Type IV P-Type ATPases: Recent Updates in Cancer Development, Progression, and Treatment
Abstract
Simple Summary
Abstract
1. Introduction
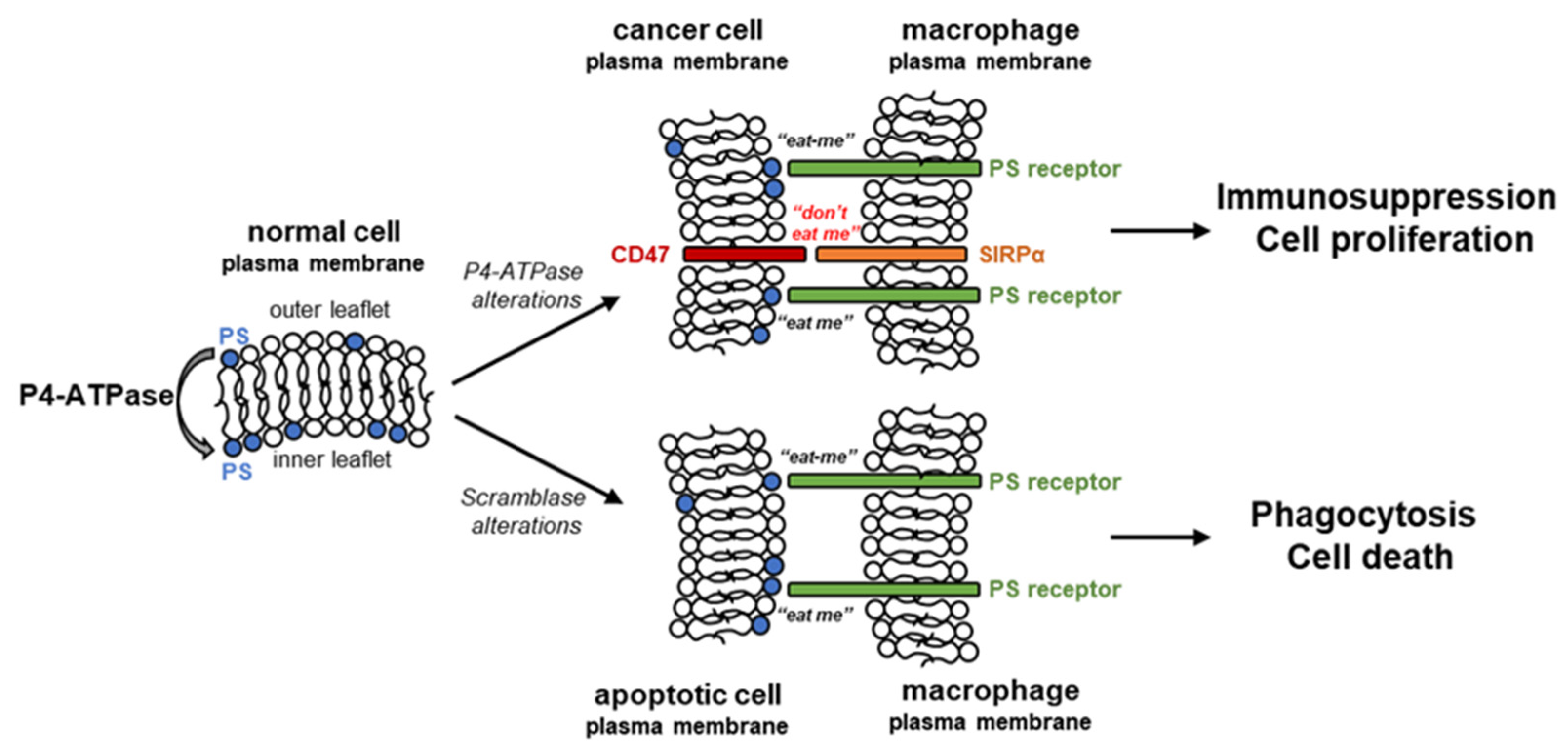
2. Flipping Phospholipids in Cancer Cells
2.1. Phosphatidylserine
2.2. Phosphatidylethanolamine
2.3. Phosphatidylcholine
3. P4-ATPases in Cancer Development and Progression
3.1. Gastrointestinal Cancers
3.2. Prostate Cancer
3.3. Endometrial, Cervical, Ovarian and Breast Cancers
3.4. Blood Cancers
3.5. Lung Cancers
3.6. Melanoma
4. P4-ATPases as Targets for Anti-Cancer Treatment
4.1. Interplay with the Small Molecule Inhibitors
4.2. Immunotherapy
5. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Clarke, R.; Hossain, K.; Cao, K. Physiological roles of transverse lipid asymmetry of animal membranes. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183382. [Google Scholar] [CrossRef]
- Nagata, S.; Segawa, K. Sensing and clearance of apoptotic cells. Curr. Opin. Immunol. 2021, 68, 1–8. [Google Scholar] [CrossRef]
- Palmgren, M.G.; Nissen, P. P-type ATPases. Annu. Rev. Biophys. 2011, 40, 243–266. [Google Scholar] [CrossRef]
- Lopez-Marques, R.L.; Theorin, L.; Palmgren, M.G.; Pomorski, T.G. P4-ATPases: Lipid flippases in cell membranes. Pflug. Arch. 2014, 466, 1227–1240. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.I.; Park, E. P5-ATPases: Structure, substrate specificities, and transport mechanisms. Curr. Opin. Struct. Biol. 2023, 79, 102531. [Google Scholar] [CrossRef]
- Sebastian, T.T.; Baldridge, R.D.; Xu, P.; Graham, T.R. Phospholipid flippases: Building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta 2012, 1821, 1068–1077. [Google Scholar] [CrossRef]
- Best, J.T.; Xu, P.; Graham, T.R. Phospholipid flippases in membrane remodeling and transport carrier biogenesis. Curr. Opin. Cell Biol. 2019, 59, 8–15. [Google Scholar] [CrossRef]
- Panatala, R.; Hennrich, H.; Holthuis, J.C.M. Inner workings and biological impact of phospholipid flippases. J. Cell Sci. 2015, 128, 2021–2032. [Google Scholar] [CrossRef] [PubMed]
- Folmer, D.E.; Elferink, R.P.O.; Paulusma, C.C. P4 ATPases—Lipid flippases and their role in disease. Biochim. Biophys. Acta 2009, 1791, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Van der Mark, V.A.; Elferink, R.P.O.; Paulusma, C.C. P4 ATPases: Flippases in health and disease. Int. J. Mol. Sci. 2013, 14, 7897–7922. [Google Scholar] [CrossRef] [PubMed]
- van der Velden, L.M.; Wichers, C.G.; Van Breevoort, A.E.; Coleman, J.A.; Molday, R.S.; Berger, R.; Klomp, L.W.; van De Graaf, S.F. Heteromeric interactions required for abundance and subcellular localization of human CDC50 proteins and class 1 P4-ATPases. J. Biol. Chem. 2010, 285, 40088–40096. [Google Scholar] [CrossRef]
- Takatsu, H.; Tanaka, G.; Segawa, K.; Suzuki, J.; Nagata, S.; Nakayama, K.; Shin, H.-W. Phospholipid flippase activities and substrate specificities of human type IV P-type ATPases localized to the plasma membrane. J. Biol. Chem. 2014, 289, 33543–33556. [Google Scholar] [CrossRef] [PubMed]
- Gong, E.-Y.; Park, E.; Lee, H.J.; Lee, K. Expression of Atp8b3 in murine testis and its characterization as a testis specific P-type ATPase. Reproduction 2009, 137, 345–351. [Google Scholar] [CrossRef]
- Wang, L.; Beserra, C.; Garbers, D.L. A novel aminophospholipid transporter exclusively expressed in spermatozoa is required for membrane lipid asymmetry and normal fertilization. Dev. Biol. 2004, 267, 203–215. [Google Scholar] [CrossRef]
- Takatsu, H.; Baba, K.; Shima, T.; Umino, H.; Kato, U.; Umeda, M.; Nakayama, K.; Shin, H.-W. ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50 protein-independent manner. J. Biol. Chem. 2011, 286, 38159–38167. [Google Scholar] [CrossRef]
- Utsugi, T.; Schroit, A.J.; Connor, J.; Bucana, C.D.; Fidler, I.J. Elevated expression of phosphatidylserine in the outer membrane leaflet of human tumor cells and recognition by activated human blood monocytes. Cancer Res. 1991, 51, 3062–3066. [Google Scholar] [PubMed]
- Tseng, D.; Volkmer, J.-P.; Willingham, S.B.; Contreras-Trujillo, H.; Fathman, J.W.; Fernhoff, N.B.; Seita, J.; Inlay, M.A.; Weiskopf, K.; Miyanishi, M.; et al. Anti-CD47 antibody-mediated phagocytosis of cancer by macrophages primes an effective antitumor T-cell response. Proc. Natl. Acad. Sci. USA 2013, 110, 11103–11108. [Google Scholar] [CrossRef]
- Huang, C.-Y.; Ye, Z.-H.; Huang, M.-Y.; Lu, J.-J. Regulation of CD47 expression in cancer cells. Transl. Oncol. 2020, 13, 100862. [Google Scholar] [CrossRef]
- Tse, E.; Kwong, Y.-L. T-cell lymphoma: Microenvironment-related biomarkers. Semin. Cancer Biol. 2015, 34, 46–51. [Google Scholar] [CrossRef]
- Vallabhapurapu, S.D.; Blanco, V.M.; Sulaiman, M.K.; Vallabhapurapu, S.L.; Chu, Z.; Franco, R.S.; Qi, X. Variation in human cancer cell external phosphatidylserine is regulated by flippase activity and intracellular calcium. Oncotarget 2015, 6, 34375–34388. [Google Scholar] [CrossRef] [PubMed]
- Levano, K.; Sobocki, T.; Jayman, F.; Debata, P.R.; Sobocka, M.B.; Banerjee, P. A genetic strategy involving a glycosyltransferase promoter and a lipid translocating enzyme to eliminate cancer cells. Glycoconj. J. 2009, 26, 739–748. [Google Scholar] [CrossRef]
- Wang, W.; Wu, S.; Cen, Z.; Zhang, Y.; Chen, Y.; Huang, Y.; Cillo, A.R.; Prokopec, J.S.; Quarato, G.; Vignali, D.A.; et al. Mobilizing phospholipids on tumor plasma membrane implicates phosphatidylserine externalization blockade for cancer immunotherapy. Cell Rep. 2022, 41, 111582. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A. Phospholipid Asymmetry in Biological Membranes: Is the Role of Phosphatidylethanolamine Underappreciated? J. Membr. Biol. 2021, 254, 127–132. [Google Scholar] [CrossRef]
- Vance, J.E. Phosphatidylserine and phosphatidylethanolamine in mammalian cells: Two metabolically related aminophospholipids. J. Lipid Res. 2008, 49, 1377–1387. [Google Scholar] [CrossRef]
- Szlasa, W.; Zendran, I.; Zalesińska, A.; Tarek, M.; Kulbacka, J. Lipid composition of the cancer cell membrane. J. Bioenergy Biomembr. 2020, 52, 321–342. [Google Scholar] [CrossRef] [PubMed]
- Broughton, L.J.; Crow, C.; Maraveyas, A.; Madden, L.A. Duramycin-induced calcium release in cancer cells. Anticancer Drugs 2016, 27, 173–182. [Google Scholar] [CrossRef]
- Stafford, J.H.; Thorpe, P.E. Increased exposure of phosphatidylethanolamine on the surface of tumor vascular endothelium. Neoplasia 2011, 13, 299–308. [Google Scholar] [CrossRef]
- Monteiro, J.P.; Oliveira, P.J.; Jurado, A.S. Mitochondrial membrane lipid remodeling in pathophysiology: A new target for diet and therapeutic interventions. Prog. Lipid Res. 2013, 52, 513–528. [Google Scholar] [CrossRef]
- Barbosa, I.A.; Machado, N.G.; Skildum, A.J.; Scott, P.M.; Oliveira, P.J. Mitochondrial remodeling in cancer metabolism and survival: Potential for new therapies. Biochim. Biophys. Acta 2012, 1826, 238–254. [Google Scholar] [CrossRef]
- Gómez-Mellado, V.E.; Chang, J.-C.; Ho-Mok, K.S.; Morcillo, C.B.; Kersten, R.H.J.; Elferink, R.P.J.O.; Verhoeven, A.J.; Paulusma, C.C. ATP8B1 Deficiency Results in Elevated Mitochondrial Phosphatidylethanolamine Levels and Increased Mitochondrial Oxidative Phosphorylation in Human Hepatoma Cells. Int. J. Mol. Sci. 2022, 23, 12344. [Google Scholar] [CrossRef] [PubMed]
- Zech, T.; Ejsing, C.S.; Gaus, K.; de Wet, B.; Shevchenko, A.; Simons, K.; Harder, T. Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signalling. EMBO J. 2009, 28, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Gottesman, M.M.; Pastan, I. Domain exchangeability between the multidrug transporter (MDR1) and phosphatidylcholine flippase (MDR2). Mol. Pharmacol. 1999, 56, 997–1004. [Google Scholar] [CrossRef]
- Zhou, Y.; Gottesman, M.M.; Pastan, I. Studies of human MDR1-MDR2 chimeras demonstrate the functional exchangeability of a major transmembrane segment of the multidrug transporter and phosphatidylcholine flippase. Mol. Cell Biol. 1999, 19, 1450–1459. [Google Scholar] [CrossRef] [PubMed]
- Eckford, P.D.W.; Sharom, F.J. The reconstituted P-glycoprotein multidrug transporter is a flippase for glucosylceramide and other simple glycosphingolipids. Biochem. J. 2005, 389 Pt 2, 517–526. [Google Scholar] [CrossRef]
- Shivakumar, M.; Miller, J.E.; Dasari, V.R.; Gogoi, R.; Kim, D. Exome-Wide Rare Variant Analysis from the DiscovEHR Study Identifies Novel Candidate Predisposition Genes for Endometrial Cancer. Front. Oncol. 2019, 9, 574. [Google Scholar] [CrossRef]
- Wittkowski, K.M.; Dadurian, C.; Seybold, M.P.; Kim, H.S.; Hoshino, A.; Lyden, D. Complex polymorphisms in endocytosis genes suggest alpha-cyclodextrin as a treatment for breast cancer. PLoS ONE 2018, 13, e0199012. [Google Scholar] [CrossRef]
- Li, D.; Xu, T.; Wang, X.; Ma, X.; Liu, T.; Wang, Y.; Jiang, S. The role of ATP8A1 in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2017, 10, 7760–7766. [Google Scholar]
- Dong, W.; Yao, C.; Teng, X.; Chai, J.; Yang, X.; Li, B. MiR-140-3p suppressed cell growth and invasion by downregulating the expression of ATP8A1 in non-small cell lung cancer. Tumour Biol. 2016, 37, 2973–2985. [Google Scholar] [CrossRef]
- Chen, Y.H.; Zhang, T.F.; Liu, Y.Y.; Zheng, J.H.; Lin, W.X.; Chen, Y.K.; Cai, J.H.; Zou, J.; Li, Z.Y. Identification of a 5-gene-risk score model for predicting luminal A-invasive lobular breast cancer survival. Genetica 2022, 150, 299–316. [Google Scholar] [CrossRef]
- Yan, H.; Guan, Q.; He, J.; Lin, Y.; Zhang, J.; Li, H.; Liu, H.; Gu, Y.; Guo, Z.; He, F. Individualized analysis reveals CpG sites with methylation aberrations in almost all lung adenocarcinoma tissues. J. Transl. Med. 2017, 15, 26. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, W.; Zhang, J.; Liu, W.; Yin, J.; Shi, D.; Ma, W. A Novel Mitochondrial-Related Nuclear Gene Signature Predicts Overall Survival of Lung Adenocarcinoma Patients. Front. Cell Dev. Biol. 2021, 9, 740487. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shi, D.; Zhao, D.; Hu, D. Aberrant Methylation and Differential Expression of SLC2A1, TNS4, GAPDH, ATP8A2, and CASZ1 Are Associated with the Prognosis of Lung Adenocarcinoma. Biomed. Res. Int. 2020, 2020, 1807089. [Google Scholar] [CrossRef] [PubMed]
- Eldai, H.; Periyasamy, S.; Al Qarni, S.; Al Rodayyan, M.; Muhammed Mustafa, S.; Deeb, A.; Al Sheikh, E.; Afzal Khan, M.; Johani, M.; Yousef, Z.; et al. Novel genes associated with colorectal cancer are revealed by high resolution cytogenetic analysis in a patient specific manner. PLoS ONE 2013, 8, e76251. [Google Scholar] [CrossRef]
- Aziz, M.A.; Periyasamy, S.; Al Yousef, Z.; AlAbdulkarim, I.; Al Otaibi, M.; Alfahed, A.; Alasiri, G. Integrated exon level expression analysis of driver genes explain their role in colorectal cancer. PLoS ONE 2014, 9, e110134. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Niu, G.-M.; Ren, J.; Ke, C.-W.; Loura, L. Identification of ATP8B1 as a Tumor Suppressor Gene for Colorectal Cancer and Its Involvement in Phospholipid Homeostasis. Biomed. Res. Int. 2020, 2020, 2015648. [Google Scholar] [CrossRef] [PubMed]
- Althenayyan, S.; AlGhamdi, A.; AlMuhanna, M.H.; Hawsa, E.; Aldeghaither, D.; Iqbal, J.; Mohammad, S.; Aziz, M.A. Modulation of ATP8B1 Gene Expression in Colorectal Cancer Cells Suggest its Role as a Tumor Suppressor. Curr. Cancer Drug Targets 2022, 22, 577–590. [Google Scholar] [CrossRef]
- Meerzaman, D.M.; Yan, C.; Chen, Q.-R.; Edmonson, M.N.; Schaefer, C.F.; Clifford, R.J.; Dunn, B.K.; Dong, L.; Finney, R.P.; Cultraro, C.M.; et al. Genome-wide transcriptional sequencing identifies novel mutations in metabolic genes in human hepatocellular carcinoma. Cancer Genom. Proteom. 2014, 11, 1–12. [Google Scholar]
- Chen, L.; Huang, S.; Shih, C.; Li, C.; Chen, Y.; Huang, C.; Yu, C.; Lin, V.C.; Lee, C.; Geng, J.; et al. ATP8B1: A prognostic prostate cancer biomarker identified via genetic analysis. Prostate 2023, 83, 602–611. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, R.; Liu, P.; Zhang, R.; Ning, J.; Ye, Y.; Yu, W.; Yu, J. ATP8B1 Knockdown Activated the Choline Metabolism Pathway and Induced High-Level Intracellular REDOX Homeostasis in Lung Squamous Cell Carcinoma. Cancers 2022, 14, 835. [Google Scholar] [CrossRef]
- Li, M.-X.; Wang, H.-Y.; Yuan, C.-H.; Ma, Z.-L.; Jiang, B.; Li, L.; Zhang, L.; Xiu, D.-R. Establishment of a Macrophage Phenotypic Switch Related Prognostic Signature in Patients with Pancreatic Cancer. Front. Oncol. 2021, 11, 619517. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Liu, J.; Xu, J.; Zhang, B.; Yu, X.; Wang, W.; Liang, C. LDHA is a prognostic biomarker on the immune response in pancreatic adenocarcinoma and associated with m6A modification. J. Cancer Res. Clin. Oncol. 2022, 149, 4853–4865. [Google Scholar] [CrossRef] [PubMed]
- Eswaran, S.; Adiga, D.; Khan, N.; Sriharikrishnaa, S.; Kabekkodu, S.P. Comprehensive analysis of the exocytosis pathway genes in cervical cancer. Am. J. Med. Sci. 2022, 363, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Carbajo-García, M.C.; Corachán, A.; Juárez-Barber, E.; Monleón, J.; Payá, V.; Trelis, A.; Quiñonero, A.; Pellicer, A.; Ferrero, H. Integrative analysis of the DNA methylome and transcriptome in uterine leiomyoma shows altered regulation of genes involved in metabolism, proliferation, extracellular matrix, and vesicles. J. Pathol. 2022, 257, 663–673. [Google Scholar] [CrossRef]
- Leonard, M.K.; Puts, G.S.; Pamidimukkala, N.; Adhikary, G.; Xu, Y.; Kwok, E.; Jin, Y.; Snyder, D.; Matsangos, N.; Novak, M.; et al. Comprehensive molecular profiling of UV-induced metastatic melanoma in Nme1/Nme2-deficient mice reveals novel markers of survival in human patients. Oncogene 2021, 40, 6329–6342. [Google Scholar] [CrossRef]
- Magi, A.; Masselli, M.; Sala, C.; Guerriero, A.; Laise, P.; Puccini, B.; Rigacci, L.; Breschi, C.; Crociani, O.; Pillozzi, S.; et al. The ion channels and transporters gene expression profile indicates a shift in excitability and metabolisms during malignant progression of Follicular Lymphoma. Sci. Rep. 2019, 9, 8586. [Google Scholar] [CrossRef]
- El-Zein, M.; Cheishvili, D.; Gotlieb, W.; Gilbert, L.; Hemmings, R.; Behr, M.A.; Szyf, M.; Franco, E.L.; for the MARKER Study Group; Bouten, S.; et al. Genome-wide DNA methylation profiling identifies two novel genes in cervical neoplasia. Int. J. Cancer 2020, 147, 1264–1274. [Google Scholar] [CrossRef]
- Olsson, L.; Castor, A.; Behrendtz, M.; Biloglav, A.; Forestier, E.; Paulsson, K.; Johansson, B. Deletions of IKZF1 and SPRED1 are associated with poor prognosis in a population-based series of pediatric B-cell precursor acute lymphoblastic leukemia diagnosed between 1992 and 2011. Leukemia 2014, 28, 302–310. [Google Scholar] [CrossRef]
- Amin, N.A.; Seymour, E.; Saiya-Cork, K.; Parkin, B.; Shedden, K.; Malek, S.N. A Quantitative Analysis of Subclonal and Clonal Gene Mutations before and after Therapy in Chronic Lymphocytic Leukemia. Clin. Cancer Res. 2016, 22, 4525–4535. [Google Scholar] [CrossRef]
- Yepes, S.; Tucker, M.A.; Koka, H.; Xiao, Y.; Zhang, T.; Jones, K.; Vogt, A.; Burdette, L.; Luo, W.; Zhu, B.; et al. Integrated Analysis of Coexpression and Exome Sequencing to Prioritize Susceptibility Genes for Familial Cutaneous Melanoma. J. Investig. Dermatol. 2022, 142, 2464–2475.e5. [Google Scholar] [CrossRef]
- Fusco, J.P.; Pita, G.; Pajares, M.J.; Andueza, M.P.; Patiño-García, A.; De-Torres, J.P.; Gurpide, A.; Zulueta, J.; Alonso, R.; Alvarez, N.; et al. Genomic characterization of individuals presenting extreme phenotypes of high and low risk to develop tobacco-induced lung cancer. Cancer Med. 2018, 7, 3474–3483. [Google Scholar] [CrossRef]
- Miyoshi, N.; Ishii, H.; Mimori, K.; Tanaka, F.; Nagai, K.; Uemura, M.; Sekimoto, M.; Doki, Y.; Mori, M. ATP11A is a novel predictive marker for metachronous metastasis of colorectal cancer. Oncol. Rep. 2010, 23, 505–510. [Google Scholar]
- Izquierdo, A.G.; Boughanem, H.; Diaz-Lagares, A.; Arranz-Salas, I.; Esteller, M.; Tinahones, F.J.; Casanueva, F.F.; Macias-Gonzalez, M.; Crujeiras, A.B. DNA methylome in visceral adipose tissue can discriminate patients with and without colorectal cancer. Epigenetics 2022, 17, 665–676. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Xiao, B.; Tang, N.; Hu, J.; Liang, S.; Pang, Y.; Xu, H.; Ao, J.; Yang, J.; et al. MiR-103a promotes tumour growth and influences glucose metabolism in hepatocellular carcinoma. Cell Death Dis. 2021, 12, 618. [Google Scholar] [CrossRef]
- Chen, L.; Tang, J.; Sheng, W.; Sun, J.; Ma, Y.; Dong, M. ATP11A promotes EMT by regulating Numb PRR(L) in pancreatic cancer cells. PeerJ 2022, 10, e13172. [Google Scholar] [CrossRef]
- Zhao, S.; Geybels, M.S.; Leonardson, A.; Rubicz, R.; Kolb, S.; Yan, Q.; Klotzle, B.; Bibikova, M.; Hurtado-Coll, A.; Troyer, D.; et al. Epigenome-Wide Tumor DNA Methylation Profiling Identifies Novel Prognostic Biomarkers of Metastatic-Lethal Progression in Men Diagnosed with Clinically Localized Prostate Cancer. Clin. Cancer Res. 2017, 23, 311–319. [Google Scholar] [CrossRef]
- Geybels, M.S.; Fang, M.; Wright, J.L.; Qu, X.; Bibikova, M.; Klotzle, B.; Fan, J.-B.; Feng, Z.; Ostrander, E.A.; Nelson, P.S.; et al. PTEN loss is associated with prostate cancer recurrence and alterations in tumor DNA methylation profiles. Oncotarget 2017, 8, 84338–84348. [Google Scholar] [CrossRef]
- Hu, L.; Gao, Y.; Shi, Z.; Liu, Y.; Zhao, J.; Xiao, Z.; Lou, J.; Xu, Q.; Tong, X. DNA methylation-based prognostic biomarkers of acute myeloid leukemia patients. Ann. Transl. Med. 2019, 7, 737. [Google Scholar] [CrossRef]
- Moreno-Smith, M.; Halder, J.B.; Meltzer, P.S.; Gonda, T.A.; Mangala, L.S.; Rupaimoole, R.; Lu, C.; Nagaraja, A.S.; Gharpure, K.M.; Kang, Y.; et al. ATP11B mediates platinum resistance in ovarian cancer. J. Clin. Investig. 2013, 123, 2119–2130. [Google Scholar] [CrossRef]
- Elsnerova, K.; Mohelnikova-Duchonova, B.; Cerovska, E.; Ehrlichova, M.; Gut, I.; Rob, L.; Skapa, P.; Hruda, M.; Bartakova, A.; Bouda, J.; et al. Gene expression of membrane transporters: Importance for prognosis and progression of ovarian carcinoma. Oncol. Rep. 2016, 35, 2159–2170. [Google Scholar] [CrossRef]
- Xu, J.; Su, S.M.; Zhang, X.; Chan, U.I.; Adhav, R.; Shu, X.; Liu, J.; Li, J.; Mo, L.; Wang, Y.; et al. ATP11B inhibits breast cancer metastasis in a mouse model by suppressing externalization of nonapoptotic phosphatidylserine. J. Clin. Investig. 2022, 132. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Lu, L.; Zou, Q. Analysis of gene expression profiles of lung cancer subtypes with machine learning algorithms. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165822. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Xiao, Y.; Huang, X.; Wu, X.; Zhao, Z.; Yang, M.; Kong, L.; Shi, D.; Chen, X.; et al. The phospholipid flippase ATP9A enhances macropinocytosis to promote nutrient starvation tolerance in hepatocellular carcinoma. J. Pathol. 2023, 260, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lyu, C.; Wang, F.; Wang, C.; Wu, F.; Li, X.; Gan, S. Identification of Potential Signatures and Their Functions for Acute Lymphoblastic Leukemia: A Study Based on the Cancer Genome Atlas. Front. Genet. 2021, 12, 656042. [Google Scholar] [CrossRef] [PubMed]
- Kap, E.J.; Seibold, P.; Scherer, D.; Habermann, N.; Balavarca, Y.; Jansen, L.; Zucknick, M.; Becker, N.; Hoffmeister, M.; Ulrich, A.; et al. SNPs in transporter and metabolizing genes as predictive markers for oxaliplatin treatment in colorectal cancer patients. Int. J. Cancer 2016, 138, 2993–3001. [Google Scholar] [CrossRef]
- Park, H.A.; Seibold, P.; Edelmann, D.; Benner, A.; Canzian, F.; Alwers, E.; Jansen, L.; Schneider, M.; Hoffmeister, M.; Brenner, H.; et al. Validation of Genetic Markers Associated with Survival in Colorectal Cancer Patients Treated with Oxaliplatin-Based Chemotherapy. Cancer Epidemiol. Biomark. Prev. 2022, 31, 352–361. [Google Scholar] [CrossRef]
- Zhang, B.; Groffen, J.; Heisterkamp, N. Resistance to farnesyltransferase inhibitors in Bcr/Abl-positive lymphoblastic leukemia by increased expression of a novel ABC transporter homolog ATP11a. Blood 2005, 106, 1355–1361. [Google Scholar] [CrossRef][Green Version]
- Tang, T.; Huang, X.; Zhang, G.; Liang, T. Oncolytic immunotherapy: Multiple mechanisms of oncolytic peptides to confer anticancer immunity. J. Immunother. Cancer 2022, 10, e005065. [Google Scholar] [CrossRef]
- Tang, T.; Huang, X.; Zhang, G.; Lu, M.; Hong, Z.; Wang, M.; Huang, J.; Zhi, X.; Liang, T. Oncolytic peptide LTX-315 induces anti-pancreatic cancer immunity by targeting the ATP11B-PD-L1 axis. J. Immunother. Cancer 2022, 10, e004129. [Google Scholar] [CrossRef]
- Genard, G.; Lucas, S.; Michiels, C. Reprogramming of Tumor-Associated Macrophages with Anticancer Therapies: Radiotherapy versus Chemo- and Immunotherapies. Front. Immunol. 2017, 8, 828. [Google Scholar] [CrossRef]
| Class | Name | Lipid Substrate | Sub-Cellular Localization | References |
|---|---|---|---|---|
| 1a | ATP8A1 | PS, PE | PM, TNG, endosome | [12] |
| 1a | ATP8A2 | PS, PE | PM, TNG, endosome | [12] |
| 1b | ATP8B1 | PS, PC, cardiolipin? | PM | [12,13] |
| 1b | ATP8B2 | PC | PM | [12,13] |
| 1b | ATP8B3 | PS? | ER, TNG | [12,14,15] |
| 1b | ATP8B4 | unknown | PM | [12] |
| 2 | ATP9A | unknown | TNG, endosome | [16] |
| 2 | ATP9B | unknown | TNG | [16] |
| 5 | ATP10A | PC, GlcCer | PM | [16] |
| 5 | ATP10B | PC, GlcCer | endosome, lysosome | [16] |
| 5 | ATP10D | GlcCer | PM | [16] |
| 6 | ATP11A | PS, PE | PM | [13,16] |
| 6 | ATP11B | PS, PE | endosome | [13,16] |
| 6 | ATP11C | PS, PE | PM | [13,16] |
| ATPase | Cancer Type | Reported Feature | Reported Effect | Citations |
|---|---|---|---|---|
| ATP8A1 | endometrial cancer | gene expression up | cancer predisposition | [36] |
| metastatic breast cancer | SNPs | cancer association | [37] | |
| non-small-cell lung cancer | protein expression up | cancer association | [38] | |
| non-small-cell lung cancer (mouse model) | protein expression up | cancer cell growth | [39] | |
| ATP8A2 | luminal A-invasive lobular breast cancer | gene expression up | poor prognosis | [40] |
| lung adenocarcinoma | gene expression down | cancer association | [41,42] | |
| lung adenocarcinoma | mRNA expression down | cancer association | [43] | |
| ATP8B1 | colorectal carcinoma | gene expression up | cancer association | [44,45,46] |
| colorectal carcinoma (cell lines) | protein expression down | increased cell growth | [47] | |
| protein expression up | decreased cell growth | |||
| hepatocellular carcinoma | gene mutation | cancer association | [48] | |
| prostate cancer | protein expression up | better prognosis | [49] | |
| metastatic breast cancer | SNPs | cancer association | [37] | |
| lung squamous cell carcinoma | protein expression down | poor prognosis | [50] | |
| ATP8B2 | pancreatic adenocarcinoma | gene expression up | poor prognosis | [51] |
| pancreatic ductal adenocarcinoma | gene expression up | poor prognosis | [52] | |
| ATP8B4 | metastasis in cervical carcinoma | gene methylation down | cancer association | [53] |
| uterine leiomyoma | gene methylation down | cancer association | [54] | |
| metastasis in melanoma (mouse model) | gene mutations | cancer progression | [55] | |
| ATP9A | hepatocellular carcinoma | protein expression up | poor outcome | [43] |
| relapsed follicular lymphoma | gene expression down | poor prognosis | [56] | |
| ATP10A | cervical cancer | gene methylation up | cancer progression | [57] |
| relapsed acute lymphoblastic leukemia | gene mutations | poor prognosis | [58] | |
| relapsed chronic lymphocytic leukemia | gene mutations | poor prognosis | [59] | |
| melanoma | gene expression up | cancer predisposition | [60] | |
| ATP10D | tobacco-induced lung cancer | mRNA expression down | poor prognosis | [61] |
| ATP11A | metastasis of colorectal carcinoma | gene expression up | cancer association | [62] |
| colorectal carcinoma | gene methylation up | cancer association | [63] | |
| hepatocellular carcinoma (mouse model) | mRNA expression down | cancer progression | [64] | |
| pancreatic cancer | protein expression up | cancer association | [65] | |
| metastatic-lethal prostate cancer | gene methylation up | poor outcome | [66,67] | |
| acute lymphoblastic leukemia | non-coding RNA up | cancer association | [23] | |
| acute myeloid leukemia | gene methylation down | poor outcome | [68] | |
| ATP11B | ovarian carcinoma | protein expression up | cancer progression | [69] |
| ovarian carcinoma | gene expression down | tumor association | [70] | |
| breast cancer | protein expression down | poor prognosis | [71] | |
| lung adenocarcinoma | gene expression up | cancer association | [72] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yazlovitskaya, E.M.; Graham, T.R. Type IV P-Type ATPases: Recent Updates in Cancer Development, Progression, and Treatment. Cancers 2023, 15, 4327. https://doi.org/10.3390/cancers15174327
Yazlovitskaya EM, Graham TR. Type IV P-Type ATPases: Recent Updates in Cancer Development, Progression, and Treatment. Cancers. 2023; 15(17):4327. https://doi.org/10.3390/cancers15174327
Chicago/Turabian StyleYazlovitskaya, Eugenia M., and Todd R. Graham. 2023. "Type IV P-Type ATPases: Recent Updates in Cancer Development, Progression, and Treatment" Cancers 15, no. 17: 4327. https://doi.org/10.3390/cancers15174327
APA StyleYazlovitskaya, E. M., & Graham, T. R. (2023). Type IV P-Type ATPases: Recent Updates in Cancer Development, Progression, and Treatment. Cancers, 15(17), 4327. https://doi.org/10.3390/cancers15174327





