Katz, J.P. Krüppel-like factors in cancer. Nat. Rev. Cancer 2013, 13, 701–713. [Google Scholar] [CrossRef]
- Limas, J.C.; Cook, J.G. Preparation for DNA replication: The key to a successful S phase. FEBS Lett. 2019, 593, 2853–2867. [Google Scholar] [CrossRef]
- Matson, J.P.; Dumitru, R.; Coryell, P.; Baxley, R.M.; Chen, W.; Twaroski, K.; Webber, B.R.; Tolar, J.; Bielinsky, A.-K.; Purvis, J.E.; et al. Rapid DNA replication origin licensing protects stem cell pluripotency. eLife 2017, 6, e30473. [Google Scholar] [CrossRef]
- Padgett, J.; Santos, S.D. From clocks to dominoes: Lessons on cell cycle remodelling from embryonic stem cells. FEBS Lett. 2020, 594, 2031–2045. [Google Scholar] [CrossRef] [PubMed]
- Soufi, A.; Dalton, S. Cycling through developmental decisions: How cell cycle dynamics control pluripotency, differentiation and reprogramming. Development 2016, 143, 4301–4311. [Google Scholar] [CrossRef]
- Dalton, S. Linking the Cell Cycle to Cell Fate Decisions. Trends Cell Biol. 2015, 25, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Roccio, M.; Schmitter, D.; Knobloch, M.; Okawa, Y.; Sage, D.; Lutolf, M.P. Predicting stem cell fate changes by differential cell cycle progression patterns. Development 2013, 140, 459–470. [Google Scholar] [CrossRef]
- Smith, Z.D.; Nachman, I.; Regev, A.; Meissner, A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat. Biotechnol. 2010, 28, 521–526. [Google Scholar] [CrossRef]
- Resnitzky, D.; Gossen, M.; Bujard, H.; Reed, S.I. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol. 1994, 14, 1669–1679. [Google Scholar] [CrossRef]
- Strom, D.K.; Nip, J.; Westendorf, J.J.; Linggi, B.; Lutterbach, B.; Downing, J.R.; Lenny, N.; Hiebert, S.W. Expression of the AML-1 Oncogene Shortens the G1Phase of the Cell Cycle. J. Biol. Chem. 2000, 275, 3438–3445. [Google Scholar] [CrossRef]
- Macheret, M.; Halazonetis, T.D. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 2018, 555, 112–116. [Google Scholar] [CrossRef]
- Gaillard, H.; García-Muse, T.; Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 2015, 15, 276–289. [Google Scholar] [CrossRef]
- Fagundes, R.; Teixeira, L.K. Cyclin E/CDK2: DNA Replication, Replication Stress and Genomic Instability. Front. Cell Dev. Biol. 2021, 9, 774845. [Google Scholar] [CrossRef] [PubMed]
- Limas, J.C.; Littlejohn, A.N.; House, A.M.; Kedziora, K.M.; Mouery, B.L.; Ma, B.; Fleifel, D.; Walens, A.; Aleman, M.M.; Dominguez, D.; et al. Quantitative profiling of adaptation to cyclin E overproduction. Life Sci. Alliance 2022, 5, e202201378. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Zhang, W.; Cun, Y.; Li, J.; Liu, Y.; Gao, J.; Zhu, H.; Zhou, H.; Zhang, R.; Zheng, P. Mouse embryonic stem cells have increased capacity for replication fork restart driven by the specific Filia-Floped protein complex. Cell Res. 2018, 28, 69–89. [Google Scholar] [CrossRef]
- Tichy, E.D.; Pillai, R.; Deng, L.; Liang, L.; Tischfield, J.; Schwemberger, S.J.; Babcock, G.F.; Stambrook, P.J.; Zimmerlin, L.; Park, T.S.; et al. Mouse Embryonic Stem Cells, but Not Somatic Cells, Predominantly Use Homologous Recombination to Repair Double-Strand DNA Breaks. Stem Cells Dev. 2010, 19, 1699–1711. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Zhang, W.-D.; Duan, Y.-L.; Lu, Y.-Q.; Cun, Y.-X.; Li, C.-H.; Guo, K.; Nie, W.-H.; Li, L.; Zhang, R.; et al. Filia Is an ESC-Specific Regulator of DNA Damage Response and Safeguards Genomic Stability. Cell Stem Cell 2015, 16, 684–698. [Google Scholar] [CrossRef]
- Dumitru, R.; Gama, V.; Fagan, B.M.; Bower, J.J.; Swahari, V.; Pevny, L.H.; Deshmukh, M. Human Embryonic Stem Cells Have Constitutively Active Bax at the Golgi and Are Primed to Undergo Rapid Apoptosis. Mol. Cell 2012, 46, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.Q.; Han, J.; Cheng, E.-C.; Yamaguchi, S.; Shima, N.; Thomas, J.-L.; Lin, H. Embryonic Stem Cells License a High Level of Dormant Origins to Protect the Genome against Replication Stress. Stem Cell Rep. 2015, 5, 185–194. [Google Scholar] [CrossRef]
- Blakemore, D.; Vilaplana-Lopera, N.; Almaghrabi, R.; Gonzalez, E.; Moya, M.; Ward, C.; Murphy, G.; Gambus, A.; Petermann, E.; Stewart, G.S.; et al. MYBL2 and ATM suppress replication stress in pluripotent stem cells. EMBO Rep. 2021, 22, e51120. [Google Scholar] [CrossRef]
- Vitale, I.; Manic, G.; De Maria, R.; Kroemer, G.; Galluzzi, L. DNA Damage in Stem Cells. Mol. Cell 2017, 66, 306–319. [Google Scholar] [CrossRef]
- Cheung, T.H.; Rando, T.A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 2013, 14, 329–340. [Google Scholar] [CrossRef]
- Soppa, U.; Schumacher, J.; Ortiz, V.F.; Pasqualon, T.; Tejedor, F.; Becker, W. The Down syndrome-related protein kinase DYRK1A phosphorylates p27Kip1 and Cyclin D1 and induces cell cycle exit and neuronal differentiation. Cell Cycle 2014, 13, 2084–2100. [Google Scholar] [CrossRef] [PubMed]
- Boni, J.; Rubio-Perez, C.; López-Bigas, N.; Fillat, C.; De La Luna, S. The DYRK Family of Kinases in Cancer: Molecular Functions and Therapeutic Opportunities. Cancers 2020, 12, 2106. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, J.; Ramos-Valdes, Y.; Perampalam, P.; Litovchick, L.; DiMattia, G.E.; Dick, F.A. A Systematic Analysis of Negative Growth Control Implicates the DREAM Complex in Cancer Cell Dormancy. Mol. Cancer Res. 2017, 15, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. The cancer stem cell: Premises, promises and challenges. Nat. Med. 2011, 17, 313–319. [Google Scholar] [CrossRef]
- Kleinsmith, L.J.; Pierce, G.B., Jr. Multipotentiality of single embryonal carcinoma cells. Cancer Res. 1964, 24, 1544–1551. [Google Scholar]
- Kim, S.; Leong, A.; Kim, M.; Yang, H.W. CDK4/6 initiates Rb inactivation and CDK2 activity coordinates cell-cycle commitment and G1/S transition. Sci. Rep. 2022, 12, 16810. [Google Scholar] [CrossRef]
- Chung, M.; Liu, C.; Yang, H.W.; Köberlin, M.S.; Cappell, S.D.; Meyer, T. Transient Hysteresis in CDK4/6 Activity Underlies Passage of the Restriction Point in G1. Mol. Cell 2019, 76, 562–573.e4. [Google Scholar] [CrossRef]
- Yang, H.W.; Cappell, S.D.; Jaimovich, A.; Liu, C.; Chung, M.; Daigh, L.H.; Pack, L.R.; Fan, Y.; Regot, S.; Covert, M.; et al. Stress-mediated exit to quiescence restricted by increasing persistence in CDK4/6 activation. eLife 2020, 9, e44571. [Google Scholar] [CrossRef]
- Cornwell, J.A.; Crncec, A.; Afifi, M.M.; Tang, K.; Amin, R.; Cappell, S.D. Loss of CDK4/6 activity in S/G2 phase leads to cell cycle reversal. Nature 2023, 619, 363–370. [Google Scholar] [CrossRef]
- Rubin, S.M.; Sage, J.; Skotheim, J.M. Integrating Old and New Paradigms of G1/S Control. Mol. Cell 2020, 80, 183–192. [Google Scholar] [CrossRef]
- Jirawatnotai, S.; Dalton, S.; Wattanapanitch, M. Role of cyclins and cyclin-dependent kinases in pluripotent stem cells and their potential as a therapeutic target. Semin. Cell Dev. Biol. 2020, 107, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Besson, A.; Dowdy, S.F.; Roberts, J.M. CDK Inhibitors: Cell Cycle Regulators and Beyond. Dev. Cell 2008, 14, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Abukhdeir, A.M.; Park, B.H. P21 and p27: Roles in carcinogenesis and drug resistance. Expert Rev. Mol. Med. 2008, 10, e19. [Google Scholar] [CrossRef] [PubMed]
- White, J.; Stead, E.; Faast, R.; Conn, S.; Cartwright, P.; Dalton, S. Developmental Activation of the Rb–E2F Pathway and Establishment of Cell Cycle-regulated Cyclin-dependent Kinase Activity during Embryonic Stem Cell Differentiation. Mol. Biol. Cell 2005, 16, 2018–2027. [Google Scholar] [CrossRef] [PubMed]
- Takeishi, S.; Nakayama, K.I. Role of Fbxw7 in the maintenance of normal stem cells and cancer-initiating cells. Br. J. Cancer 2014, 111, 1054–1059. [Google Scholar] [CrossRef]
- Gonnot, F.; Langer, D.; Bourillot, P.-Y.; Doerflinger, N.; Savatier, P. Regulation of Cyclin E by transcription factors of the naïve pluripotency network in mouse embryonic stem cells. Cell Cycle 2019, 18, 2697–2712. [Google Scholar] [CrossRef]
- Pauklin, S.; Vallier, L. The Cell-Cycle State of Stem Cells Determines Cell Fate Propensity. Cell 2014, 156, 1338. [Google Scholar] [CrossRef]
- Liu, L.; Michowski, W.; Inuzuka, H.; Shimizu, K.; Nihira, N.T.; Chick, J.M.; Li, N.; Geng, Y.; Meng, A.Y.; Ordureau, A.; et al. G1 cyclins link proliferation, pluripotency and differentiation of embryonic stem cells. Nat. Cell Biol. 2017, 19, 177–188. [Google Scholar] [CrossRef]
- Ouyang, J.; Yu, W.; Liu, J.; Zhang, N.; Florens, L.; Chen, J.; Liu, H.; Washburn, M.; Pei, D.; Xie, T. Cyclin-dependent Kinase-mediated Sox2 Phosphorylation Enhances the Ability of Sox2 to Establish the Pluripotent State. J. Biol. Chem. 2015, 290, 22782–22794. [Google Scholar] [CrossRef]
- Neganova, I.; Zhang, X.; Atkinson, S.; Lako, M. Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene 2009, 28, 20–30. [Google Scholar] [CrossRef]
- Zhan, Z.; Song, L.; Zhang, W.; Gu, H.; Cheng, H.; Zhang, Y.; Yang, Y.; Ji, G.; Feng, H.; Cheng, T.; et al. Absence of cyclin-dependent kinase inhibitor p27 or p18 increases efficiency of iPSC generation without induction of iPSC genomic instability. Cell Death Dis. 2019, 10, 271. [Google Scholar] [CrossRef]
- Hwang, H.C.; Clurman, B.E. Cyclin E in normal and neoplastic cell cycles. Oncogene 2005, 24, 2776–2786. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, L.K.; Reed, S.I. Cyclin E Deregulation and Genomic Instability. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2017; pp. 527–547. [Google Scholar] [CrossRef]
- Caldon, C.E.; Sergio, C.M.; Burgess, A.; Deans, A.J.; Sutherland, R.L.; Musgrove, E.A. Cyclin E2 induces genomic instability by mechanisms distinct from cyclin E1. Cell Cycle 2013, 12, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Fernández, M.; Malumbres, M. Mechanisms of Sensitivity and Resistance to CDK4/6 Inhibition. Cancer Cell 2020, 37, 514–529. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.C.H.; Peng, Q.; Fong, S.W.; Lee, K.C.; Yeung, W.S.B.; Lee, Y.L. DNA Damage Response and Cell Cycle Regulation in Pluripotent Stem Cells. Genes 2021, 12, 1548. [Google Scholar] [CrossRef]
- Ballabeni, A.; Park, I.-H.; Zhao, R.; Wang, W.; Lerou, P.H.; Daley, G.Q.; Kirschner, M.W. Cell cycle adaptations of embryonic stem cells. Proc. Natl. Acad. Sci. USA 2011, 108, 19252–19257. [Google Scholar] [CrossRef]
- Egozi, D.; Shapira, M.; Paor, G.; Ben-Izhak, O.; Skorecki, K.; Hershko, D.D. Regulation of the cell cycle inhibitor p27 and its ubiquitin ligase Skp2 in differentiation of human embryonic stem cells. FASEB J. 2007, 21, 2807–2817. [Google Scholar] [CrossRef]
- Bar-On, O.; Shapira, M.; Skorecki, K.; Hershko, A.; Hershko, D.D. Regulation of APC/CCdh1 ubiquitin ligase in differentiation of human embryonic stem cells. Cell Cycle 2010, 9, 1986–1989. [Google Scholar] [CrossRef]
- Cai, Z.; Moten, A.; Peng, D.; Hsu, C.-C.; Pan, B.-S.; Manne, R.; Li, H.-Y.; Lin, H.-K. The Skp2 Pathway: A Critical Target for Cancer Therapy. Semin. Cancer Biol. 2020, 67, 16–33. [Google Scholar] [CrossRef]
- Chrysanthou, S.; Flores, J.C.; Dawlaty, M.M. Tet1 Suppresses p21 to Ensure Proper Cell Cycle Progression in Embryonic Stem Cells. Cells 2022, 11, 1366. [Google Scholar] [CrossRef]
- Itahana, Y.; Zhang, J.; Göke, J.; Vardy, L.A.; Han, R.; Iwamoto, K.; Cukuroglu, E.; Robson, P.; Pouladi, M.A.; Colman, A.; et al. Histone modifications and p53 binding poise the p21 promoter for activation in human embryonic stem cells. Sci. Rep. 2016, 6, 28112. [Google Scholar] [CrossRef]
- Engeland, K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022, 29, 946–960. [Google Scholar] [CrossRef]
- Remus, D.; Diffley, J.F. Eukaryotic DNA replication control: Lock and load, then fire. Curr. Opin. Cell Biol. 2009, 21, 771–777. [Google Scholar] [CrossRef]
- Das, S.P.; Rhind, N. How and why multiple MCMs are loaded at origins of DNA replication. BioEssays 2016, 38, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Pozo, P.N.; Cook, J.G. Regulation and Function of Cdt1; A Key Factor in Cell Proliferation and Genome Stability. Genes 2016, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, C.; Saxena, S.; Jeon, Y.; Lee, C.; Murata, K.; Machida, Y.; Wagle, N.; Hwang, D.S.; Dutta, A. A p53-Dependent Checkpoint Pathway Prevents Rereplication. Mol. Cell 2003, 11, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Varma, D.; Chandrasekaran, S.; Sundin, L.J.R.; Reidy, K.T.; Wan, X.; Chasse, D.A.D.; Nevis, K.R.; DeLuca, J.G.; Salmon, E.D.; Cook, J.G. Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore–microtubule attachment. Nat. Cell Biol. 2012, 14, 593–603. [Google Scholar] [CrossRef]
- Petropoulou, C.; Kotantaki, P.; Karamitros, D.; Taraviras, S. Cdt1 and Geminin in cancer: Markers or triggers of malignant transformation? Front. Biosci. 2008, 13, 4485–4494. [Google Scholar] [CrossRef]
- Kent, L.N.; Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 2019, 19, 326–338. [Google Scholar] [CrossRef]
- Liontos, M.; Koutsami, M.; Sideridou, M.; Evangelou, K.; Kletsas, D.; Levy, B.; Kotsinas, A.; Nahum, O.; Zoumpourlis, V.; Kouloukoussa, M.; et al. Deregulated Overexpression of hCdt1 and hCdc6 Promotes Malignant Behavior. Cancer Res. 2007, 67, 10899–10909. [Google Scholar] [CrossRef]
- Galanos, P.; Vougas, K.; Walter, D.; Polyzos, A.; Maya-Mendoza, A.; Haagensen, E.J.; Kokkalis, A.; Roumelioti, F.-M.; Gagos, S.; Tzetis, M.; et al. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat. Cell Biol. 2016, 18, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, S.; Búa, S.; Rodríguez-Acebes, S.; Megías, D.; Ortega, S.; de Martino, A.; Méndez, J. In Vivo DNA Re-replication Elicits Lethal Tissue Dysplasias. Cell Rep. 2017, 19, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Feng, H.; Santiago, F.E.; Kipreos, E.T. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 2003, 423, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Petersen, B.O.; Wagener, C.; Marinoni, F.; Kramer, E.R.; Melixetian, M.; Denchi, E.L.; Gieffers, C.; Matteucci, C.; Peters, J.-M.; Helin, K. Cell cycle– and cell growth–regulated proteolysis of mammalian CDC6 is dependent on APC–CDH1. Genes Dev. 2000, 14, 2330–2343. [Google Scholar] [CrossRef]
- Hsu, J.Y.; Reimann, J.D.; Sørensen, C.S.; Lukas, J.; Jackson, P.K. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APCCdh1. Nat. Cell Biol. 2002, 4, 358–366. [Google Scholar] [CrossRef]
- Lehman, N.L.; Tibshirani, R.; Hsu, J.Y.; Natkunam, Y.; Harris, B.T.; West, R.B.; Masek, M.A.; Montgomery, K.; van de Rijn, M.; Jackson, P.K. Oncogenic Regulators and Substrates of the Anaphase Promoting Complex/Cyclosome Are Frequently Overexpressed in Malignant Tumors. Am. J. Pathol. 2007, 170, 1793–1805. [Google Scholar] [CrossRef][Green Version]
- Niimi, S.; Arakawa-Takeuchi, S.; Uranbileg, B.; Park, J.-H.; Jinno, S.; Okayama, H. Cdc6 Protein Obstructs Apoptosome Assembly and Consequent Cell Death by Forming Stable Complexes with Activated Apaf-1 Molecules. J. Biol. Chem. 2012, 287, 18573–18583. [Google Scholar] [CrossRef]
- Sideridou, M.; Zakopoulou, R.; Evangelou, K.; Liontos, M.; Kotsinas, A.; Rampakakis, E.; Gagos, S.; Kahata, K.; Grabusic, K.; Gkouskou, K.; et al. Cdc6 expression represses E-cadherin transcription and activates adjacent replication origins. J. Cell Biol. 2011, 195, 1123–1140. [Google Scholar] [CrossRef]
- Petrakis, T.G.; Vougas, K.; Gorgoulis, V.G. Cdc6: A multi-functional molecular switch with critical role in carcinogenesis. Transcription 2012, 3, 124–129. [Google Scholar] [CrossRef]
- Ishimi, Y.; Irie, D. G364R mutation of MCM4 detected in human skin cancer cells affects DNA helicase activity of MCM4/6/7 complex. J. Biochem. 2015, 157, 561–569. [Google Scholar] [CrossRef]
- Tatsumi, R.; Ishimi, Y. An MCM4 mutation detected in cancer cells affects MCM4/6/7 complex formation. J. Biochem. 2017, 161, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, A.; Schwob, E.; Méndez, J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl. Acad. Sci. USA 2008, 105, 8956–8961. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.Q.; Jackson, D.A.; Blow, J.J. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev. 2007, 21, 3331–3341. [Google Scholar] [CrossRef] [PubMed]
- Shima, N.; Pederson, K.D. Dormant origins as a built-in safeguard in eukaryotic DNA replication against genome instability and disease development. DNA Repair 2017, 56, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, K.M.; Jones, R.M.; Petermann, E.; Jeggo, P.A. Diminished Origin-Licensing Capacity Specifically Sensitizes Tumor Cells to Replication Stress. Mol. Cancer Res. 2013, 11, 370–380. [Google Scholar] [CrossRef]
- Ahuja, A.K.; Jodkowska, K.; Teloni, F.; Bizard, A.H.; Zellweger, R.; Herrador, R.; Ortega, S.; Hickson, I.D.; Altmeyer, M.; Mendez, J.; et al. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun. 2016, 7, 10660. [Google Scholar] [CrossRef]
- Böhly, N.; Schmidt, A.-K.; Zhang, X.; Slusarenko, B.O.; Hennecke, M.; Kschischo, M.; Bastians, H. Increased replication origin firing links replication stress to whole chromosomal instability in human cancer. Cell Rep. 2022, 41, 111836. [Google Scholar] [CrossRef]
- Mei, L.; Cook, J.G. Efficiency and equity in origin licensing to ensure complete DNA replication. Biochem. Soc. Trans. 2021, 49, 2133–2141. [Google Scholar] [CrossRef]
- Fu, H.; Redon, C.E.; Thakur, B.L.; Utani, K.; Sebastian, R.; Jang, S.-M.; Gross, J.M.; Mosavarpour, S.; Marks, A.B.; Zhuang, S.Z.; et al. Dynamics of replication origin over-activation. Nat. Commun. 2021, 12, 3448. [Google Scholar] [CrossRef]
- Paniza, T.; Deshpande, M.; Wang, N.; O’neil, R.; Zuccaro, M.V.; Smith, M.E.; Madireddy, A.; James, D.; Ecker, J.; Rosenwaks, Z.; et al. Pluripotent stem cells with low differentiation potential contain incompletely reprogrammed DNA replication. J. Cell Biol. 2020, 219, e201909163. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, L.; An, K.; Cai, J.; Li, G.; Yang, C.; Liu, H.; Du, F.; Han, X.; Zhang, Z.; et al. Lower genomic stability of induced pluripotent stem cells reflects increased non-homologous end joining. Cancer Commun. 2018, 38, 49. [Google Scholar] [CrossRef] [PubMed]
- Araki, R.; Hoki, Y.; Suga, T.; Obara, C.; Sunayama, M.; Imadome, K.; Fujita, M.; Kamimura, S.; Nakamura, M.; Wakayama, S.; et al. Genetic aberrations in iPSCs are introduced by a transient G1/S cell cycle checkpoint deficiency. Nat. Commun. 2020, 11, 197. [Google Scholar] [CrossRef] [PubMed]
- Flach, J.; Bakker, S.T.; Mohrin, M.; Conroy, P.C.; Pietras, E.M.; Reynaud, D.; Alvarez, S.; Diolaiti, M.E.; Ugarte, F.; Forsberg, E.C.; et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 2014, 512, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Tomasetti, C.; Vogelstein, B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 2015, 347, 78–81. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Guan, J.; Wang, G.; Wang, J.; Zhang, Z.; Fu, Y.; Cheng, L.; Meng, G.; Lyu, Y.; Zhu, J.; Li, Y.; et al. Chemical reprogramming of human somatic cells to pluripotent stem cells. Nature 2022, 605, 325–331. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, W.; Li, Y. Stemness-related markers in cancer. Cancer Transl. Med. 2017, 3, 87–95. [Google Scholar] [CrossRef]
- Zhang, S.; Xiong, X.; Sun, Y. Functional characterization of SOX2 as an anticancer target. Signal Transduct. Target. Ther. 2020, 5, 1–17. [Google Scholar] [CrossRef]
- Hagey, D.W.; Muhr, J. Sox2 Acts in a Dose-Dependent Fashion to Regulate Proliferation of Cortical Progenitors. Cell Rep. 2014, 9, 1908–1920. [Google Scholar] [CrossRef]
- Liu, Z.; Walters, B.J.; Owen, T.; Brimble, M.A.; Steigelman, K.A.; Zhang, L.; Lagarde, M.M.M.; Valentine, M.B.; Yu, Y.; Cox, B.C.; et al. Regulation of p27Kip1 by Sox2 Maintains Quiescence of Inner Pillar Cells in the Murine Auditory Sensory Epithelium. J. Neurosci. 2012, 32, 10530–10540. [Google Scholar] [CrossRef]
- Otsubo, T.; Akiyama, Y.; Yanagihara, K.; Yuasa, Y. SOX2 is frequently downregulated in gastric cancers and inhibits cell growth through cell-cycle arrest and apoptosis. Br. J. Cancer 2008, 98, 824–831. [Google Scholar] [CrossRef] [PubMed]
- Al Mamun, M.; Mannoor, K.; Cao, J.; Qadri, F.; Song, X. SOX2 in cancer stemness: Tumor malignancy and therapeutic potentials. J. Mol. Cell Biol. 2020, 12, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-C.; Chou, Y.-T.; Jiang, S.S.; Chang, J.-L.; Chung, C.-H.; Kao, Y.-R.; Chang, I.-S.; Wu, C.-W. Epigenetic Switch between SOX2 and SOX9 Regulates Cancer Cell Plasticity. Cancer Res. 2016, 76, 7036–7048. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; He, B.; Huang, C.; Yang, H.; Cao, L.; Huang, J.; Hu, C.; Cai, T.; Wu, X.-Y.; Zhang, X.-Q.; et al. Sex-Determining Region Y-box 2 Promotes Growth of Lung Squamous Cell Carcinoma and Directly Targets Cyclin D1. DNA Cell Biol. 2017, 36, 264–272. [Google Scholar] [CrossRef]
- Lee, S.H.; Oh, S.-Y.; Do, S.I.; Lee, H.J.; Kang, H.J.; Rho, Y.S.; Bae, W.J.; Lim, Y.C. SOX2 regulates self-renewal and tumorigenicity of stem-like cells of head and neck squamous cell carcinoma. Br. J. Cancer 2014, 111, 2122–2130. [Google Scholar] [CrossRef]
- Fukazawa, T.; Guo, M.; Ishida, N.; Yamatsuji, T.; Takaoka, M.; Yokota, E.; Haisa, M.; Miyake, N.; Ikeda, T.; Okui, T.; et al. SOX2 suppresses CDKN1A to sustain growth of lung squamous cell carcinoma. Sci. Rep. 2016, 6, 20113. [Google Scholar] [CrossRef]
- Metz, E.P.; Rizzino, A. Sox2 dosage: A critical determinant in the functions of Sox2 in both normal and tumor cells. J. Cell. Physiol. 2019, 234, 19298–19306. [Google Scholar] [CrossRef]
- Kopp, J.L.; Ormsbee, B.D.; Desler, M.; Rizzino, A. Small Increases in the Level of Sox2 Trigger the Differentiation of Mouse Embryonic Stem Cells. Stem Cells 2008, 26, 903–911. [Google Scholar] [CrossRef]
- Wuebben, E.L.; Wilder, P.J.; Cox, J.L.; Grunkemeyer, J.A.; Caffrey, T.; Hollingsworth, M.A.; Rizzino, A. SOX2 functions as a molecular rheostat to control the growth, tumorigenicity and drug responses of pancreatic ductal adenocarcinoma cells. Oncotarget 2016, 7, 34890–34906. [Google Scholar] [CrossRef]
- Gao, Z.; Cox, J.L.; Gilmore, J.M.; Ormsbee, B.D.; Mallanna, S.K.; Washburn, M.P.; Rizzino, A. Determination of Protein Interactome of Transcription Factor Sox2 in Embryonic Stem Cells Engineered for Inducible Expression of Four Reprogramming Factors. J. Biol. Chem. 2012, 287, 11384–11397. [Google Scholar] [CrossRef]
- Fang, X.; Yoon, J.-G.; Li, L.; Tsai, Y.S.; Zheng, S.; Hood, L.; Goodlett, D.R.; Foltz, G.; Lin, B. Landscape of the SOX2 protein-protein interactome. Proteomics 2011, 11, 921–934. [Google Scholar] [CrossRef]
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef]
- She, S.; Wei, Q.; Kang, B.; Wang, Y.-J. Cell cycle and pluripotency: Convergence on octamer-binding transcription factor 4. Mol. Med. Rep. 2017, 16, 6459–6466. [Google Scholar] [CrossRef]
- Zeineddine, D.; Abou Hammoud, A.; Mortada, M.; Boeuf, H. The Oct4 protein: More than a magic stemness marker. Am. J. Stem Cells 2014, 3, 74–82. [Google Scholar] [PubMed]
- Li, Z.; Li, X.; Li, C.; Su, Y.; Fang, W.; Zhong, C.; Ji, W.; Zhang, Q.; Su, C. Transcription factor OCT4 promotes cell cycle progression by regulating CCND1 expression in esophageal carcinoma. Cancer Lett. 2014, 354, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-M.; Han, S.-H.; Coh, Y.-R.; Jang, G.; Ra, J.C.; Kang, S.-K.; Lee, H.-W.; Youn, H.-Y. Enhanced proliferation and differentiation of Oct4- and Sox2-overexpressing human adipose tissue mesenchymal stem cells. Exp. Mol. Med. 2014, 46, e101. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Yuan, M.; Liao, H.; Chen, J.; Xie, B.; Yan, D.; Xi, X.; Xu, X.; Zhang, Z.; Feng, Y. OCT4 pseudogene 5 upregulates OCT4 expression to promote proliferation by competing with miR-145 in endometrial carcinoma. Oncol. Rep. 2015, 33, 1745–1752. [Google Scholar] [CrossRef] [PubMed]
- Su, C. Survivin in survival of hepatocellular carcinoma. Cancer Lett. 2016, 379, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Koo, B.S.; Lee, S.H.; Kim, J.M.; Huang, S.; Kim, S.H.; Rho, Y.S.; Bae, W.J.; Kang, H.J.; Kim, Y.S.; Moon, J.H.; et al. Oct4 is a critical regulator of stemness in head and neck squamous carcinoma cells. Oncogene 2015, 34, 2317–2324. [Google Scholar] [CrossRef]
- Kanai, D.; Ueda, A.; Akagi, T.; Yokota, T.; Koide, H. Oct3/4 directly regulates expression of E2F3a in mouse embryonic stem cells. Biochem. Biophys. Res. Commun. 2015, 459, 374–378. [Google Scholar] [CrossRef]
- Lu, Y.; Qu, H.; Qi, D.; Xu, W.; Liu, S.; Jin, X.; Song, P.; Guo, Y.; Jia, Y.; Wang, X.; et al. OCT4 maintains self-renewal and reverses senescence in human hair follicle mesenchymal stem cells through the downregulation of p21 by DNA methyltransferases. Stem Cell Res. Ther. 2019, 10, 28. [Google Scholar] [CrossRef]
- Lee, J.; Go, Y.; Kang, I.; Han, Y.-M.; Kim, J. Oct-4 controls cell-cycle progression of embryonic stem cells. Biochem. J. 2010, 426, 171–181. [Google Scholar] [CrossRef]
- Comisso, E.; Scarola, M.; Rosso, M.; Piazza, S.; Marzinotto, S.; Ciani, Y.; Orsaria, M.; Mariuzzi, L.; Schneider, C.; Schoeftner, S.; et al. OCT4 controls mitotic stability and inactivates the RB tumor suppressor pathway to enhance ovarian cancer aggressiveness. Oncogene 2017, 36, 4253–4266. [Google Scholar] [CrossRef]
- Schoeftner, S.; Scarola, M.; Comisso, E.; Schneider, C.; Benetti, R. An Oct4-pRb Axis, Controlled by MiR-335, Integrates Stem Cell Self-Renewal and Cell Cycle Control. Stem Cells 2013, 31, 717–728. [Google Scholar] [CrossRef]
- Zhao, R.; Deibler, R.W.; Lerou, P.H.; Ballabeni, A.; Heffner, G.C.; Cahan, P.; Unternaehrer, J.J.; Kirschner, M.W.; Daley, G.Q. A nontranscriptional role for Oct4 in the regulation of mitotic entry. Proc. Natl. Acad. Sci. USA 2014, 111, 15768–15773. [Google Scholar] [CrossRef]
- Shields, J.M.; Christy, R.J.; Yang, V.W. Identification and Characterization of a Gene Encoding a Gut-enriched Krüppel-like Factor Expressed during Growth Arrest. J. Biol. Chem. 1996, 271, 20009–20017. [Google Scholar] [CrossRef]
- Chen, X.; Johns, D.C.; Geiman, D.E.; Marban, E.; Dang, D.T.; Hamlin, G.; Sun, R.; Yang, V.W. Krüppel-like Factor 4 (Gut-enriched Krüppel-like Factor) Inhibits Cell Proliferation by Blocking G1/S Progression of the Cell Cycle. J. Biol. Chem. 2001, 276, 30423–30428. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.S.; Chen, X.; Yang, V.W. Krüppel-like Factor 4 Mediates p53-dependent G1/S Cell Cycle Arrest in Response to DNA Damage. J. Biol. Chem. 2003, 278, 2101–2105. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.-D.; Zhao, F. KLF4 suppresses the proliferation and metastasis of NSCLC cells via inhibition of MSI2 and regulation of the JAK/STAT3 signaling pathway. Transl. Oncol. 2022, 22, 101396. [Google Scholar] [CrossRef] [PubMed]
- Rowland, B.D.; Bernards, R.; Peeper, D.S. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat. Cell Biol. 2005, 7, 1074–1082. [Google Scholar] [CrossRef]
- Ganguly, K.; Krishn, S.R.; Rachagani, S.; Jahan, R.; Shah, A.; Nallasamy, P.; Rauth, S.; Atri, P.; Cox, J.L.; Pothuraju, R.; et al. Secretory Mucin 5AC Promotes Neoplastic Progression by Augmenting KLF4-Mediated Pancreatic Cancer Cell Stemness. Cancer Res. 2021, 81, 91–102. [Google Scholar] [CrossRef]
- Wei, D.; Kanai, M.; Jia, Z.; Le, X.; Xie, K. Krüppel-like Factor 4 Induces p27Kip1 Expression in and Suppresses the Growth and Metastasis of Human Pancreatic Cancer Cells. Cancer Res. 2008, 68, 4631–4639. [Google Scholar] [CrossRef]
- Ghaleb, A.M.; Yang, V.W. Krüppel-like factor 4 (KLF4): What we currently know. Gene 2017, 611, 27–37. [Google Scholar] [CrossRef]
- Nandan, M.O.; Yang, V.W. The role of Krüppel-like factors in the reprogramming of somatic cells to induced pluripotent stem cells. Histol. Histopathol. 2009, 24, 1343–1355. [Google Scholar] [CrossRef]
- Yamanaka, S. Strategies and New Developments in the Generation of Patient-Specific Pluripotent Stem Cells. Cell Stem Cell 2007, 1, 39–49. [Google Scholar] [CrossRef]
- Singh, A.M.; Dalton, S. The Cell Cycle and Myc Intersect with Mechanisms that Regulate Pluripotency and Reprogramming. Cell Stem Cell 2009, 5, 141–149. [Google Scholar] [CrossRef]
- Knoepfler, P.S. Why Myc? An Unexpected Ingredient in the Stem Cell Cocktail. Cell Stem Cell 2008, 2, 18–21. [Google Scholar] [CrossRef]
- Meyer, N.; Penn, L.Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 2008, 8, 976–990. [Google Scholar] [CrossRef]
- Kaczmarek, L.; Hyland, J.K.; Watt, R.; Rosenberg, M.; Baserga, R. Microinjected c-myc as a Competence Factor. Science 1985, 228, 1313–1315. [Google Scholar] [CrossRef]
- Eilers, M.; Picard, D.; Yamamoto, K.R.; Bishop, J.M. Chimaeras of Myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature 1989, 340, 66–68. [Google Scholar] [CrossRef]
- Bretones, G.; Delgado, M.D.; León, J. Myc and cell cycle control. Biochim. Biophys. Acta-Gene Regul. Mech. 2015, 1849, 506–516. [Google Scholar] [CrossRef]
- García-Gutiérrez, L.; Delgado, M.D.; León, J. MYC Oncogene Contributions to Release of Cell Cycle Brakes. Genes 2019, 10, 244. [Google Scholar] [CrossRef]
- Dominguez-Sola, D.; Ying, C.Y.; Grandori, C.; Ruggiero, L.; Chen, B.; Li, M.; Galloway, D.A.; Gu, W.; Gautier, J.; Dalla-Favera, R. Non-transcriptional control of DNA replication by c-Myc. Nature 2007, 448, 445–451. [Google Scholar] [CrossRef]
- Das, S.K.; Lewis, B.A.; Levens, D. MYC: A complex problem. Trends Cell Biol. 2023, 33, 235–246. [Google Scholar] [CrossRef]
- Mei, L.; Kedziora, K.M.; Song, E.-A.; Purvis, J.E.; Cook, J.G. The consequences of differential origin licensing dynamics in distinct chromatin environments. Nucleic Acids Res. 2022, 50, 9601–9620. [Google Scholar] [CrossRef]
- Huangfu, D.; Maehr, R.; Guo, W.; Eijkelenboom, A.; Snitow, M.; Chen, A.E.; Melton, D.A. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008, 26, 795–797. [Google Scholar] [CrossRef]
- Poole, C.J.; Van Riggelen, J. MYC—Master Regulator of the Cancer Epigenome and Transcriptome. Genes 2017, 8, 142. [Google Scholar] [CrossRef]
- Carey, B.W.; Markoulaki, S.; Hanna, J.H.; Faddah, D.A.; Buganim, Y.; Kim, J.; Ganz, K.; Steine, E.J.; Cassady, J.P.; Creyghton, M.P.; et al. Reprogramming Factor Stoichiometry Influences the Epigenetic State and Biological Properties of Induced Pluripotent Stem Cells. Cell Stem Cell 2011, 9, 588–598. [Google Scholar] [CrossRef]
- Papapetrou, E.P.; Tomishima, M.J.; Chambers, S.M.; Mica, Y.; Reed, E.; Menon, J.; Tabar, V.; Mo, Q.; Studer, L.; Sadelain, M. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 12759–12764. [Google Scholar] [CrossRef]
- Ruiz, S.; Panopoulos, A.D.; Herrerías, A.; Bissig, K.-D.; Lutz, M.; Berggren, W.T.; Verma, I.M.; Belmonte, J.C.I. A High Proliferation Rate Is Required for Cell Reprogramming and Maintenance of Human Embryonic Stem Cell Identity. Curr. Biol. 2011, 21, 45–52. [Google Scholar] [CrossRef]
- Tanabe, K.; Nakamura, M.; Narita, M.; Takahashi, K.; Yamanaka, S. Maturation, not initiation, is the major roadblock during reprogramming toward pluripotency from human fibroblasts. Proc. Natl. Acad. Sci. USA 2013, 110, 12172–12179. [Google Scholar] [CrossRef]

| Cell Cycle Regulator | Stem Cells versus Somatic Cells | Some Cancers versus Somatic Cells | Section |
|---|---|---|---|
| Cyclins | Section 3.1 | ||
| Cyclin E | Higher levels | Higher levels | |
| Cyclin D | Intermediate levels | Higher levels | |
| Cyclin A | Higher levels | Higher levels | |
| Cyclin-dependent kinases (CDKs) | |||
| CDK2 | Higher activity | Higher activity | |
| CDK4/6 | Intermediate activity | Higher activity | |
| CDK1 | Higher activity | Higher activity | |
| CDK inhibitor proteins | Section 3.2 | ||
| p27 | Lower levels | Lower levels | |
| p21 | Lower levels | Lower levels | |
| Origin licensing factors | Section 4.1 | ||
| CDC6 | Higher levels | Higher levels | |
| CDT1 | Higher levels | Higher levels | |
| MCM loading | Faster loading in G1 | ? * | |
| Pluripotency Factor | Stem Cells versus Somatic Cells | Some Cancers versus Somatic Cells | Section 5 |
| SOX2 | Higher levels | Higher levels in a subset of cancer cells | Section 5.1 |
| OCT4 | Higher levels | Higher levels in a subset of cancer cells | Section 5.2 |
| KLF4 | Higher levels | Expression levels are cancer-specific | Section 5.3 |
| c-MYC ** | ? | Higher levels in most cancers | Section 5.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fleifel, D.; Cook, J.G. G1 Dynamics at the Crossroads of Pluripotency and Cancer. Cancers 2023, 15, 4559. https://doi.org/10.3390/cancers15184559
Fleifel D, Cook JG. G1 Dynamics at the Crossroads of Pluripotency and Cancer. Cancers. 2023; 15(18):4559. https://doi.org/10.3390/cancers15184559
Chicago/Turabian StyleFleifel, Dalia, and Jeanette Gowen Cook. 2023. "G1 Dynamics at the Crossroads of Pluripotency and Cancer" Cancers 15, no. 18: 4559. https://doi.org/10.3390/cancers15184559
APA StyleFleifel, D., & Cook, J. G. (2023). G1 Dynamics at the Crossroads of Pluripotency and Cancer. Cancers, 15(18), 4559. https://doi.org/10.3390/cancers15184559






