Automatic Detection of Brain Metastases in T1-Weighted Construct-Enhanced MRI Using Deep Learning Model
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
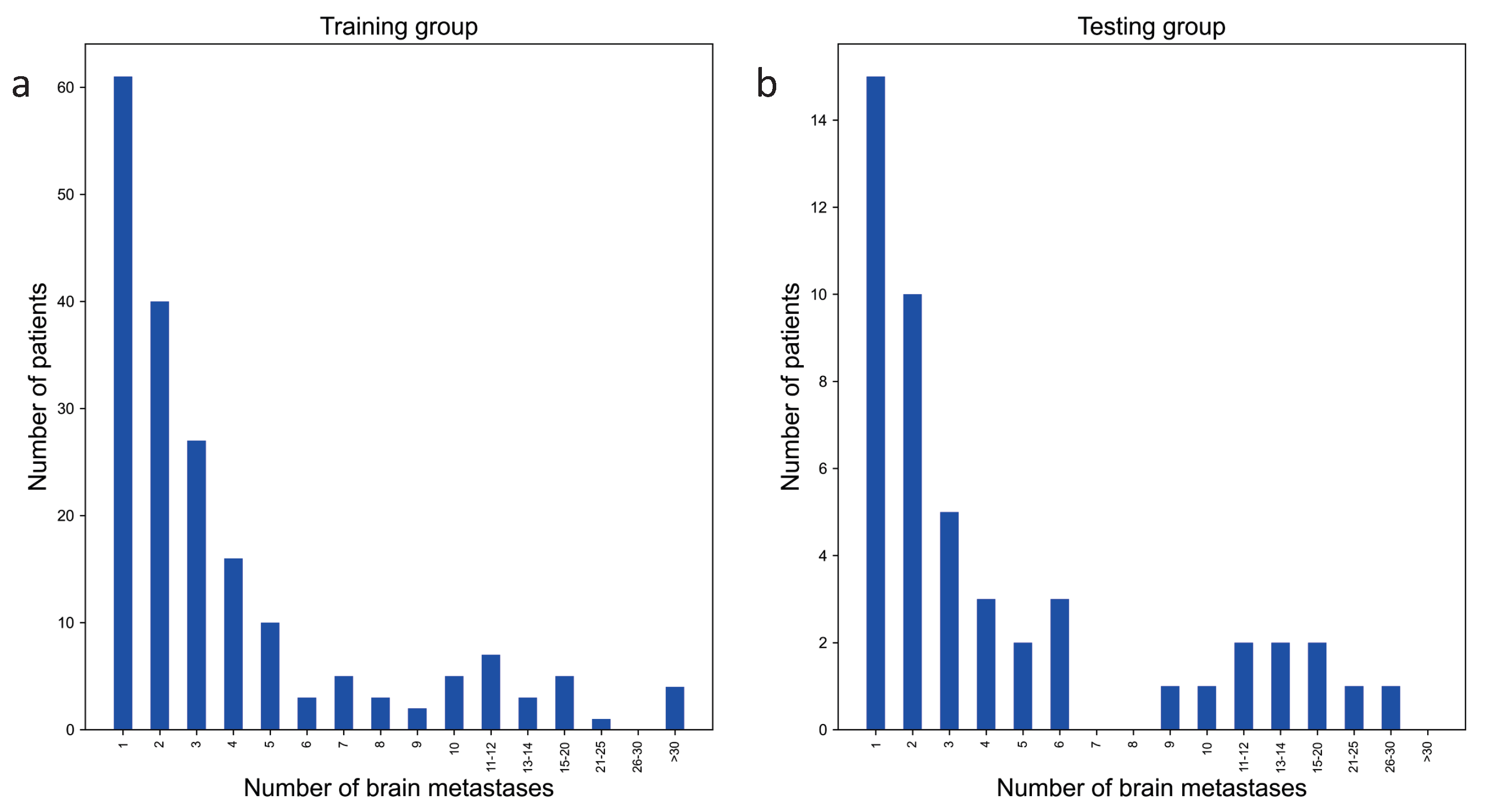
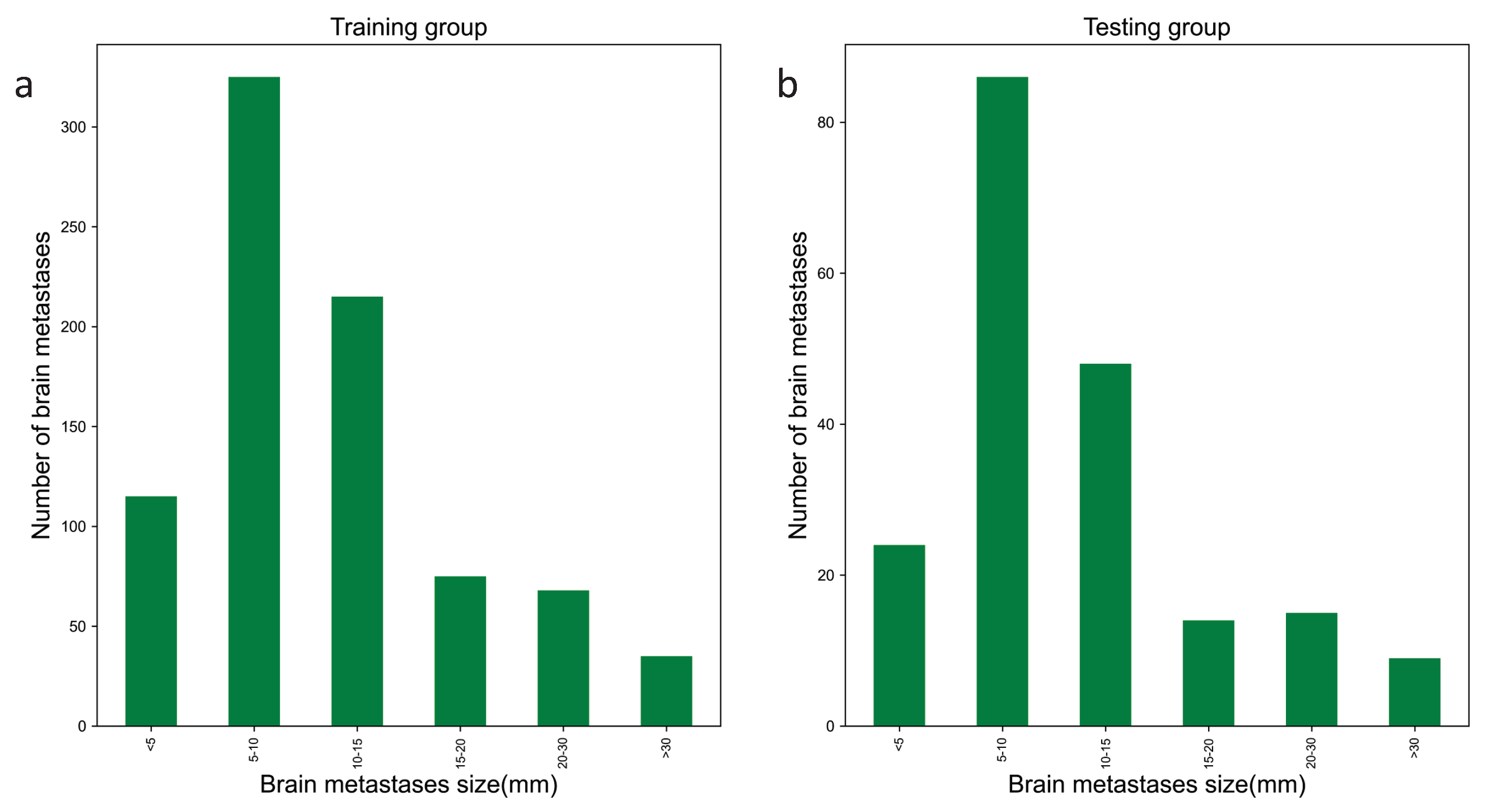
2.2. Dataset Construction
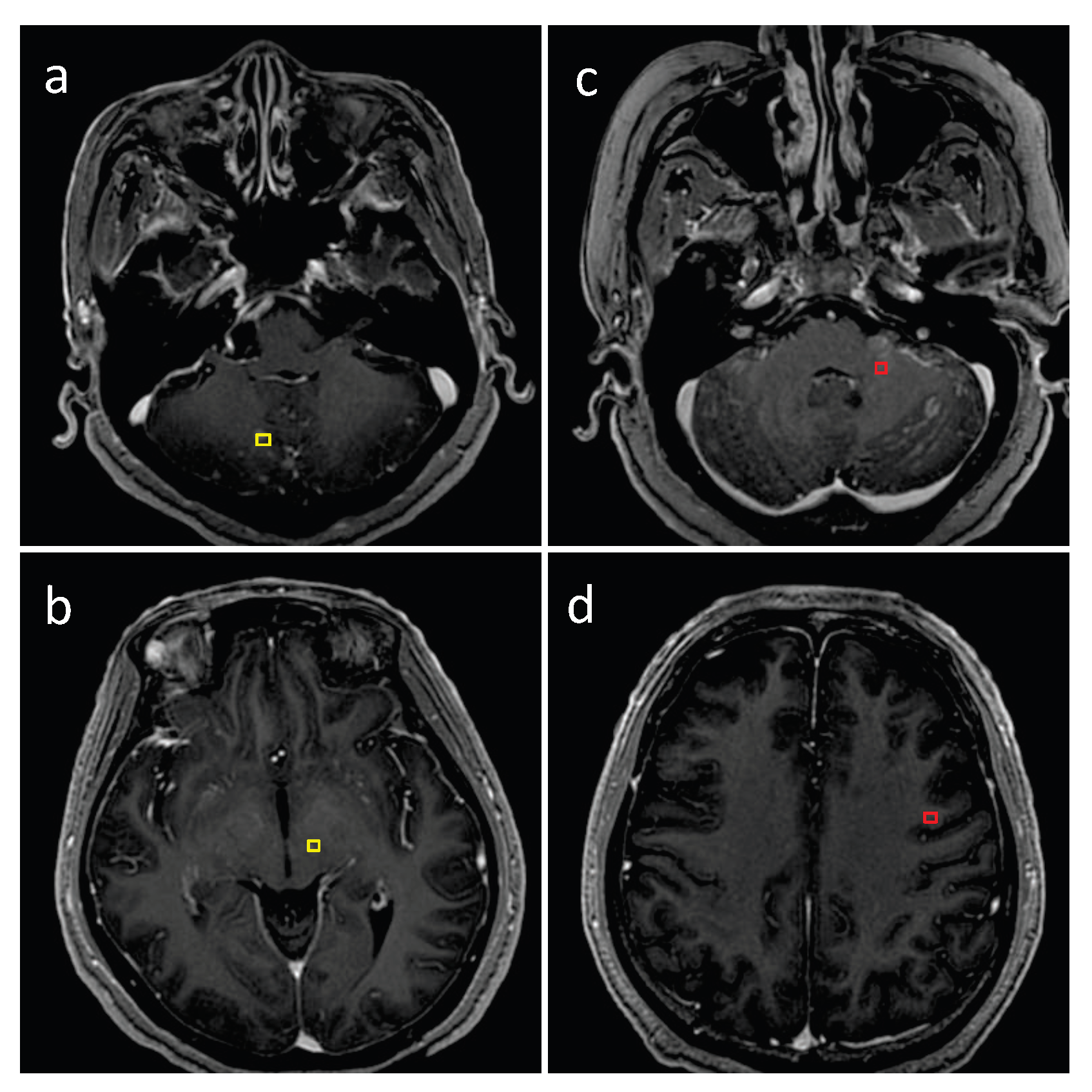
2.3. Dataset Ground Truth
2.4. Image Preprocessing
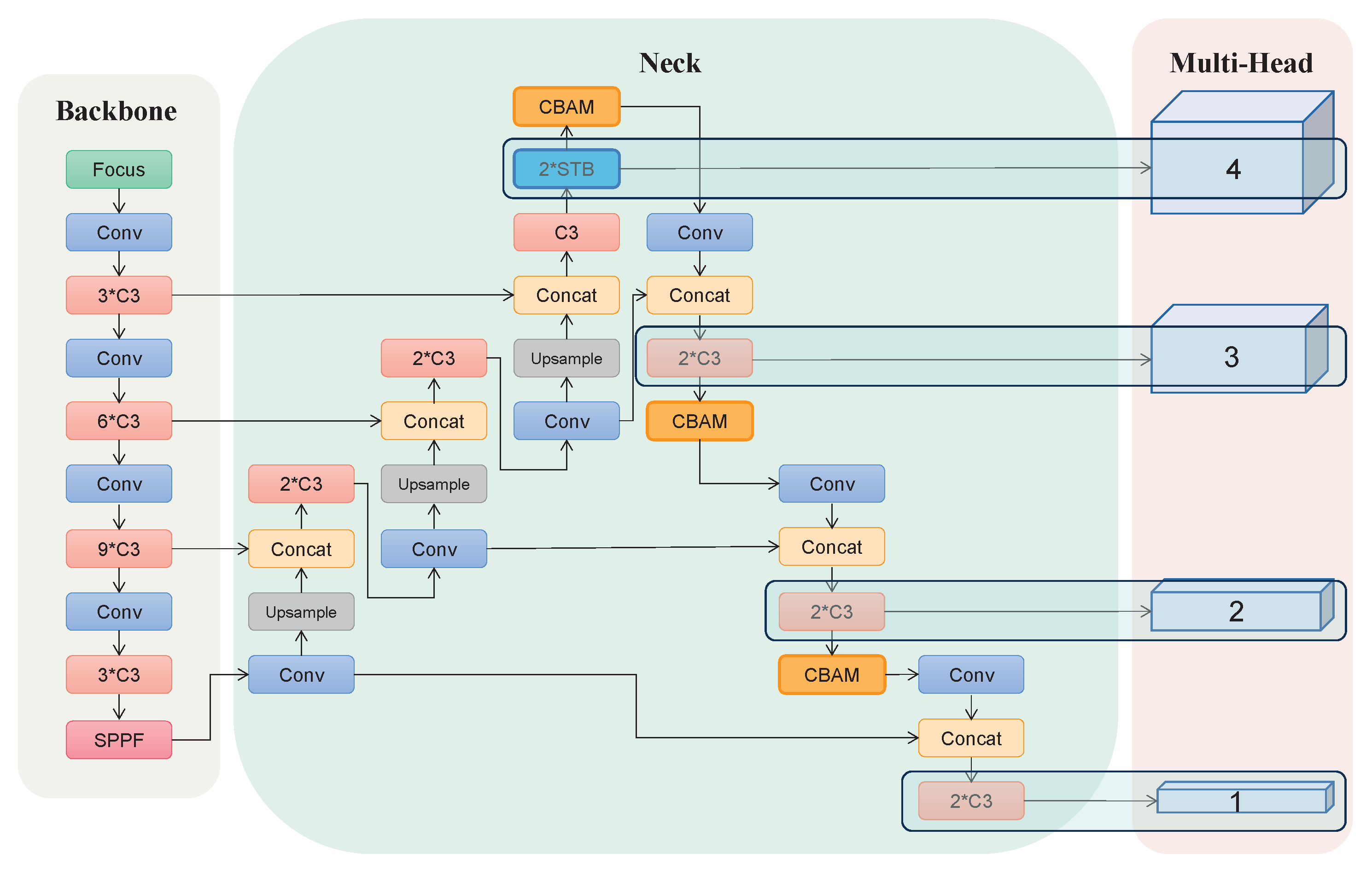
2.5. Architecture of SA-YOLOv5
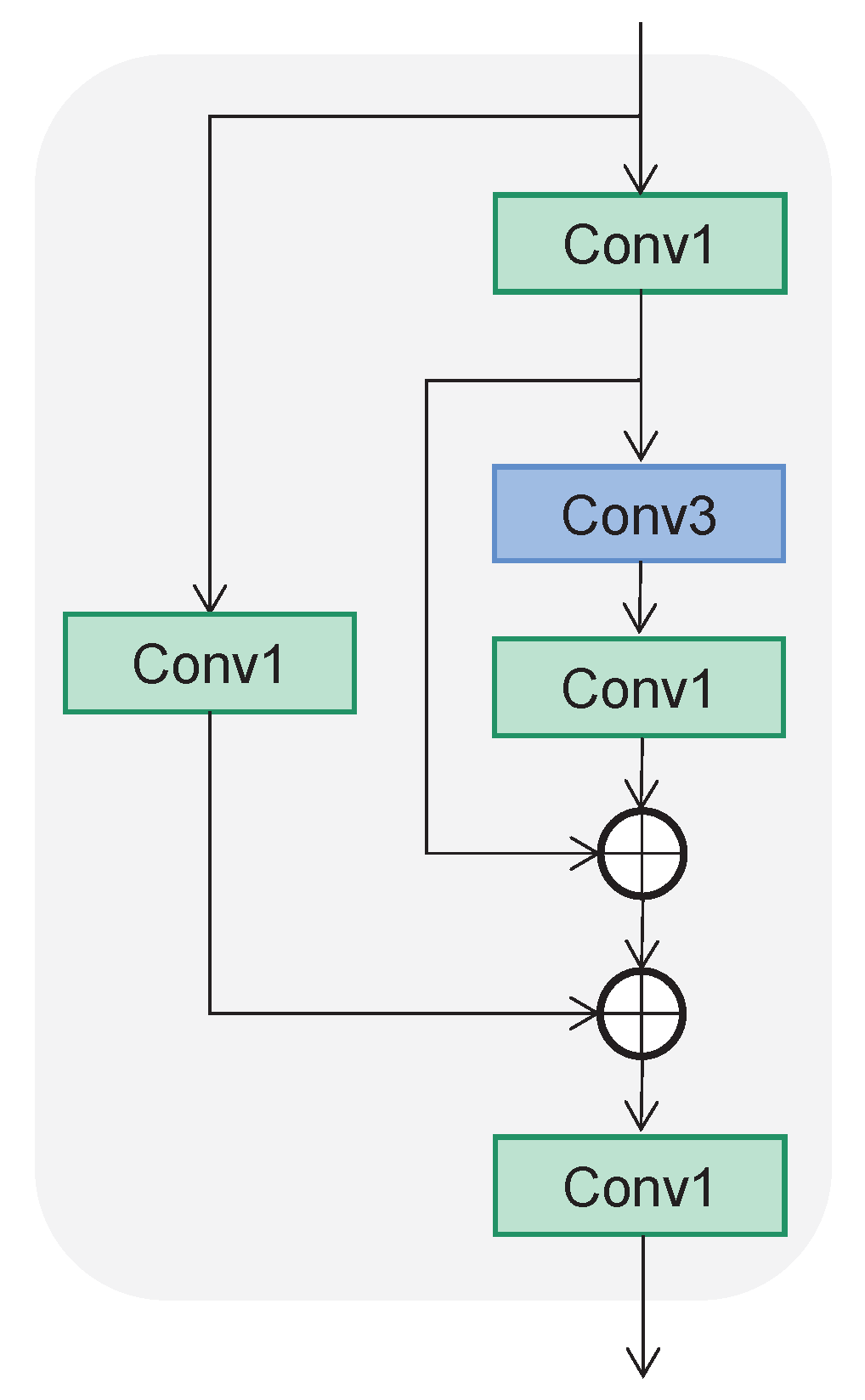
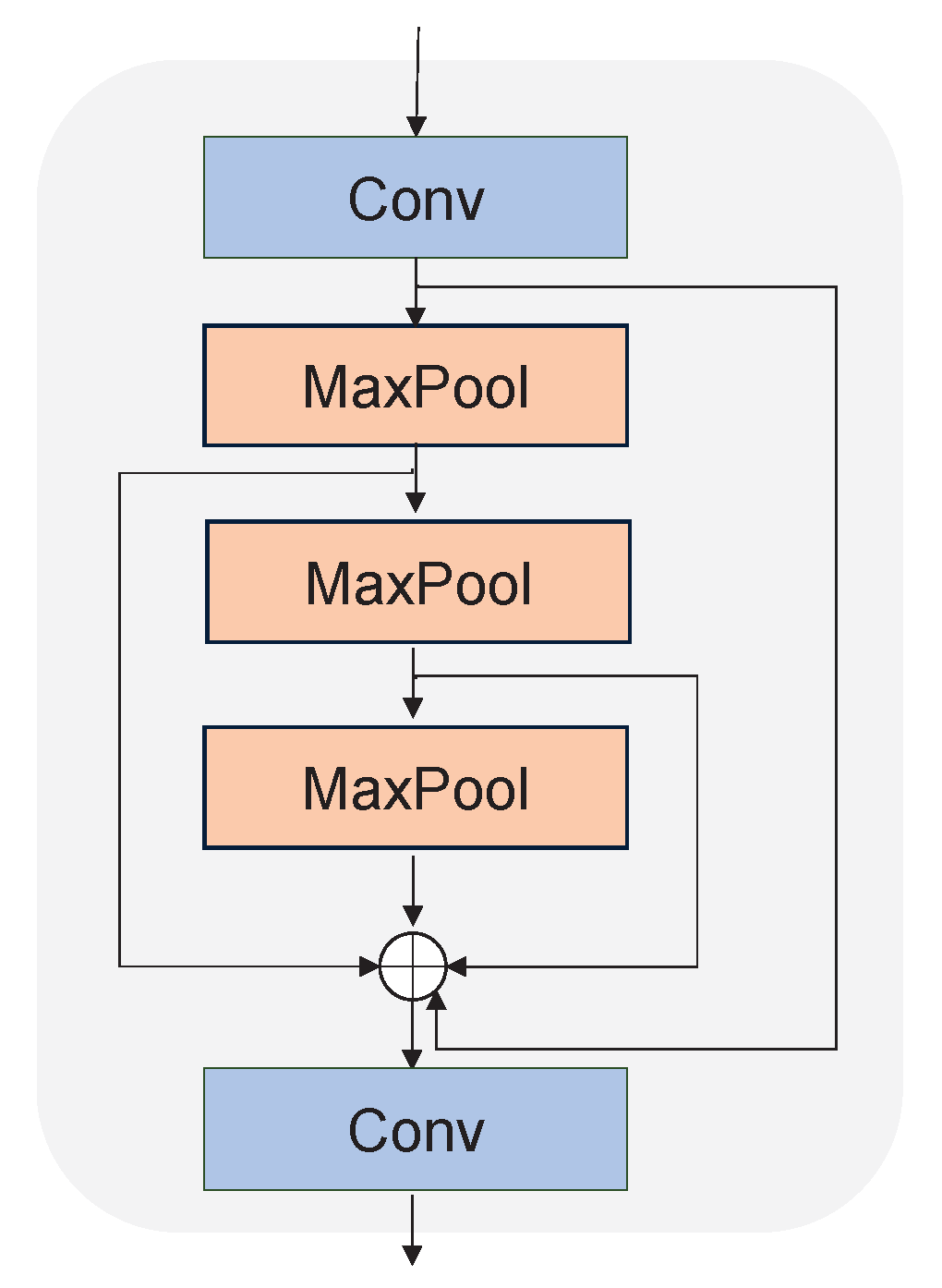
2.5.1. Backbone
2.5.2. Neck
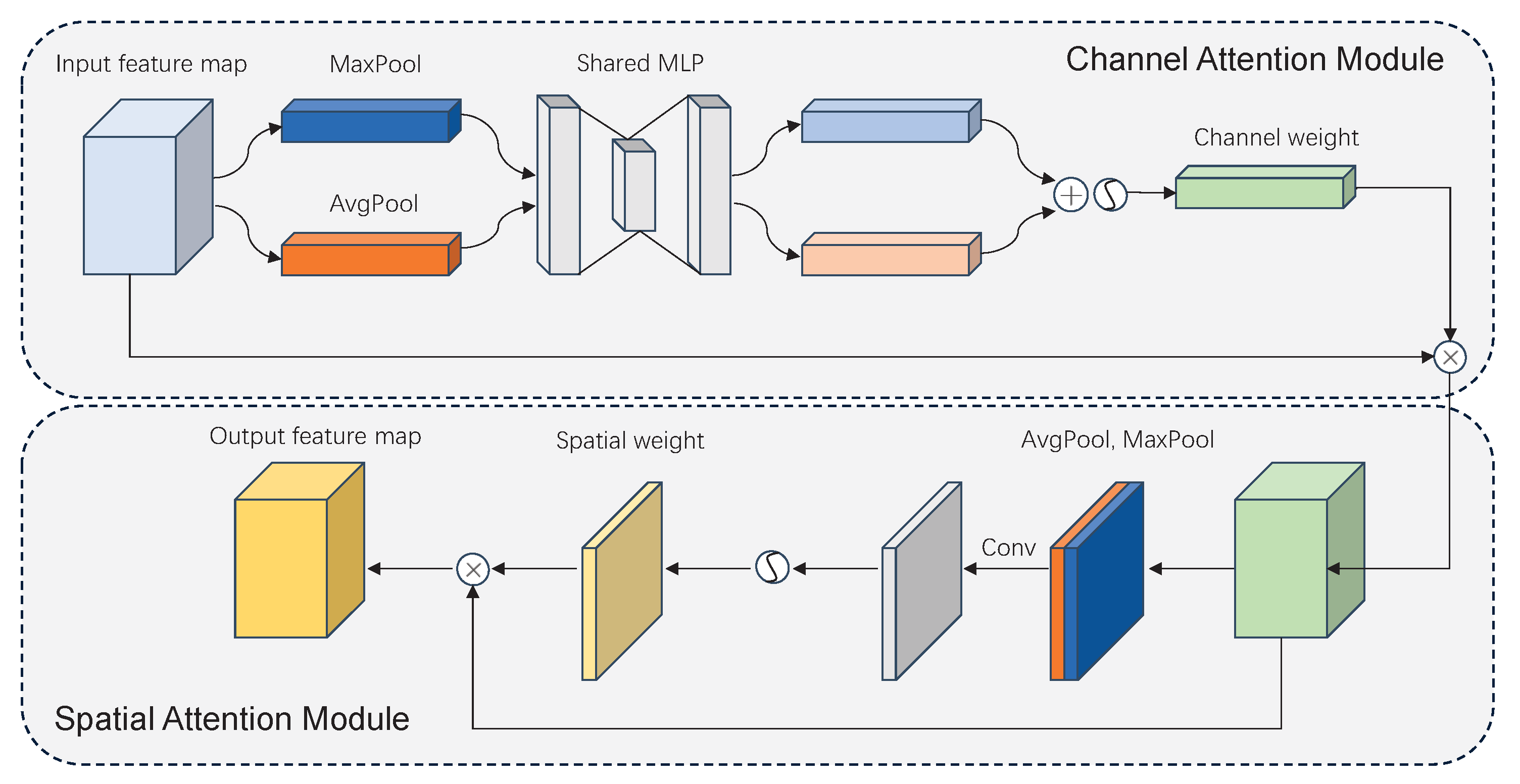
2.5.3. Multi-Head Attention
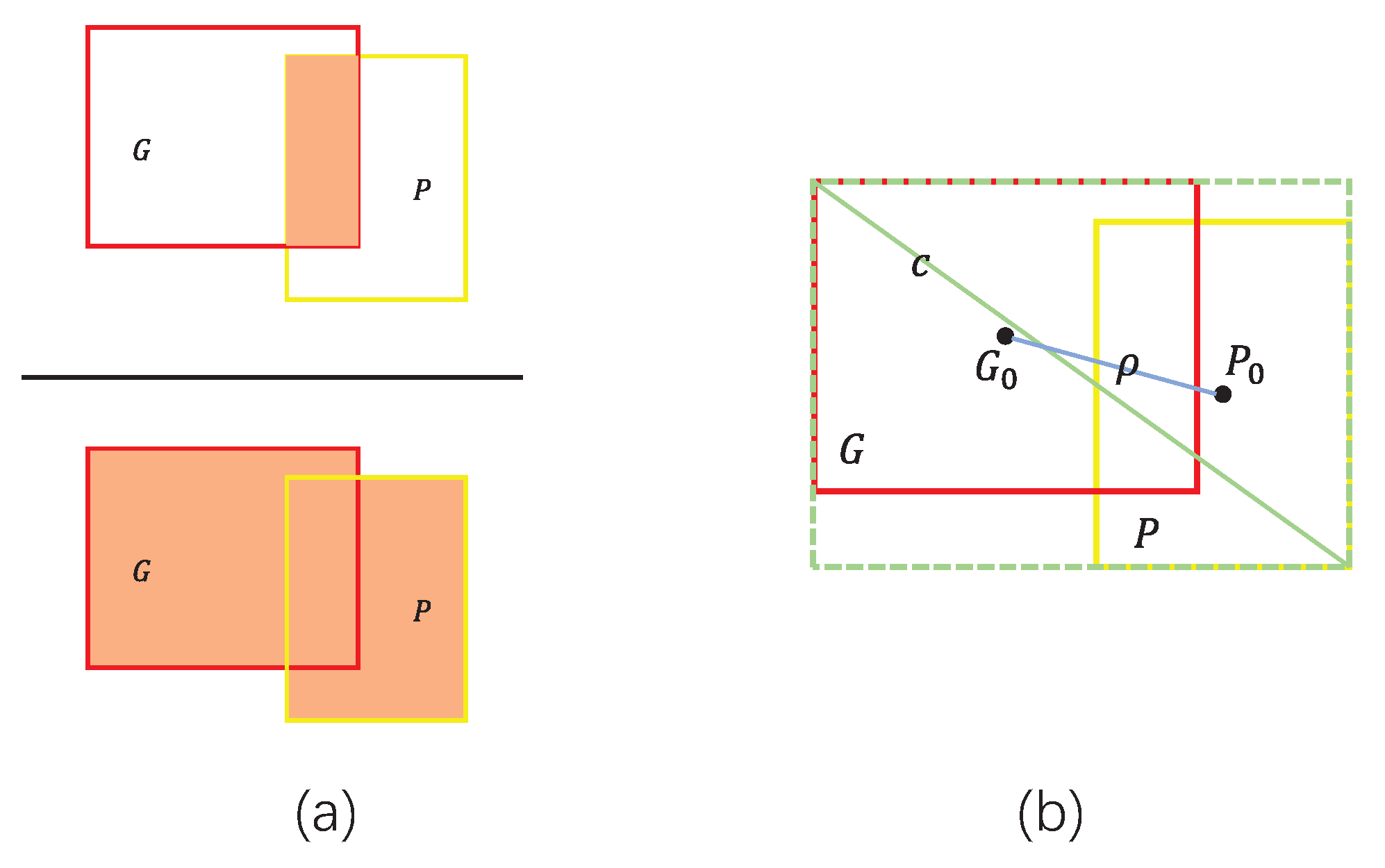
2.6. Boundary Loss Function
2.7. Training Configuration and Procedure
2.8. Postprocessing
2.9. Model Evaluation
3. Results
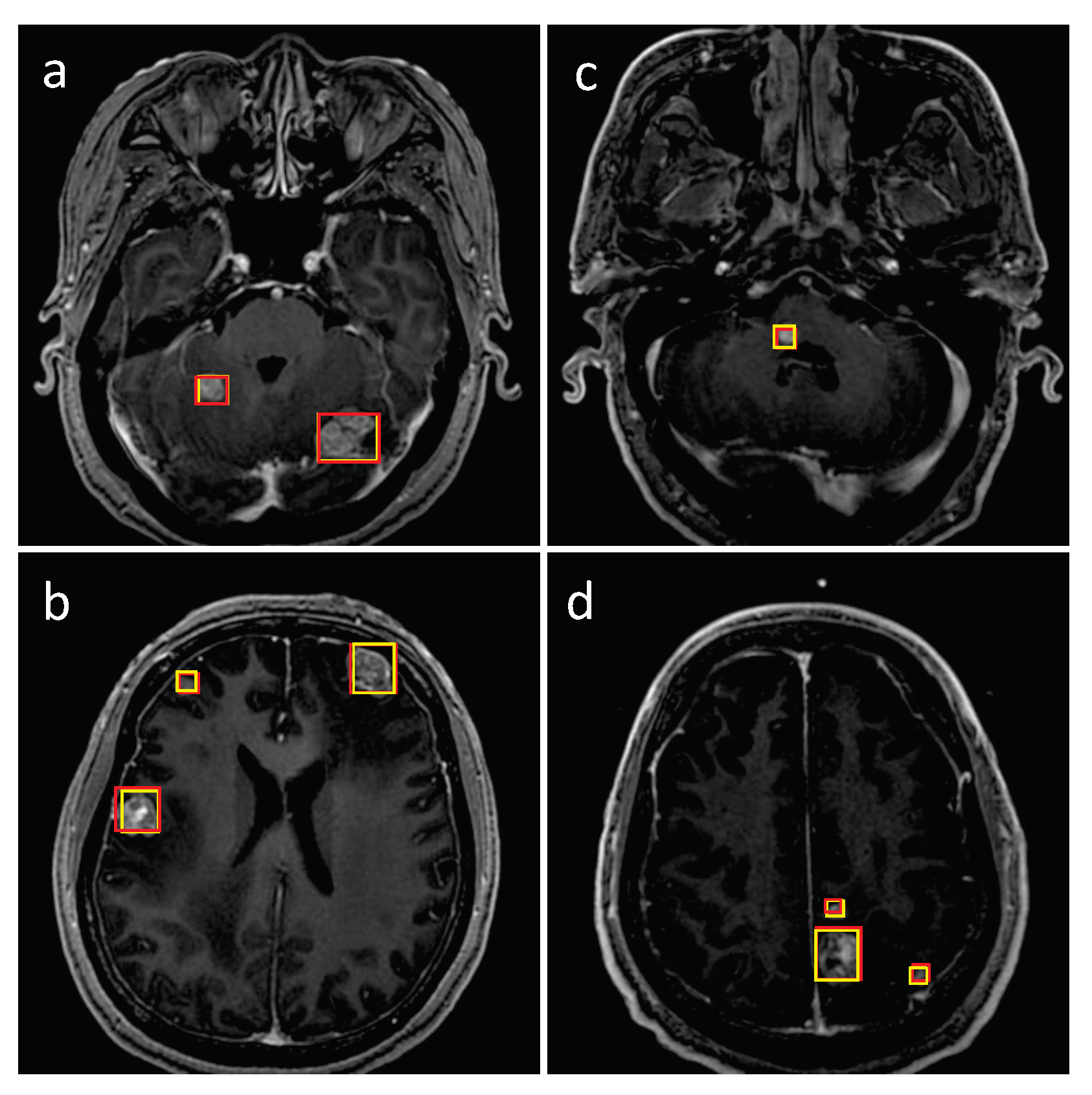
3.1. Detection Performance of SA-YOLOv5
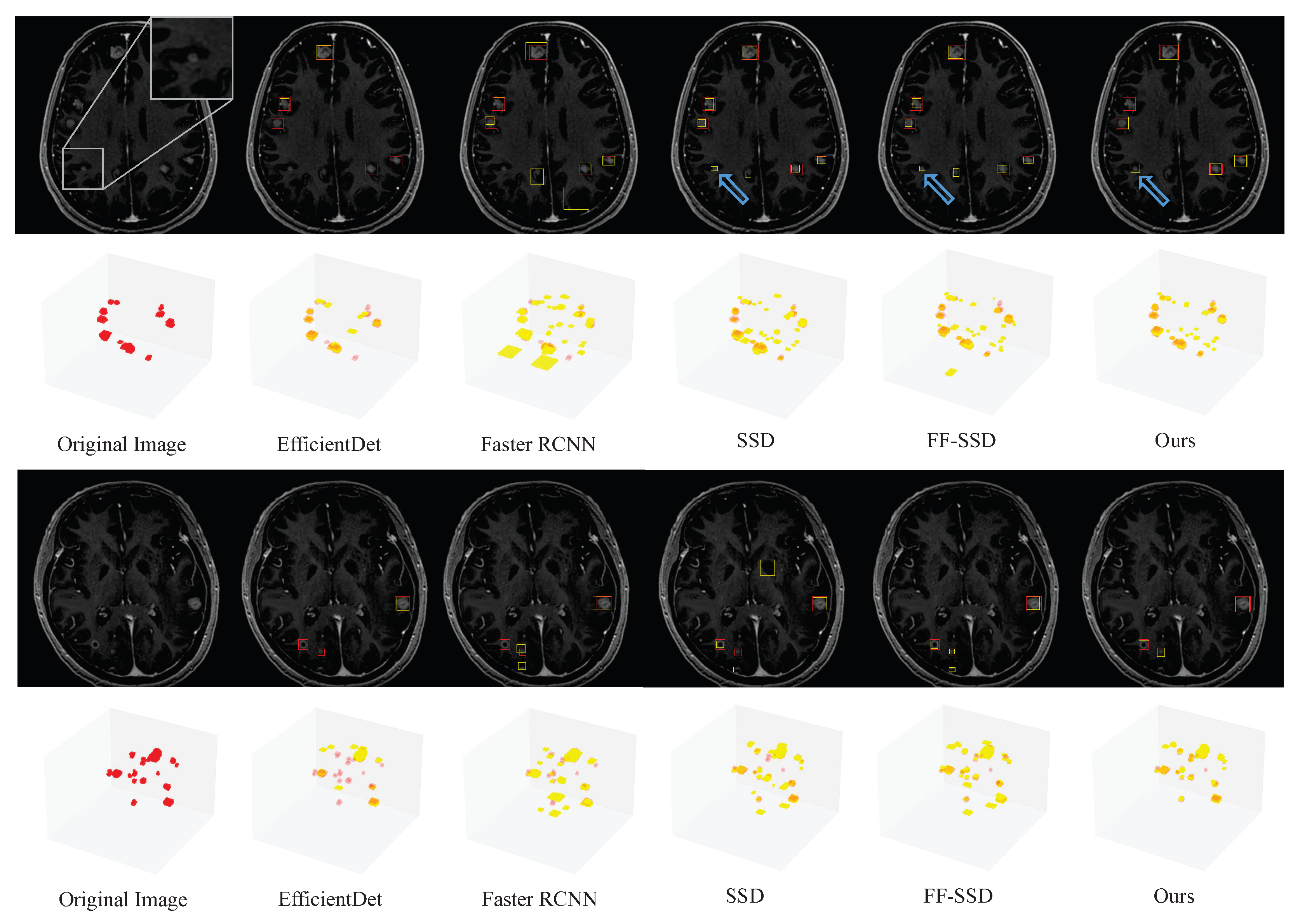
3.2. Comparison with Existing Detection Methods
3.3. Effectiveness Analysis of the Improvements Made in SA-YOLOv5
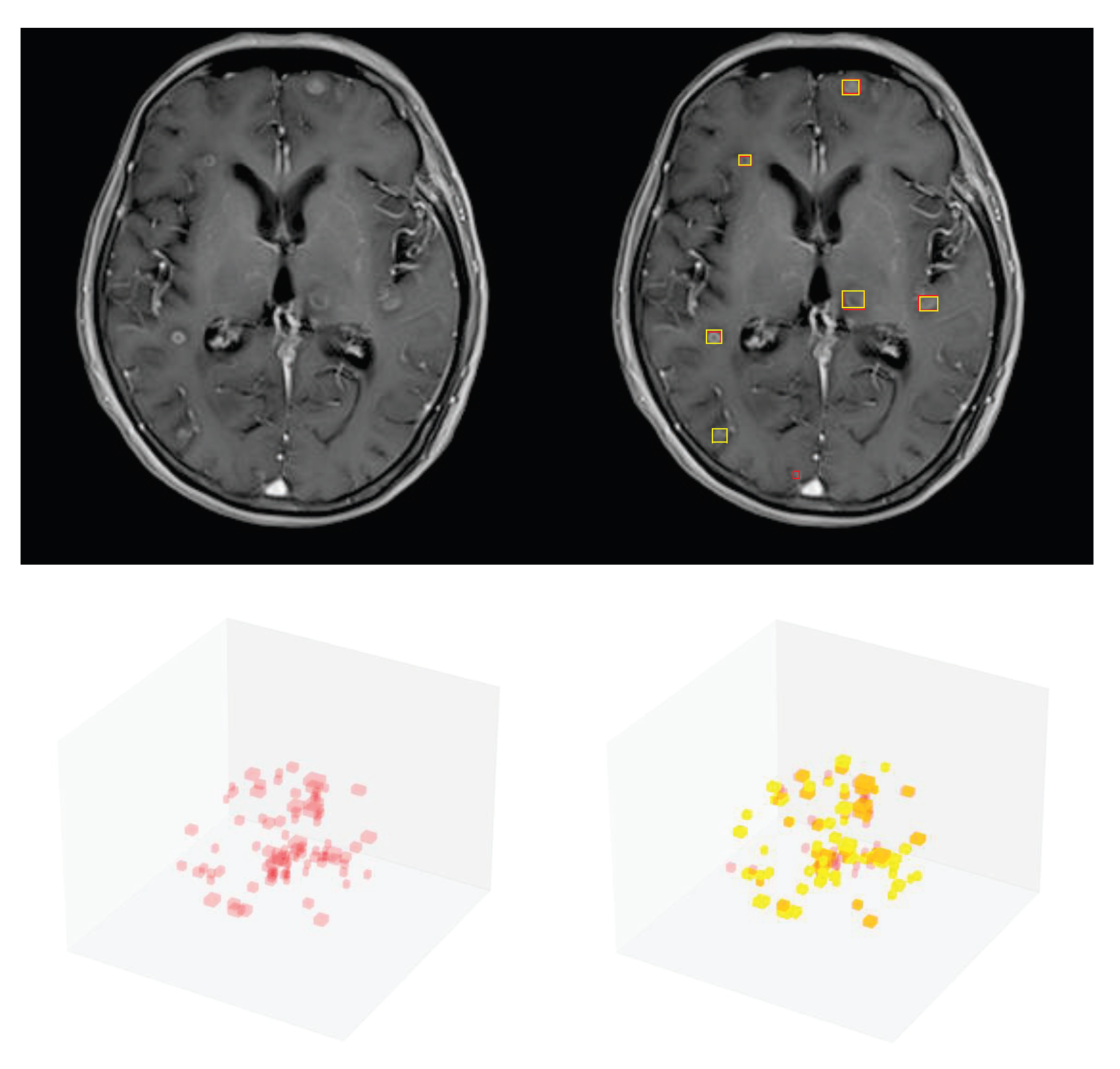
3.4. Detection Performance on the External Testing Set
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BM | Brain metastasis |
| DL | Deep learning |
| NSCLC | Non-small-cell lung cancer |
| SRS | Stereotactic radiosurgery |
| ROI | Region of interest |
| SSD | Single-shot detector |
| PPV | Positive predictive value |
| FF | Feature fusion |
| YOLOv5 | You only look once version 5 |
| SA-YOLOv5 | Self-attention YOLOv5 |
| T1ce | T1-weighted contrast-enhanced |
| P | Precision |
| R | Recall |
| CBAM | Convolutional block attention module |
| CAM | Channel Attention Module |
| SAM | Spatial Attention Module |
| STB | Swin transformer block |
| STPH | Swin transformer prediction head |
| SW-MSA | Shifted windows multi-head self-attention |
| W-MSA | Windows multi-head self-attention |
| NMS | Non-maximum suppression |
| IoU | Intersection over union |
| SGD | Stochastic gradient descent |
References
- Achrol, A.S.; Rennert, R.C.; Anders, C.; Soffietti, R.; Ahluwalia, M.S.; Nayak, L.; Peters, S.; Arvold, N.D.; Harsh, G.R.; Steeg, P.S.; et al. Brain metastases. Nat. Rev. Dis. Prim. 2019, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Page, S.; Milner-Watts, C.; Perna, M.; Janzic, U.; Vidal, N.; Kaudeer, N.; Ahmed, M.; McDonald, F.; Locke, I.; Minchom, A.; et al. Systemic treatment of brain metastases in non-small cell lung cancer. Eur. J. Cancer 2020, 132, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, S.B.; Contessa, J.N.; Omay, S.B.; Chiang, V. Lung Cancer Brain Metastases. Cancer J. 2015, 21, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Sperduto, P.W.; Mesko, S.; Li, J.; Cagney, D.; Aizer, A.; Lin, N.U.; Nesbit, E.; Kruser, T.J.; Chan, J.; Braunstein, S.; et al. Incidence and prognosis of patients with brain metastases at diagnosis of systemic malignancy: A population-based study. J. Clin. Oncol. 2020, 38, 3773–3784. [Google Scholar] [CrossRef] [PubMed]
- Cagney, D.N.; Martin, A.M.; Catalano, P.J.; Redig, A.J.; Lin, N.U.; Lee, E.Q.; Wen, P.Y.; Dunn, I.F.; Bi, W.L.; Weiss, S.E.; et al. Incidence and prognosis of patients with brain metastases at diagnosis of systemic malignancy: A population-based study. Neuro-oncology 2017, 19, 1511–1521. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.H.; Kotecha, R.; Chao, S.T.; Ahluwalia, M.S.; Sahgal, A.; Chang, E.L. Current approaches to the management of brain metastases. Nat. Rev. Clin. Oncol. 2020, 17, 279–299. [Google Scholar] [CrossRef]
- Yamamoto, M.; Serizawa, T.; Shuto, T.; Akabane, A.; Higuchi, Y.; Kawagishi, J.; Yamanaka, K.; Sato, Y.; Jokura, H.; Yomo, S.; et al. Stereotactic radiosurgery for patients with multiple brain metastases (JLGK0901): A multi-institutional prospective observational study. Lancet Oncol. 2014, 15, 387–395. [Google Scholar] [CrossRef]
- Niranjan, A.; Monaco, E.; Flickinger, J.; Lunsford, L.D. Guidelines for Multiple Brain Metastases Radiosurgery. Prog. Neurol. Surg. 2019, 34, 100–109. [Google Scholar]
- Zhou, Z.; Sanders, J.W.; Johnson, J.M.; Gule-Monroe, M.K.; Chen, M.M.; Briere, T.M.; Wang, Y.; Son, J.B.; Pagel, M.D.; Li, J.; et al. Computer-aided Detection of Brain Metastases in T1-weighted MRI for Stereotactic Radiosurgery Using Deep Learning Single-Shot Detectors. Radiology 2020, 295, 407–415. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single Shot MultiBox Detector. In Proceedings of the Computer Vision—ECCV 2016, Amsterdam, The Netherlands, 11–14 October 2016; Leibe, B., Matas, J., Sebe, N., Welling, M., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 21–37. [Google Scholar]
- Amemiya, S.; Takao, H.; Kato, S.; Yamashita, H.; Sakamoto, N.; Abe, O. Feature-fusion improves MRI single-shot deep learning detection of small brain metastases. J. Neuroimaging 2022, 32, 111–119. [Google Scholar] [CrossRef]
- Cao, Y.; Vassantachart, A.; Ye, J.C.; Yu, C.; Ruan, D.; Sheng, K.; Lao, Y.; Shen, Z.L.; Balik, S.; Bian, S.; et al. Automatic detection and segmentation of multiple brain metastases on magnetic resonance image using asymmetric UNet architecture. Phys. Med. Biol. 2021, 66, 015003. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Guo, Y.; Zhao, Z.; Chen, M.; Liu, X.; Gong, G.; Wang, L. MRI-based two-stage deep learning model for automatic detection and segmentation of brain metastases. Eur. Radiol. 2023, 33, 3521–3531. [Google Scholar] [CrossRef] [PubMed]
- Jocher, G.; Chaurasia, A.; Stoken, A.; Borovec, J.; NanoCode012; Kwon, Y.; TaoXie; Michael, K.; Fang, J.; Imyhxy; et al. ultralytics/yolov5: v6.2 - YOLOv5 Classification Models, Apple M1, Reproducibility, ClearML and Deci.ai Integrations. 2022. Available online: https://doi.org/10.5281/zenodo.7002879 (accessed on 1 October 2022).
- Chen, S.; Duan, J.; Wang, H.; Wang, R.; Li, J.; Qi, M.; Duan, Y.; Qi, S. Automatic detection of stroke lesion from diffusion-weighted imaging via the improved YOLOv5. Comput. Biol. Med. 2022, 150, 106120. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Chen, B.; Yu, Y. Polyp Detection from Colorectum Images by Using Attentive YOLOv5. Diagnostics 2021, 11, 2264. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Y.; Hsiao, Y.P.; Mukundan, A.; Tsao, Y.M.; Chang, W.Y.; Wang, H.C. Classification of Skin Cancer Using Novel Hyperspectral Imaging Engineering via YOLOv5. J. Clin. Med. 2023, 12, 1134. [Google Scholar] [CrossRef]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. Adv. Neural Inf. Process. Syst. 2019, 32, 8024–8035. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Proceedings of the Computer Vision—ECCV 2018, Munich, Germany, 8–14 September 2018; Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 3–19. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, BC, Canada, 11–17 October 2021; pp. 9992–10002. [Google Scholar]
- Ioffe, S.S.C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Elfwing, S.; Uchibe, E.; Doya, K. Sigmoid-weighted linear units for neural network function approximation in reinforcement learning. Neural Netw. 2018, 107, 3–11. [Google Scholar] [CrossRef]
- Wang, W.; Xie, E.; Song, X.; Zang, Y.; Wang, W.; Lu, T.; Yu, G.; Shen, C. Efficient and Accurate Arbitrary-Shaped Text Detection With Pixel Aggregation Network. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 8439–8448. [Google Scholar]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 936–944. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like Pure Transformer for Medical Image Segmentation. In Proceedings of the ECCV Workshops, Tel Aviv, Israel, 23 October 2022. [Google Scholar]
- Yu, J.; Jiang, Y.; Wang, Z.; Cao, Z.; Huang, T.S. UnitBox: An Advanced Object Detection Network. In Proceedings of the 24th ACM international conference on Multimedia, Amsterdam, The Netherlands, 15–19 October 2016. [Google Scholar]
- Zheng, Z.; Wang, P.; Liu, W.; Li, J.; Ye, R.; Ren, D. Distance-IoU Loss: Faster and Better Learning for Bounding Box Regression. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January 2019–1 February 2019. [Google Scholar]
- Sinha, N.K.; Griscik, M.P. A Stochastic Approximation Method. IEEE Trans. Syst. Man Cybern. 1971, SMC-1, 338–344. [Google Scholar] [CrossRef]
- Neubeck, A.; Gool, L.V. Efficient Non-Maximum Suppression. In Proceedings of the 18th International Conference on Pattern Recognition (ICPR’06), Washington, DC, USA, 20–24 August 2006; Volume 3, pp. 850–855. [Google Scholar]
- Kato, S.; Amemiya, S.; Takao, H.; Yamashita, H.; Sakamoto, N.; Abe, O. Automated detection of brain metastases on non-enhanced CT using single-shot detectors. Neuroradiology 2021, 63, 1995–2004. [Google Scholar] [CrossRef]
- Takao, H.; Amemiya, S.; Kato, S.; Yamashita, H.; Sakamoto, N.; Abe, O. Deep-learning 2.5-dimensional single-shot detector improves the performance of automated detection of brain metastases on contrast-enhanced CT. Neuroradiology 2022, 64, 1511–1518. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Zhang, Y.; Lang, S.; Gong, Y. Swin-Transformer-Based YOLOv5 for Small-Object Detection in Remote Sensing Images. Sensors 2023, 23, 3634. [Google Scholar] [CrossRef]
- Dai, G.; Hu, L.; Fan, J. DA-ActNN-YOLOV5: Hybrid YOLO v5 Model with Data Augmentation and Activation of Compression Mechanism for Potato Disease Identification. Comput. Intell. Neurosci. 2022, 2022, 6114061. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, L.; Luo, W. PDNet: Improved YOLOv5 Nondeformable Disease Detection Network for Asphalt Pavement. Comput. Intell. Neurosci. 2022, 2022, 5133543. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef]
- Tan, M.; Pang, R.; Le, Q.V. EfficientDet: Scalable and Efficient Object Detection. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 10778–10787. [Google Scholar]



| Method | Recall | Precision | F2-Score | FN/Patient | FP/Patient |
|---|---|---|---|---|---|
| Faster R-CNN [36] | 0.690(136/197) | 0.342(136/398) | 0.573 | 1.271 | 5.458 |
| EfficientDet [37] | 0.614(121/197) | 0.669(121/181) | 0.624 | 1.583 | 1.250 |
| SSD [9] | 0.822(162/197) | 0.369(162/439) | 0.660 | 0.729 | 5.771 |
| FF-SSD [11] | 0.827(163/197) | 0.397(163/411) | 0.680 | 0.708 | 5.167 |
| Ours | 0.904(178/197) | 0.612(178/291) | 0.825 | 0.396 | 2.354 |
| Model | Recall | Precision | F2-Score | FN/Patient | FP/Patient |
|---|---|---|---|---|---|
| YOLOv5 | 0.883(174/197) | 0.611(174/285) | 0.812 | 0.479 | 2.313 |
| YOLOv5+CBAM | 0.898(177/197) | 0.598(177/296) | 0.816 | 0.417 | 2.479 |
| YOLOv5+CBAM+PH 1 | 0.898(177/197) | 0.586(177/302) | 0.812 | 0.417 | 2.604 |
| YOLOv5+CBAM+STPH | 0.904(178/197) | 0.612(178/291) | 0.825 | 0.396 | 2.354 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Z.; Qiu, Q.; Liu, H.; Ge, X.; Li, T.; Xing, L.; Yang, R.; Yin, Y. Automatic Detection of Brain Metastases in T1-Weighted Construct-Enhanced MRI Using Deep Learning Model. Cancers 2023, 15, 4443. https://doi.org/10.3390/cancers15184443
Zhou Z, Qiu Q, Liu H, Ge X, Li T, Xing L, Yang R, Yin Y. Automatic Detection of Brain Metastases in T1-Weighted Construct-Enhanced MRI Using Deep Learning Model. Cancers. 2023; 15(18):4443. https://doi.org/10.3390/cancers15184443
Chicago/Turabian StyleZhou, Zichun, Qingtao Qiu, Huiling Liu, Xuanchu Ge, Tengxiang Li, Ligang Xing, Runtao Yang, and Yong Yin. 2023. "Automatic Detection of Brain Metastases in T1-Weighted Construct-Enhanced MRI Using Deep Learning Model" Cancers 15, no. 18: 4443. https://doi.org/10.3390/cancers15184443
APA StyleZhou, Z., Qiu, Q., Liu, H., Ge, X., Li, T., Xing, L., Yang, R., & Yin, Y. (2023). Automatic Detection of Brain Metastases in T1-Weighted Construct-Enhanced MRI Using Deep Learning Model. Cancers, 15(18), 4443. https://doi.org/10.3390/cancers15184443






