Circulating Tumour Cell Associated MicroRNA Profiles Change during Chemoradiation and Are Predictive of Response in Locally Advanced Rectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection and Sample Time-Points
2.2. TRG Scoring
2.3. CTC Sample Processing
2.4. miRNA Candidate Selection
2.5. RNA Extraction and miRNA OpenArray Profiling
2.6. Data Analyses
3. Results
3.1. CTC Enumeration
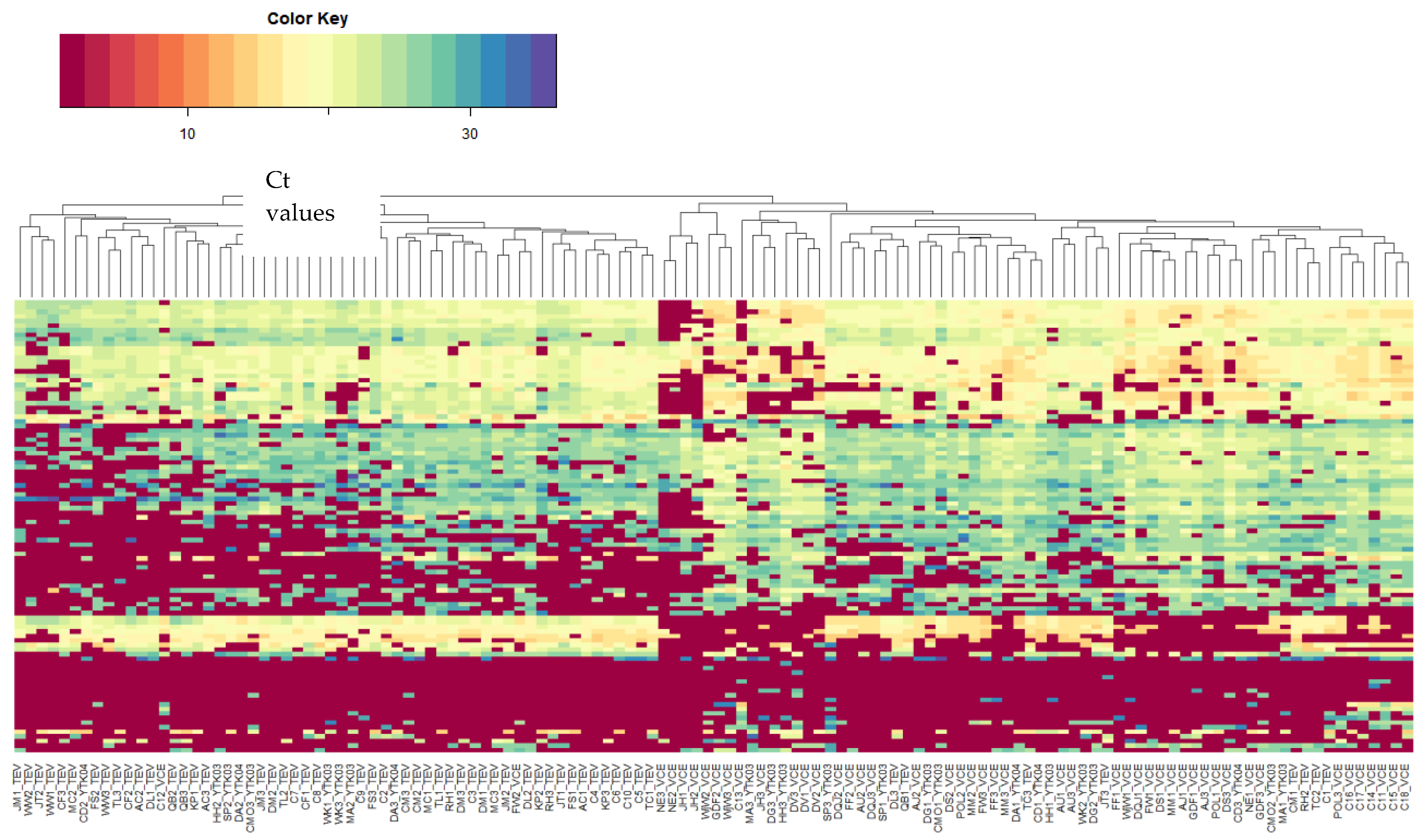
3.2. miRNA Candidate Selection and miRNA Expression Profiling
3.3. Change in CTC and Lymphocyte miRNA Expression during Neoadjuvantchemoradiation
3.4. Correlation of miRNA Expression with Tumour Response
3.5. Correlation of miRNA Changes with Tumour Response
3.6. Network Analysis and Gene Ontology Annotation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sauer, R.; Liersch, T.; Merkel, S.; Fietkau, R.; Hohenberger, W.; Hess, C.; Becker, H.; Raab, H.R.; Villanueva, M.T.; Witzigmann, H.; et al. Preoperative versus postoperative chemoradiotherapy for locally advanced rectal cancer: Results of the German CAO/ARO/AIO-94 randomized phase III trial after a median follow-up of 11 years. J. Clin. Oncol. 2012, 30, 1926–1933. [Google Scholar] [CrossRef] [PubMed]
- Conroy, T.; Bosset, J.F.; Etienne, P.L.; Rio, E.; Francois, E.; Mesgouez-Nebout, N.; Vendrely, V.; Artignan, X.; Bouche, O.; Gargot, D.; et al. Neoadjuvant chemotherapy with FOLFIRINOX and preoperative chemoradiotherapy for patients with locally advanced rectal cancer (UNICANCER-PRODIGE 23): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2021, 22, 702–715. [Google Scholar] [PubMed]
- Garcia-Aguilar, J.; Patil, S.; Gollub, M.J.; Kim, J.K.; Yuval, J.B.; Thompson, H.M.; Verheij, F.S.; Omer, D.M.; Lee, M.; Dunne, R.F.; et al. Organ Preservation in Patients With Rectal Adenocarcinoma Treated With Total Neoadjuvant Therapy. J. Clin. Oncol. 2022, 40, 2546–2556. [Google Scholar] [CrossRef] [PubMed]
- Fokas, E.; Liersch, T.; Fietkau, R.; Hohenberger, W.; Beissbarth, T.; Hess, C.; Becker, H.; Ghadimi, M.; Mrak, K.; Merkel, S.; et al. Tumor Regression Grading After Preoperative Chemoradiotherapy for Locally Advanced Rectal Carcinoma Revisited: Updated Results of the CAO/ARO/AIO-94 Trial. J. Clin. Oncol. 2014, 32, 1554–1562. [Google Scholar] [CrossRef]
- Parkinson, D.R.; Dracopoli, N.; Petty, B.G.; Compton, C.; Cristofanilli, M.; Deisseroth, A.; Hayes, D.F.; Kapke, G.; Kumar, P.; Lee, J.; et al. Considerations in the development of circulating tumor cell technology for clinical use. J. Transl. Med. 2012, 10, 138. [Google Scholar] [CrossRef] [PubMed]
- Azizian, A.; Gruber, J.; Ghadimi, B.M.; Gaedcke, J. MicroRNA in rectal cancer. World J. Gastrointest. Oncol. 2016, 8, 416–426. [Google Scholar] [CrossRef]
- Gaedcke, J.; Grade, M.; Camps, J.; Sokilde, R.; Kaczkowski, B.; Schetter, A.J.; Difilippantonio, M.J.; Harris, C.C.; Ghadimi, B.M.; Moller, S.; et al. The rectal cancer microRNAome–microRNA expression in rectal cancer and matched normal mucosa. Clin. Cancer Res. 2012, 18, 4919–4930. [Google Scholar]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.; Gibbons, D.; Hyland, J.M.; Treanor, D.; White, A.; Mulcahy, H.E.; O’Donoghue, D.P.; Moriarty, M.; Fennelly, D.; Sheahan, K. Pathological response following long-course neoadjuvant chemoradiotherapy for locally advanced rectal cancer. Histopathology 2005, 47, 141–146. [Google Scholar] [CrossRef]
- Crt, a Relative Threshold Method for qPCR Data Analysis on the QuantStudio™ 12K Flex System with OpenArray® Technology; Applied Biosystems; Thermo Fisher Scientific Inc.: Waltham, MA, USA, 2014.
- Raftery, A.; Hoeting, J.; Volinsky, C.; Painter, I.; Yeung, K. BMA: Bayesian Model Averaging. R package version 3.18.12. 2020. Available online: https://cran.r-project.org/web/packages/BMA/BMA.pdf (accessed on 22 June 2023).
- Dvinge, H.; Bertone, P. HTqPCR: High-throughput analysis and visualization of quantitative real-time PCR data in R. Bioinformatics 2009, 25, 3325–3326. [Google Scholar] [CrossRef]
- R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022.
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef]
- Ru, Y.; Kechris, K.J.; Tabakoff, B.; Hoffman, P.; Radcliffe, R.A.; Bowler, R.; Mahaffey, S.; Rossi, S.; Calin, G.A.; Bemis, L.; et al. The multiMiR R package and database: Integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 2014, 42, e133. [Google Scholar] [CrossRef] [PubMed]
- Novotna, E.; Bukum, N.; Hofman, J.; Flaxova, M.; Kouklikova, E.; Louvarova, D.; Wsol, V. Aldo-keto reductase 1C3 (AKR1C3): A missing piece of the puzzle in the dinaciclib interaction profile. Arch. Toxicol. 2018, 92, 2845–2857. [Google Scholar] [CrossRef] [PubMed]
- High Sensitivity Immunomagnetic CTC Isolation as Compared to Alternative Isolation Methods; Fluxion Biosciences: Oakland, CA, USA, 2013.
- Baek, D.H.; Kim, G.H.; Song, G.A.; Han, I.S.; Park, E.Y.; Kim, H.S.; Jo, H.J.; Ko, S.H.; Park, D.Y.; Cho, Y.K. Clinical Potential of Circulating Tumor Cells in Colorectal Cancer: A Prospective Study. Clin. Transl. Gastroenterol. 2019, 10, e00055. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Li, G.; Wan, J.; Zhu, J.; Shen, W.; Zhang, Z. Circulating tumor cells: A promising marker of predicting tumor response in rectal cancer patients receiving neoadjuvant chemo-radiation therapy. Oncotarget 2016, 7, 69507–69517. [Google Scholar] [CrossRef]
- Galizia, G.; Gemei, M.; Orditura, M.; Romano, C.; Zamboli, A.; Castellano, P.; Mabilia, A.; Auricchio, A.; De Vita, F.; Del Vecchio, L.; et al. Postoperative detection of circulating tumor cells predicts tumor recurrence in colorectal cancer patients. J. Gastrointest. Surg. 2013, 17, 1809–1818. [Google Scholar] [CrossRef]
- Hinz, S.; Roder, C.; Tepel, J.; Hendricks, A.; Schafmayer, C.; Becker, T.; Kalthoff, H. Cytokeratin 20 positive circulating tumor cells are a marker for response after neoadjuvant chemoradiation but not for prognosis in patients with rectal cancer. BMC Cancer 2015, 15, 953. [Google Scholar] [CrossRef]
- Kienle, P.; Koch, M.; Autschbach, F.; Benner, A.; Treiber, M.; Wannenmacher, M.; von Knebel Doeberitz, M.; Buchler, M.; Herfarth, C.; Weitz, J. Decreased detection rate of disseminated tumor cells of rectal cancer patients after preoperative chemoradiation: A first step towards a molecular surrogate marker for neoadjuvant treatment in colorectal cancer. Ann. Surg. 2003, 238, 324–330; discussion 330–331. [Google Scholar] [CrossRef]
- Magni, E.; Botteri, E.; Ravenda, P.S.; Cassatella, M.C.; Bertani, E.; Chiappa, A.; Luca, F.; Zorzino, L.; Bianchi, P.P.; Adamoli, L.; et al. Detection of circulating tumor cells in patients with locally advanced rectal cancer undergoing neoadjuvant therapy followed by curative surgery. Int. J. Colorectal Dis. 2014, 29, 1053–1059. [Google Scholar] [CrossRef]
- Bork, U.; Rahbari, N.N.; Scholch, S.; Reissfelder, C.; Kahlert, C.; Buchler, M.W.; Weitz, J.; Koch, M. Circulating tumour cells and outcome in non-metastatic colorectal cancer: A prospective study. Br. J. Cancer 2015, 112, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.H.; Spring, K.J.; de Souza, P.; MacKenzie, S.; Bokey, L. Circulating tumour cells and circulating nucleic acids as a measure of tumour dissemination in non-metastatic colorectal cancer surgery. Eur. J. Surg. Oncol. 2015, 41, 309–314. [Google Scholar]
- Huang, Z.; Huang, S.; Wang, Q.; Liang, L.; Ni, S.; Wang, L.; Sheng, W.; He, X.; Du, X. MicroRNA-95 promotes cell proliferation and targets sorting Nexin 1 in human colorectal carcinoma. Cancer Res. 2011, 71, 2582–2589. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.G.; Huang, Z.P.; Liu, Q.Z.; Ji-Fu, E.; Gao, X.H.; Xin, C.; Zhang, W.; Li, P.; Hao, L.Q. MicroRNA-95–3p inhibits cell proliferation and metastasis in colorectal carcinoma by HDGF. Biomed. J. 2020, 43, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, G.; Luo, F.; Ruan, J.; Huang, D.; Feng, D.; Xiao, D.; Zeng, Z.; Chen, X.; Wu, W. Identification of aberrantly expressed miRNAs in rectal cancer. Oncol. Rep. 2012, 28, 77–84. [Google Scholar]
- Kheirelseid, E.A.; Miller, N.; Chang, K.H.; Curran, C.; Hennessey, E.; Sheehan, M.; Newell, J.; Lemetre, C.; Balls, G.; Kerin, M.J. miRNA expressions in rectal cancer as predictors of response to neoadjuvant chemoradiation therapy. Int. J. Colorectal Dis. 2013, 28, 247–260. [Google Scholar] [CrossRef]
- arcia-Foncillas, J. Low MicroRNA-19b Expression Shows a Promising Clinical Impact in Locally Advanced Rectal Cancer. Cancers 2021, 13, 1456. [Google Scholar]
- Santos, A.; Cristobal, I.; Rubio, J.; Carames, C.; Luque, M.; Sanz-Alvarez, M.; Zazo, S.; Madoz-Gurpide, J.; Rojo, F.; Garcia-Foncillas, J. MicroRNA-19b Plays a Key Role in 5-Fluorouracil Resistance and Predicts Tumor Progression in Locally Advanced Rectal Cancer Patients. Int. J. Mol. Sci. 2022, 23, 12447. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Z.; Han, X.; Jiang, L.; Ge, R.; Wang, X.; Li, J. Up-regulation of microRNA-19b is associated with metastasis and predicts poor prognosis in patients with colorectal cancer. Int. J. Clin. Exp. Pathol. 2018, 11, 3952–3960. [Google Scholar]
- Sun, T.; Yin, Y.F.; Jin, H.G.; Liu, H.R.; Tian, W.C. Exosomal microRNA-19b targets FBXW7 to promote colorectal cancer stem cell stemness and induce resistance to radiotherapy. Kaohsiung J. Med. Sci. 2022, 38, 108–119. [Google Scholar] [CrossRef]
- Huang, L.; Cai, J.L.; Huang, P.Z.; Kang, L.; Huang, M.J.; Wang, L.; Wang, J.P. miR19b-3p promotes the growth and metastasis of colorectal cancer via directly targeting ITGB8. Am. J. Cancer Res. 2017, 7, 1996–2008. [Google Scholar] [PubMed]
- Della Vittoria Scarpati, G.; Falcetta, F.; Carlomagno, C.; Ubezio, P.; Marchini, S.; De Stefano, A.; Singh, V.K.; D’Incalci, M.; De Placido, S.; Pepe, S. A specific miRNA signature correlates with complete pathological response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Int. J. Radiat. Oncol. Biol. Phys. 2012, 83, 1113–1119. [Google Scholar]
- Zhu, Y.; Peng, Q.; Lin, Y.; Zou, L.; Shen, P.; Chen, F.; Min, M.; Shen, L.; Chen, J.; Shen, B. Identification of biomarker microRNAs for predicting the response of colorectal cancer to neoadjuvant chemoradiotherapy based on microRNA regulatory network. Oncotarget 2017, 8, 2233–2248. [Google Scholar]
- Cui, H.; Liu, Y.; Jiang, J.; Liu, Y.; Yang, Z.; Wu, S.; Cao, W.; Cui, I.H.; Yu, C. IGF2-derived miR-483 mediated oncofunction by suppressing DLC-1 and associated with colorectal cancer. Oncotarget 2016, 7, 48456–48466.39. [Google Scholar] [CrossRef] [PubMed]
- Bandres, E.; Arias, F.; Guerrero, D.; Lopez, I.; Gonzalez-Huarriz, M.; Dorronsoro, M.L.G.; Montes, M.; Monzon, F.; Torrea, N.; Armendariz, P.; et al. Association between a specific miRNA signature and pathological response to neoadjuvant chemoradiotherapy (CRT) in locally advanced rectal cancer (LARC) patients. J. Clin. Oncol. 2012, 30, e14057. [Google Scholar] [CrossRef]
- D’angelo, E.; Fassan, M.; Maretto, I.; Pucciarelli, S.; Zanon, C.; Digito, M.; Rugge, M.; Nitti, D.; Agostini, M. Serum miR-125b is a non-invasive predictive biomarker of the pre-operative chemoradiotherapy responsiveness in patients with rectal adenocarcinoma. Oncotarget 2016, 7, 28647–28657. [Google Scholar] [CrossRef]
- Drebber, U.; Lay, M.; Wedemeyer, I.; Vallböhmer, D.; Bollschweiler, E.; Brabender, J.; Mönig, S.P.; Hölscher, A.H.; Dienes, H.P.; Odenthal, M. Altered levels of the onco-microRNA 21 and the tumor-supressor microRNAs 143 and 145 in advanced rectal cancer indicate successful neoadjuvant chemoradiotherapy. Int. J. Oncol. 2011, 39, 409–415. [Google Scholar] [CrossRef]
- Hotchi, M.; Shimada, M.; Kurita, N.; Iwata, T.; Sato, H.; Morimoto, S.; Yoshikawa, K.; Higashijima, J.; Miyatani, T. microRNA expression is able to predict response to chemoradiotherapy in rectal cancer. Mol. Clin. Oncol. 2012, 1, 137–142. [Google Scholar] [CrossRef]
- Liu, F.; Liu, S.; Ai, F.; Zhang, D.; Xiao, Z.; Nie, X.; Fu, Y. miR-107 Promotes Proliferation and Inhibits Apoptosis of Colon Cancer Cells by Targeting Prostate Apoptosis Response-4 (Par4). Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 967–974. [Google Scholar] [CrossRef]
- Metheetrairut, C.; Slack, F.J. MicroRNAs in the ionizing radiation response and in radiotherapy. Curr. Opin. Genet. Dev. 2013, 23, 12–19. [Google Scholar] [CrossRef]
- Millino, C.; Maretto, I.; Pacchioni, B.; Digito, M.; De Paoli, A.; Canzonieri, V.; D’Angelo, E.; Agostini, M.; Rizzolio, F.; Giordano, A.; et al. Gene and MicroRNA Expression Are Predictive of Tumor Response in Rectal Adenocarcinoma Patients Treated With Preoperative Chemoradiotherapy. J. Cell. Physiol. 2016, 232, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Motoyama, K.; Inoue, H.; Takatsuno, Y.; Tanaka, F.; Mimori, K.; Uetake, H.; Sugihara, K.; Mori, M. Over- and under-expressed microRNAs in human colorectal cancer. Int. J. Oncol. 2009, 34, 1069–1075. [Google Scholar] [PubMed]
- O Ng, E.K.; Chong, W.W.S.; Jin, H.; Lam, E.K.Y.; Shin, V.Y.; Yu, J.; Poon, T.C.W.; Ng, S.S.M.; Sung, J.J.Y. Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut 2009, 58, 1375–1381. [Google Scholar] [CrossRef] [PubMed]
- Pichler, M.; Stiegelbauer, V.; Vychytilova-Faltejskova, P.; Ivan, C.; Ling, H.; Winter, E.; Zhang, X.; Goblirsch, M.; Wulf-Goldenberg, A.; Ohtsuka, M.; et al. Genome-Wide miRNA Analysis Identifies miR-188-3p as a Novel Prognostic Marker and Molecular Factor Involved in Colorectal Carcinogenesis. Clin. Cancer Res. 2017, 23, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Pu, X.-X.; Huang, G.-L.; Guo, H.-Q.; Guo, C.-C.; Li, H.; Ye, S.; Ling, S.; Jiang, L.; Tian, Y.; Lin, T.-Y. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J. Gastroenterol. Hepatol. 2010, 25, 1674–1680. [Google Scholar] [CrossRef]
- Salendo, J.; Spitzner, M.; Kramer, F.; Zhang, X.; Jo, P.; Wolff, H.A.; Kitz, J.; Kaulfuß, S.; Beißbarth, T.; Dobbelstein, M.; et al. Identification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a, -224, -132 and let7g. Radiother. Oncol. 2013, 108, 451–457. [Google Scholar] [CrossRef]
- Song, Y.; Xu, Y.; Wang, Z.; Chen, Y.; Yue, Z.; Gao, P.; Xing, C.; Xu, H. MicroRNA-148b suppresses cell growth by targeting cholecystokinin-2 receptor in colorectal cancer. Int. J. Cancer 2011, 131, 1042–1051. [Google Scholar] [CrossRef]
- Svoboda, M.; Sana, J.; Fabian, P.; Kocakova, I.; Gombosova, J.; Nekvindova, J.; Radova, L.; Vyzula, R.; Slaby, O. MicroRNA expression profile associated with response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer patients. Radiat. Oncol. 2012, 7, 1–7. [Google Scholar] [CrossRef]
- Wang, S.; Xiang, J.; Li, Z.; Lu, S.; Hu, J.; Gao, X.; Yu, L.; Wang, L.; Wang, J.; Wu, Y.; et al. A plasma microRNA panel for early detection of colorectal cancer. Int. J. Cancer 2013, 136, 152–161. [Google Scholar] [CrossRef]





| Patient and Tumour Characteristics | n | Percent (%) |
|---|---|---|
| Age | ||
| ˂65 | 30 | 58 |
| ≥65 | 22 | 42 |
| Gender | ||
| Male | 38 | 73 |
| Female | 14 | 27 |
| Ethnicity | ||
| Caucasian | 37 | 71 |
| Asian | 15 | 29 |
| Tumour stage * | ||
| T2 | 5 | 10 |
| T3 | 42 | 80 |
| T4 | 5 | 10 |
| Nodal stage * | ||
| N0 | 8 | 15 |
| N1 | 20 | 38 |
| N2 | 24 | 47 |
| Smoking status | ||
| Yes | 22 | 42 |
| No | 30 | 58 |
| ECOG performance status | ||
| 0 | 37 | 71 |
| 1 | 15 | 29 |
| Tumour regression grade ^ | ||
| 0 | 9 | 18 |
| 1 | 15 | 31 |
| 2 | 21 | 43 |
| 3 | 4 | 8 |
| Recurrence | ||
| Yes | 11 | 21 |
| No | 41 | 79 |
| miRNA | Log2 Fold Change | Adjusted p-Value |
|---|---|---|
| Baseline and during treatment | ||
| hsa-miR-95 | 12.65 | 0.00031 |
| hsa-miR-10a | 9.94 | 0.0045 |
| hsa-miR-16-1* | 9.46 | 0.0087 |
| hsa-miR-210 | 9.31 | 0.0045 |
| hsa-miR-194 | 9.086 | 0.016 |
| hsa-miR-101 | 8.76 | 0.038 |
| hsa-miR-29b | 8.56 | 0.0046 |
| hsa-miR-19b | −4.60 | 0.016 |
| hsa-miR-342−3p | −4.55 | 0.021 |
| hsa-miR-16 | −4.19 | 0.014 |
| hsa-miR-92a | −3.25 | 0.034 |
| hsa-miR-26b | −2.88 | 0.038 |
| During treatment and post-treatment | ||
| hsa-let-7e | 4.32 | 0.020 |
| hsa-miR-95 | −12.03 | 0.00086 |
| hsa-miR-10a | −9.40 | 0.010 |
| hsa-miR-16-1* | −8.67 | 0.020 |
| hsa-miR-200b | −7.95 | 0.010 |
| hsa-miR-127 | −4.45 | 0.020 |
| p! = 0 | EV | SD | Model 1 | Model 2 | Model 3 | Model 4 | Model 5 | |
|---|---|---|---|---|---|---|---|---|
| Intercept | 100 | 6.97 | 5.094072 | 9.30181 | 3.34419 | 2.89069 | 4.60466 | 12.00436 |
| * hsa-miR-483-5p | 100 | −1.21 × 10−1 | 0.035075 | −0.12257 | −0.12391 | −0.11371 | −0.12577 | −0.12061 |
| hsa-miR-10a | 4.5 | 1.65 × 10−3 | 0.012055 | |||||
| hsa-miR-16 | 4 | −1.96 × 10−3 | 0.047242 | |||||
| * hsa-miR-19b | 82.7 | −3.65 × 10−1 | 0.286371 | −0.36668 | −0.6098 | −0.57137 | −0.32431 | |
| hsa-miR-26b | 10.5 | 3.48 × 10−2 | 0.147071 | 0.40815 | ||||
| hsa-miR-29b | 5.9 | −2.25 × 10−3 | 0.013898 | |||||
| hsa-miR-92a | 19.1 | 9.45 × 10−2 | 0.232413 | 0.50124 | ||||
| hsa-miR-95 | 7.9 | 4.54 × 10−3 | 0.021901 | |||||
| hsa-miR-127 | 11.7 | −1.12 × 10−2 | 0.039798 | −0.09133 | ||||
| hsa-miR-194 | 2.5 | −4.01 × 10−4 | 0.005315 | |||||
| hsa-miR-210 | 2.2 | 7.53 × 10−5 | 0.004704 | |||||
| hsa-miR-200b | 2.2 | −3.92 × 10−4 | 0.013972 | |||||
| hsa-miR-101 | 2.3 | 4.21 × 10−4 | 0.009652 | |||||
| hsa-miR-342-3p | 2.2 | −2.66 × 10−4 | 0.012814 | |||||
| hsa-let-7e | 3.4 | −4.97 × 10−4 | 0.014303 | |||||
| hsa-miR-16-1 | 2.2 | 9.23 × 10−5 | 0.00867 | |||||
| nVar | 2 | 3 | 1 | 3 | 3 | |||
| BIC | −353.513 | −352.046 | −351.483 | −350.8 | −350.573 | |||
| post prob | 0.222 | 0.107 | 0.08 | 0.057 | 0.051 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, S.H.; Chua, W.; Ng, W.; Ip, E.; Marques, T.M.; Tran, N.T.; Gama-Carvalho, M.; Asghari, R.; Henderson, C.; Ma, Y.; et al. Circulating Tumour Cell Associated MicroRNA Profiles Change during Chemoradiation and Are Predictive of Response in Locally Advanced Rectal Cancer. Cancers 2023, 15, 4184. https://doi.org/10.3390/cancers15164184
Lim SH, Chua W, Ng W, Ip E, Marques TM, Tran NT, Gama-Carvalho M, Asghari R, Henderson C, Ma Y, et al. Circulating Tumour Cell Associated MicroRNA Profiles Change during Chemoradiation and Are Predictive of Response in Locally Advanced Rectal Cancer. Cancers. 2023; 15(16):4184. https://doi.org/10.3390/cancers15164184
Chicago/Turabian StyleLim, Stephanie H., Wei Chua, Weng Ng, Emilia Ip, Tania M. Marques, Nham T. Tran, Margarida Gama-Carvalho, Ray Asghari, Christopher Henderson, Yafeng Ma, and et al. 2023. "Circulating Tumour Cell Associated MicroRNA Profiles Change during Chemoradiation and Are Predictive of Response in Locally Advanced Rectal Cancer" Cancers 15, no. 16: 4184. https://doi.org/10.3390/cancers15164184
APA StyleLim, S. H., Chua, W., Ng, W., Ip, E., Marques, T. M., Tran, N. T., Gama-Carvalho, M., Asghari, R., Henderson, C., Ma, Y., de Souza, P., & Spring, K. J. (2023). Circulating Tumour Cell Associated MicroRNA Profiles Change during Chemoradiation and Are Predictive of Response in Locally Advanced Rectal Cancer. Cancers, 15(16), 4184. https://doi.org/10.3390/cancers15164184







