Unraveling the Role of EV-Derived miR-150-5p in Prostate Cancer Metastasis and Its Association with High-Grade Gleason Scores: Implications for Diagnosis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue and Plasma Collection
2.2. Cell Culture and Conditioned Media Collection
2.3. Microarrays from Paraffin Tissues and Plasma Samples
2.4. Ultracentrifugation for EV Isolation
2.5. Aqueous Two-Phase Separation for EV Isolation
2.6. Western Blot for EV Markers
2.7. Electron Microscopy for EVs
2.8. Nanoscale Flow Cytometry
2.9. RT-qPCR for miRNA Validation
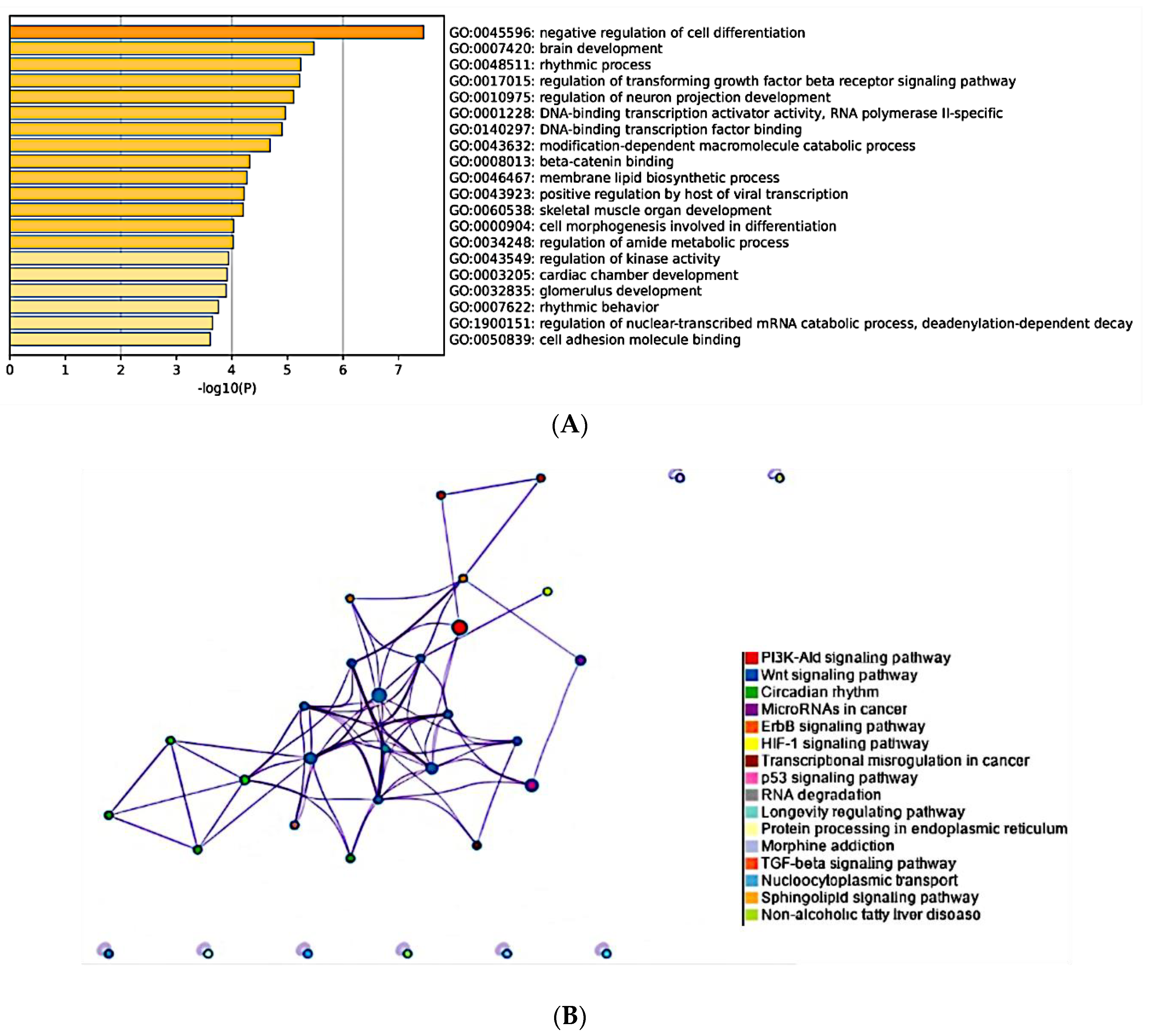
2.10. Prediction of Target Genes and Cellular Pathways of Differentially Expressed miRNAs
3. Results
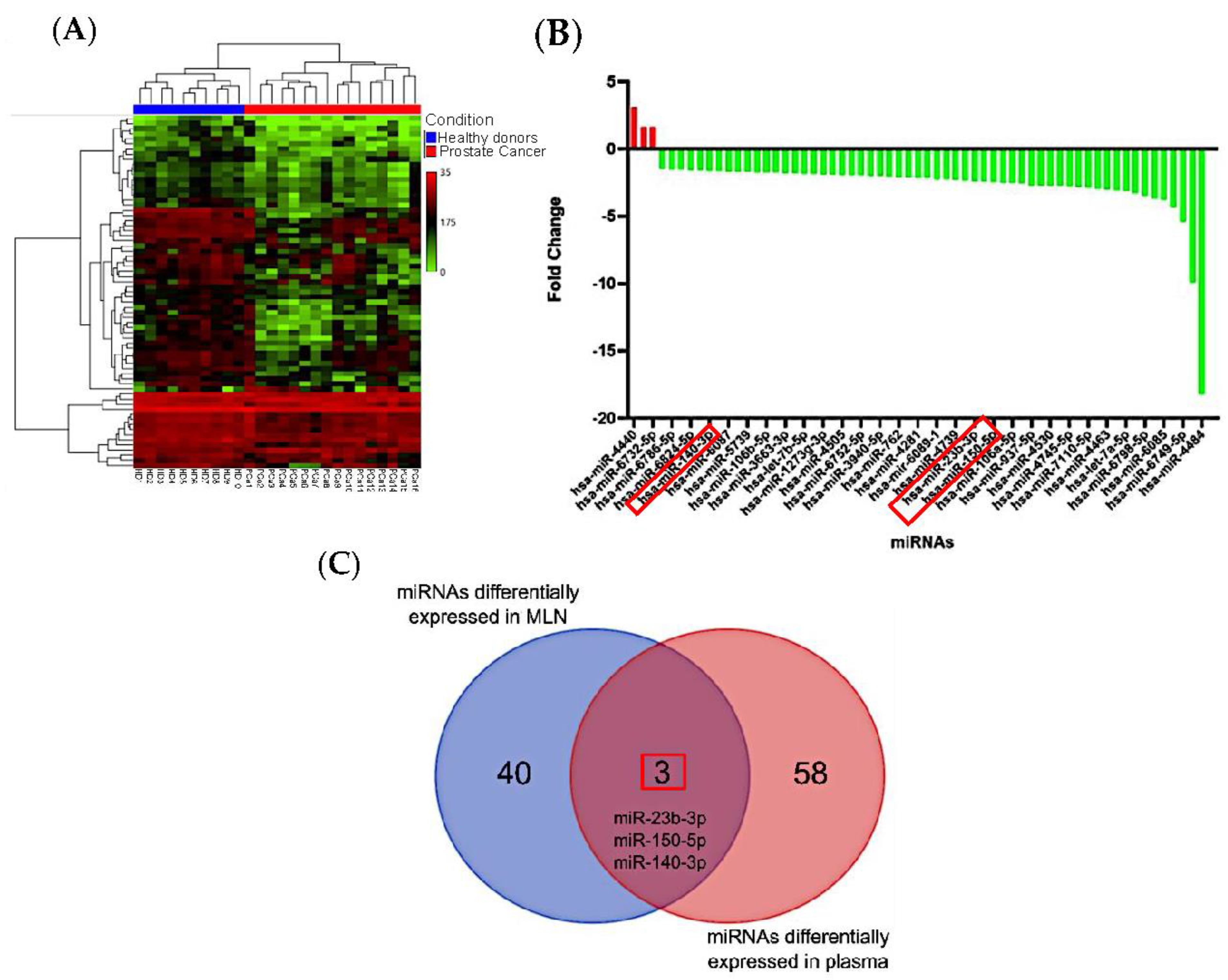
3.1. Differentially Expressed miRNAs in Metastatic Lymph Nodes
3.2. miRNAs from Metastatic Lymph Nodes Are Found in Plasma of PCa Patients
3.3. Validation of EVs Isolated from Conditioned Media
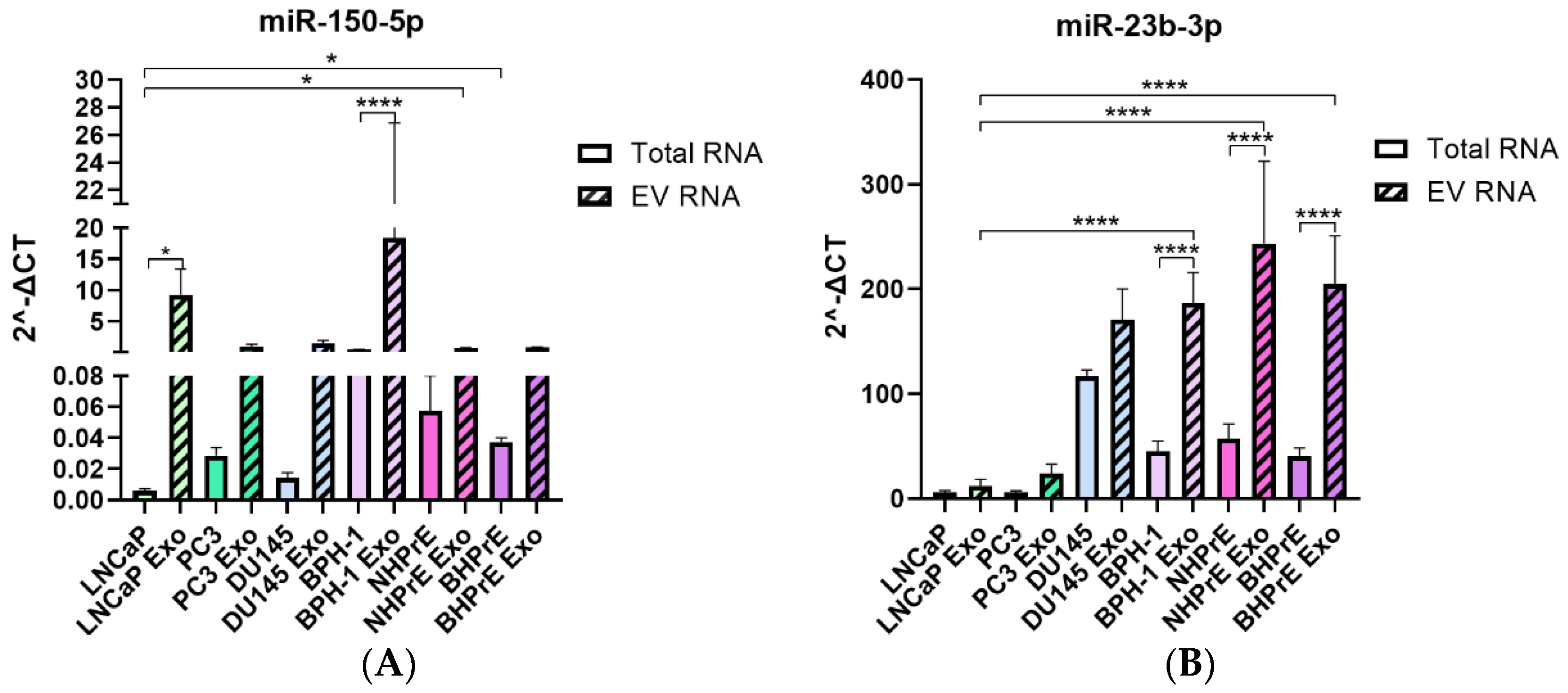
3.4. miRNAs Are Enriched in EVs from Tumoral and Non-Tumoral PCa Cell Lines
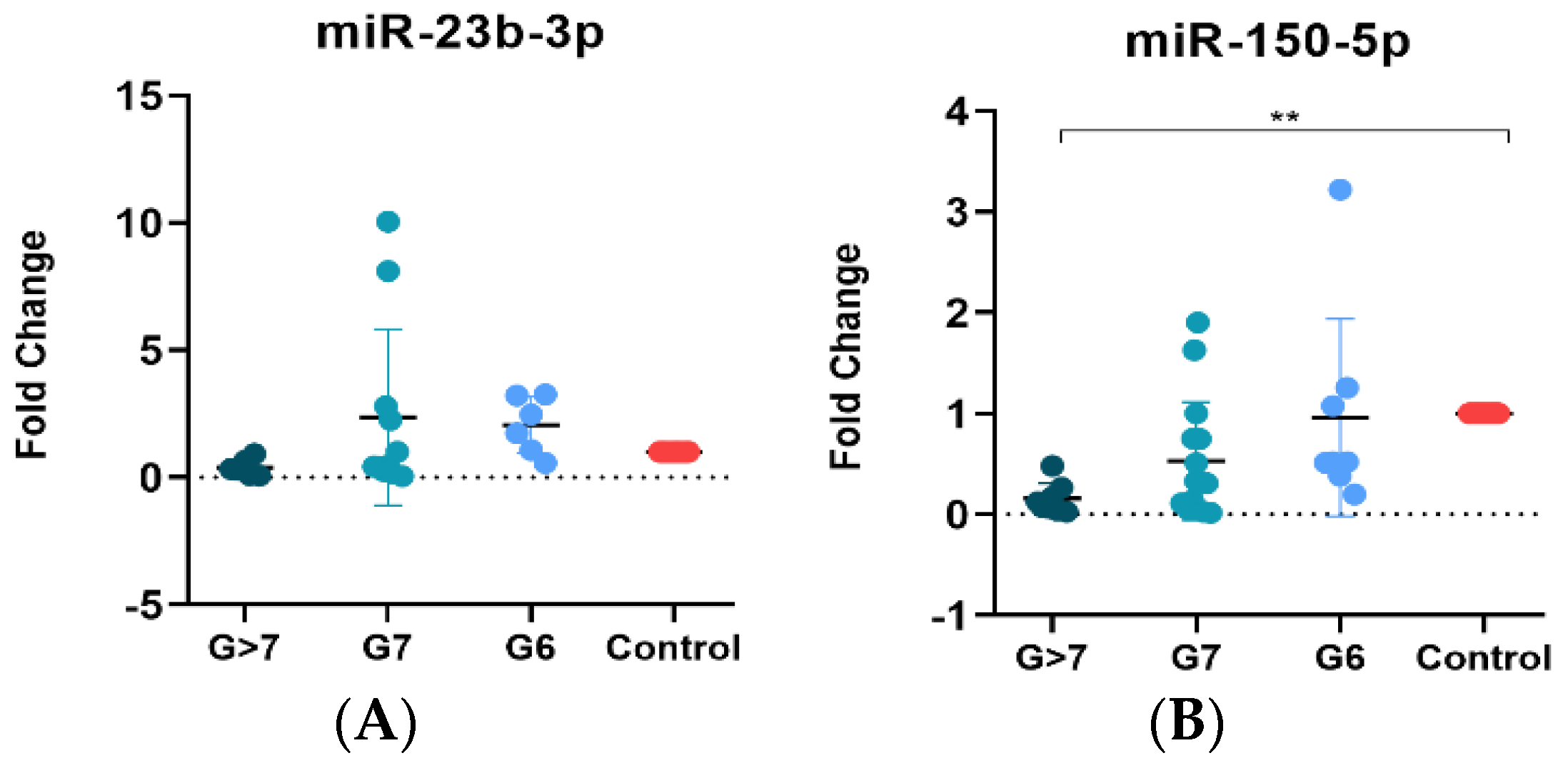
3.5. MiR-150-5p Is Associated with a High Gleason Score
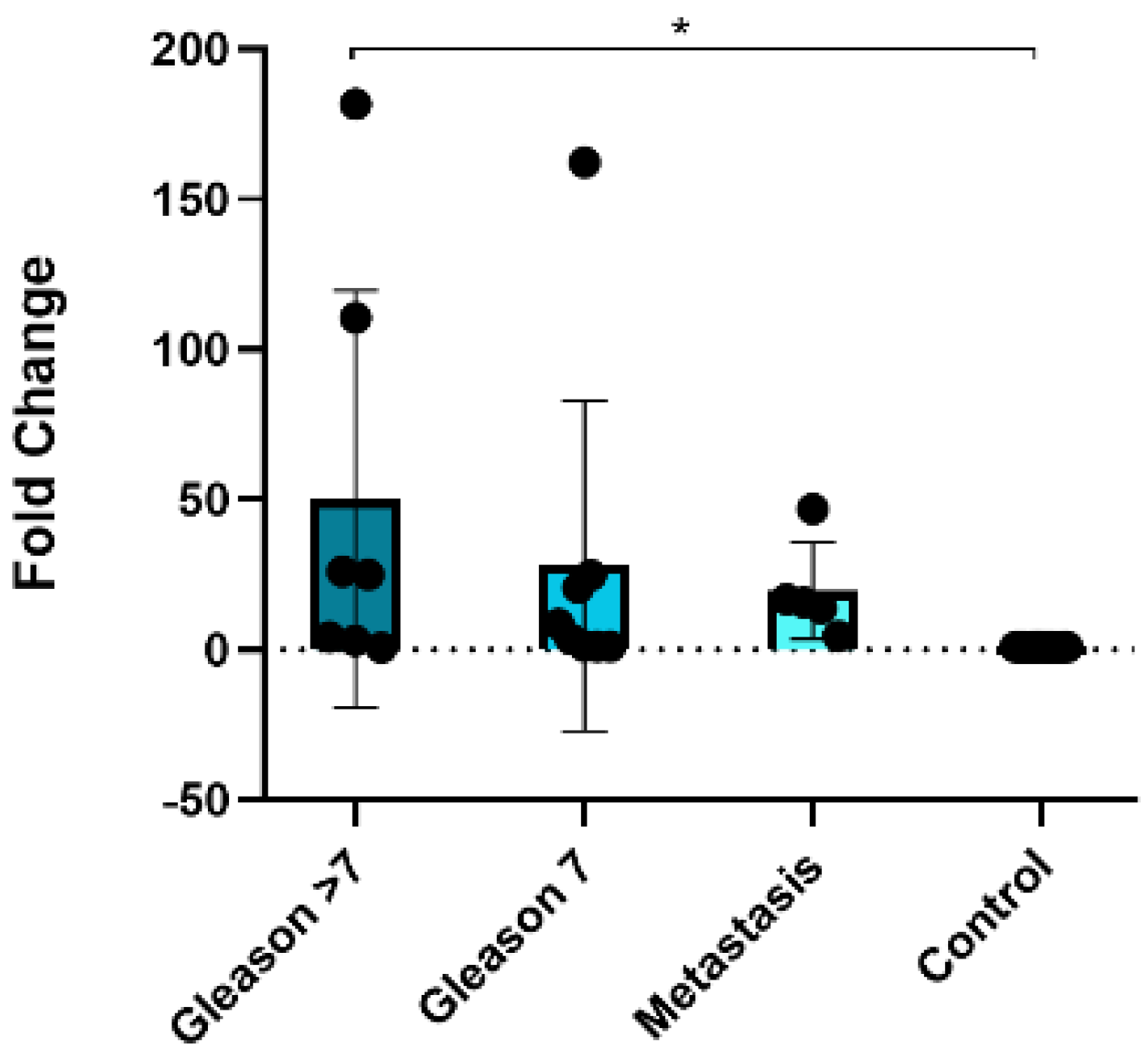
3.6. MiR-150-5p Is Potentially Associated with Bone Metastasis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Kilpeläinen, T.P.; Tammela, T.L.; Roobol, M.; Hugosson, J.; Ciatto, S.; Nelen, V.; Moss, S.; Määttänen, L.; Auvinen, A. False-positive screening results in the European randomized study of screening for prostate cancer. Eur. J. Cancer 2011, 47, 2698–2705. [Google Scholar] [CrossRef] [PubMed]
- Grossman, D.C.; Curry, S.J.; Owens, D.K.; Bibbins-Domingo, K.; Caughey, A.B.; Davidson, K.W.; Doubeni, C.A.; Ebell, M.; Epling, J.W.; Kemper, A.R.; et al. Screening for Prostate Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2018, 319, 1901–1913. [Google Scholar]
- Sekhoacha, M.; Riet, K.; Motloung, P.; Gumenku, L.; Adegoke, A.; Mashele, S. Prostate Cancer Review: Genetics, Diagnosis, Treatment Options, and Alternative Approaches. Molecules 2022, 27, 5730. [Google Scholar] [CrossRef]
- EAU. Prostate Cancer [Internet]. EAU Risk Groups of Biochemical Recurrence of Localised and Locally-Advanced Prostate Cancer. 2023. Available online: https://uroweb.org/guidelines/prostate-cancer/summary-of-changes (accessed on 11 August 2023).
- Schmanke, K.; Okut, H.; Ablah, E. Trends for Stage and Grade Group of Prostate Cancer in the US (2010–2016). Urology 2020, 149, 110–116. [Google Scholar] [CrossRef]
- Sorce, G.; Hoeh, B.; Flammia, R.S.; Chierigo, F.; Hohenhorst, L.; Panunzio, A.; Nimer, N.; Tian, Z.; Gandaglia, G.; Tilki, D.; et al. Rates of metastatic prostate cancer in newly diagnosed patients: Numbers needed to image according to risk level. Prostate 2022, 82, 1210–1218. [Google Scholar] [CrossRef]
- Landgraf, M.; Lahr, C.A.; Sanchez-Herrero, A.; Meinert, C.; Shokoohmand, A.; Pollock, P.M.; Hutmacher, D.W.; Shafiee, A.; McGovern, J.A. Humanized bone facilitates prostate cancer metastasis and recapitulates therapeutic effects of zoledronic acid in vivo. Bone Res. 2019, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Gulati, R.; Psutka, S.P.; Etzioni, R. Personalized Risks of Over Diagnosis for Screen Detected Prostate Cancer Incorporating Patient Comorbidities: Estimation and Communication. J. Urol. 2019, 202, 936–943. [Google Scholar] [CrossRef]
- Bubendorf, L.; Schöpfer, A.; Wagner, U.; Sauter, G.; Moch, H.; Willi, N.; Gasser, T.C.; Mihatsch, M.J. Metastatic patterns of prostate cancer: An autopsy study of 1,589 patients. Hum. Pathol. 2000, 31, 578–583. [Google Scholar] [CrossRef]
- Yu, J.; Du, W.; Yan, F.; Wang, Y.; Li, H.; Cao, S.; Yu, W.; Shen, C.; Liu, J.; Ren, X. Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J. Immunol. 2013, 190, 3783–3797. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Lai, Y.-S.; Chu, P.-Y.; Chan, S.-H.; Wang, L.-H.; Hung, W.-C. Cancer-Derived VEGF-C Increases Chemokine Production in Lymphatic Endothelial Cells to Promote CXCR2-Dependent Cancer Invasion and MDSC Recruitment. Cancers 2019, 11, 1120. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Deguchi, K.; Zheng, R.; Tamai, H.; Wang, L.-X.; Cohen, P.A.; Shu, S. Tumor-Induced CD11b+Gr-1+ Myeloid Cells Suppress T Cell Sensitization in Tumor-Draining Lymph Nodes. J. Immunol. 2008, 181, 3291–3300. [Google Scholar] [CrossRef] [PubMed]
- Cochran, A.J.; Huang, R.-R.; Su, A.; Itakura, E.; Wen, D.-R. Is sentinel node susceptibility to metastases related to nodal immune modulation? Cancer J. 2015, 21, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Stacker, S.A.; Williams, S.P.; Karnezis, T.; Shayan, R.; Fox, S.B.; Achen, M.G. Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nat. Rev. Cancer 2014, 14, 159–172. [Google Scholar] [CrossRef]
- Cho, J.-K.; Hyun, S.H.; Choi, N.; Kim, M.-J.; Padera, T.P.; Choi, J.Y.; Jeong, H.-S. Significance of Lymph Node Metastasis in Cancer Dissemination of Head and Neck Cancer. Transl. Oncol. 2015, 8, 119–125. [Google Scholar] [CrossRef]
- Akhtar, M.; Haider, A.; Rashid, S.; Al-Nabet, A.D.M.H. Paget’s “seed and Soil” Theory of Cancer Metastasis: An Idea Whose Time has Come. Adv. Anat. Pathol. 2019, 26, 69–74. [Google Scholar] [CrossRef]
- Pereira, E.R.; Kedrin, D.; Seano, G.; Gautier, O.; Meijer, E.F.J.; Jones, D.; Chin, S.M.; Kitahara, S.; Bouta, E.M.; Chang, J.; et al. Lymph node metastases can invade local blood vessels, exit the node, and colonize distant organs in mice. Science 2018, 359, 1403–1407. [Google Scholar] [CrossRef]
- Sleeman, J.P. The lymph node pre-metastatic niche. J. Mol. Med. 2015, 93, 1173–1184. [Google Scholar] [CrossRef]
- Peinado, H.; Zhang, H.; Matei, I.R.; Costa-Silva, B.; Hoshino, A.; Rodrigues, G.; Psaila, B.; Kaplan, R.N.; Bromberg, J.F.; Kang, Y.; et al. Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 2017, 17, 302–317. [Google Scholar] [CrossRef]
- Kaplan, R.N.; Riba, R.D.; Zacharoulis, S.; Bramley, A.H.; Vincent, L.; Costa, C.; MacDonald, D.D.; Jin, D.K.; Shido, K.; Kerns, S.A.; et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 2005, 438, 820–827. [Google Scholar] [CrossRef]
- Yu, D.; Li, Y.; Wang, M.; Gu, J.; Xu, W.; Cai, H.; Fang, X.; Zhang, X. Exosomes as a new frontier of cancer liquid biopsy. Mol. Cancer 2022, 21, 56. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Mazumdar, A.; Urdinez, J.; Boro, A.; Migliavacca, J.; Arlt, M.J.E.; Muff, R.; Fuchs, B.; Snedeker, J.G.; Gvozdenovic, A. Osteosarcoma-Derived Extracellular Vesicles Induce Lung Fibroblast Reprogramming. Int. J. Mol. Sci. 2020, 21, 5451. [Google Scholar] [CrossRef]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.F.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef]
- Zhao, Y.; Jin, L.-J.; Zhang, X.-Y. Exosomal miRNA-205 promotes breast cancer chemoresistance and tumorigenesis through E2F1. Aging 2021, 13, 18498–18514. [Google Scholar] [CrossRef]
- Wong, G.L.; Abu Jalboush, S.; Lo, H.-W. Exosomal MicroRNAs and Organotropism in Breast Cancer Metastasis. Cancers 2020, 12, 1827. [Google Scholar] [CrossRef]
- Ma, Y.-S.; Liu, J.-B.; Lin, L.; Zhang, H.; Wu, J.-J.; Shi, Y.; Jia, C.-Y.; Zhang, D.-D.; Yu, F.; Wang, H.-M.; et al. Exosomal microRNA-15a from mesenchymal stem cells impedes hepatocellular carcinoma progression via downregulation of SALL4. Cell Death Discov. 2021, 7, 224. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Li, Y.; Pan, Y.; Lan, X.; Song, F.; Sun, J.; Zhou, K.; Liu, X.; Ren, X.; Wang, F.; et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018, 9, 5395. [Google Scholar] [CrossRef]
- Yuan, X.; Qian, N.; Ling, S.; Li, Y.; Sun, W.; Li, J.; Du, R.; Zhong, G.; Liu, C.; Yu, G.; et al. Breast cancer exosomes contribute to pre-metastatic niche formation and promote bone metastasis of tumor cells. Theranostics 2021, 11, 1429–1445. [Google Scholar] [CrossRef]
- Fang, T.; Lv, H.; Lv, G.; Li, T.; Wang, C.; Han, Q.; Yu, L.; Su, B.; Guo, L.; Huang, S.; et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 2018, 9, 191. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Cui, Q. The relationship of human tissue microRNAs with those from body fluids. Sci. Rep. 2020, 10, 5644. [Google Scholar] [CrossRef] [PubMed]
- Azzalini, E.; De Martino, E.; Fattorini, P.; Canzonieri, V.; Stanta, G.; Bonin, S. Reliability of miRNA Analysis from Fixed and Paraffin-Embedded Tissues. Int. J. Mol. Sci. 2019, 20, 4819. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Xu, X. MicroRNA Isolation from Formalin-Fixed, Paraffin-Embedded Tissues. In Formalin-Fixed Paraffin-Embedded Tissues; Methods in Molecular Biology; Springer: Cham, Switzerland, 2011; Volume 724, pp. 259–267. [Google Scholar] [CrossRef]
- Hafron, J.M.; Yu, H.; Juang, A.; Vuong, D.; Kamer, S.; Carbonell, L.; Singh, S.; Reinhard, R.; Arthurs, J.; Ohrnberger, J.; et al. New developments in prostate cancer screening using a novel cancer-specific, non-PSA biomarker assay derived from autoantibody signatures. J. Med. Ther. 2017, 1, 192–203. [Google Scholar] [CrossRef]
- Aveta, A.; Cilio, S.; Contieri, R.; Spena, G.; Napolitano, L.; Manfredi, C.; Franco, A.; Crocerossa, F.; Cerrato, C.; Ferro, M.; et al. Urinary MicroRNAs as Biomarkers of Urological Cancers: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 10846. [Google Scholar] [CrossRef]
- Sun, G.; Sun, Y.; Zou, Z.; Xu, S. Analysis of Segmental Lymph Node Metastasis and Clinical Features in cT1N0M0 Lung Adenocarcinoma. BioMed Res. Int. 2020, 2020, 2842604. [Google Scholar] [CrossRef]
- To, B.; Isaac, D.; Andrechek, E.R. Studying Lymphatic Metastasis in Breast Cancer: Current Models, Strategies, and Clinical Perspectives. J. Mammary Gland. Biol. Neoplasia 2020, 25, 191–203. [Google Scholar] [CrossRef]
- Datta, K.; Muders, M.; Zhang, H.; Tindall, D.J.; Novelli, G.; Margiotti, K.; Chiocca, A.M.; Spera, E.; Micali, F.; Reichardt, J.K.; et al. Mechanism of Lymph Node Metastasis in Prostate Cancer. Futur. Oncol. 2010, 6, 823–836. Available online: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2892838/%5Cn (accessed on 6 March 2023). [CrossRef]
- Brown, M.; Assen, F.P.; Leithner, A.; Abe, J.; Schachner, H.; Asfour, G.; Bago-Horvath, Z.; Stein, J.V.; Uhrin, P.; Sixt, M.; et al. Lymph node blood vessels provide exit routes for metastatic tumor cell dissemination in mice. Science 2018, 359, 1408–1411. [Google Scholar] [CrossRef]
- Jo, P.; Azizian, A.; Salendo, J.; Kramer, F.; Bernhardt, M.; Wolff, H.A.; Gruber, J.; Grade, M.; Beißbarth, T.; Ghadimi, B.M.; et al. Changes of Microrna Levels in Plasma of Patients with Rectal Cancer during Chemoradiotherapy. Int. J. Mol. Sci. 2017, 18, 1140. [Google Scholar] [CrossRef]
- Pigati, L.; Yaddanapudi, S.C.S.; Iyengar, R.; Kim, D.-J.; Hearn, S.A.; Danforth, D.; Hastings, M.L.; Duelli, D.M. Selective Release of MicroRNA Species from Normal and Malignant Mammary Epithelial Cells. PLoS ONE 2010, 5, e13515. [Google Scholar] [CrossRef] [PubMed]
- Wulfken, L.M.; Moritz, R.; Ohlmann, C.; Holdenrieder, S.; Jung, V.; Becker, F.; Herrmann, E.; Walgenbach-Brünagel, G.; von Ruecker, A.; Müller, S.C.; et al. MicroRNAs in Renal Cell Carcinoma: Diagnostic Implications of Serum miR-1233 Levels. PLoS ONE 2011, 6, e25787. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, M.; Wang, M.; Yan, D.; Feng, G.; An, G. The expression of microRNA-375 in plasma and tissue is matched in human colorectal cancer. BMC Cancer 2014, 14, 714. [Google Scholar] [CrossRef]
- Wang, S.; Yin, J.; Li, T.; Yuan, L.; Wang, D.; He, J.; Du, X.; Lu, J. Upregulated circulating miR-150 is associated with the risk of intrahepatic cholangiocarcinoma. Oncol. Rep. 2014, 33, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.J.; Martinet, L.; Gilfillan, S.; Souza-Fonseca-Guimaraes, F.; Chow, M.T.; Town, L.; Ritchie, D.S.; Colonna, M.; Andrews, D.M.; Smyth, M.J. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat. Immunol. 2014, 15, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Wang, R.; Xie, H.; Hu, L.; Wang, C.; Xu, J.; Zhu, C.; Liu, Y.; Gao, F.; Li, X.; et al. Single-cell RNA sequencing reveals cell heterogeneity and transcriptome profile of breast cancer lymph node metastasis. Oncogenesis 2021, 10, 66. [Google Scholar] [CrossRef]
- Dong, Q.; Liu, X.; Cheng, K.; Sheng, J.; Kong, J.; Liu, T. Pre-metastatic Niche Formation in Different Organs Induced by Tumor Extracellular Vesicles. Front. Cell Dev. Biol. 2021, 9, 733627. [Google Scholar] [CrossRef]
- Zhao, J.; Schlößer, H.A.; Wang, Z.; Qin, J.; Li, J.; Popp, F.; Popp, M.C.; Alakus, H.; Chon, S.-H.; Hansen, H.P.; et al. Tumor-derived extracellular vesicles inhibit natural killer cell function in pancreatic cancer. Cancers 2019, 11, 874. [Google Scholar] [CrossRef]
- Ji, Q.; Zhou, L.; Sui, H.; Yang, L.; Wu, X.; Song, Q.; Jia, R.; Li, R.; Sun, J.; Wang, Z.; et al. Primary tumors release ITGBL1-rich extracellular vesicles to promote distal metastatic tumor growth through fibroblast-niche formation. Nat. Commun. 2020, 11, 1211. [Google Scholar] [CrossRef]
- Maus, R.L.; Jakub, J.W.; Hieken, T.J.; Nevala, W.K.; Christensen, T.A.; Sutor, S.L.; Flotte, T.J.; Markovic, S.N. Identification of novel, immune-mediating extracellular vesicles in human lymphatic effluent draining primary cutaneous melanoma. Oncoimmunology 2019, 8, e1667742. [Google Scholar] [CrossRef] [PubMed]
- Broggi, M.A.; Maillat, L.; Clement, C.C.; Bordry, N.; Corthésy, P.; Auger, A.; Matter, M.; Hamelin, R.; Potin, L.; Demurtas, D.; et al. Tumor-associated factors are enriched in lymphatic exudate compared to plasma in metastatic melanoma patients. J. Exp. Med. 2019, 216, 1091–1107. [Google Scholar] [CrossRef] [PubMed]
- Jankovičová, J.; Sečová, P.; Michalková, K.; Antalíková, J. Tetraspanins, More than Markers of Extracellular Vesicles in Reproduction. Int. J. Mol. Sci. 2020, 21, 7568. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; De Wever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3, 24858. [Google Scholar] [CrossRef]
- Kırbaş, O.K.; Bozkurt, B.T.; Asutay, A.B.; Mat, B.; Ozdemir, B.; Öztürkoğlu, D.; Ölmez, H.; İşlek, Z.; Şahin, F.; Taşlı, P.N. Optimized Isolation of Extracellular Vesicles From Various Organic Sources Using Aqueous Two-Phase System. Sci. Rep. 2019, 9, 19159. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 2020, 182, 1044–1061.e18. [Google Scholar] [CrossRef]
- Groot, M.; Lee, H. Sorting Mechanisms for MicroRNAs into Extracellular Vesicles and Their Associated Diseases. Cells 2020, 9, 1044. [Google Scholar] [CrossRef]
- Garcia-Martin, R.; Wang, G.; Brandão, B.B.; Zanotto, T.M.; Shah, S.; Patel, S.K.; Schilling, B.; Kahn, C.R. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2021, 601, 446–451. [Google Scholar] [CrossRef]
- Dillekås, H.; Rogers, M.S.; Straume, O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019, 8, 5574–5576. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, D. Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; García, V.; Zaballos, Á.; Provencio, M.; Lombardía, L.; Almonacid, L.; García, J.M.; Domínguez, G.; Peña, C.; Diaz, R.; et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur. Respir. J. 2011, 37, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Goldie, B.J.; Dun, M.D.; Lin, M.; Smith, N.D.; Verrills, N.M.; Dayas, C.V.; Cairns, M.J. Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 2014, 42, 9195–9208. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The Majority of MicroRNAs Detectable in Serum and Saliva Is Concentrated in Exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef] [PubMed]
- Hessvik, N.P.; Phuyal, S.; Brech, A.; Sandvig, K.; Llorente, A. Profiling of microRNAs in exosomes released from PC-3 prostate cancer cells. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2012, 1819, 1154–1163. [Google Scholar] [CrossRef]
- Liang, M.; Yu, S.; Tang, S.; Bai, L.; Cheng, J.; Gu, Y.; Li, S.; Zheng, X.; Duan, L.; Wang, L.; et al. A Panel of Plasma Exosomal miRNAs as Potential Biomarkers for Differential Diagnosis of Thyroid Nodules. Front. Genet. 2020, 11, 449. [Google Scholar] [CrossRef]
- Sueta, A.; Yamamoto, Y.; Tomiguchi, M.; Takeshita, T.; Yamamoto-Ibusuki, M.; Iwase, H. Differential expression of exosomal miRNAs between breast cancer patients with and without recurrence. Oncotarget 2017, 8, 69934–69944. [Google Scholar] [CrossRef]
- Martellucci, S.; Orefice, N.S.; Angelucci, A.; Luce, A.; Caraglia, M.; Zappavigna, S. Extracellular Vesicles: New Endogenous Shuttles for miRNAs in Cancer Diagnosis and Therapy? Int. J. Mol. Sci. 2020, 21, 6486. [Google Scholar] [CrossRef]
- Ramirez-Garrastacho, M.; Bajo-Santos, C.; Line, A.; Martens-Uzunova, E.S.; de la Fuente, J.M.; Moros, M.; Soekmadji, C.; Tasken, K.A.; Llorente, A. Extracellular vesicles as a source of prostate cancer biomarkers in liquid biopsies: A decade of research. Br. J. Cancer 2021, 126, 331–350. [Google Scholar] [CrossRef]
- McNally, C.J.; Ruddock, M.W.; Moore, T.; McKenna, D.J. Biomarkers That Differentiate Benign Prostatic Hyperplasia from Prostate Cancer: A Literature Review. Cancer Manag. Res. 2020, 12, 5225–5241. [Google Scholar] [CrossRef]
- Vafaizadeh, V.; Buechel, D.; Rubinstein, N.; Kalathur, R.K.R.; Bazzani, L.; Saxena, M.; Valenta, T.; Hausmann, G.; Cantù, C.; Basler, K.; et al. The interactions of Bcl9/Bcl9L with β-catenin and Pygopus promote breast cancer growth, invasion, and metastasis. Oncogene 2021, 40, 6195–6209. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhou, W.; Cheng, M.; Wang, J.; Liu, Z.; He, S.; Luo, X.; Huang, W.; Chen, T.; Yan, W.; et al. Hypoxia activates Wnt/β-catenin signaling by regulating the expression of BCL9 in human hepatocellular carcinoma. Sci. Rep. 2017, 7, 40446. [Google Scholar] [CrossRef] [PubMed]
- Kasoha, M.; Bohle, R.M.; Seibold, A.; Gerlinger, C.; Juhasz-Böss, I.; Solomayer, E.-F. Dickkopf-1 (Dkk1) protein expression in breast cancer with special reference to bone metastases. Clin. Exp. Metastasis 2018, 35, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ouyang, R.; Wang, Z.; Zhou, W.; Chen, H.; Jiang, Y.; Zhang, Y.; Li, H.; Liao, M.; Wang, W.; et al. MiR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4. Sci. Rep. 2016, 6, 39001. [Google Scholar] [CrossRef]
- Chang, W.-A.; Tsai, M.-J.; Hung, J.-Y.; Wu, K.-L.; Tsai, Y.-M.; Huang, Y.-C.; Chang, C.-Y.; Tsai, P.-H.; Hsu, Y.-L. miR-150-5p-Containing Extracellular Vesicles Are a New Immunoregulator That Favor the Progression of Lung Cancer in Hypoxic Microenvironments by Altering the Phenotype of NK Cells. Cancers 2021, 13, 6252. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Song, X.; Niu, L.; Tang, Y.; Song, X.; Xie, L. Circulating Exosomal miR-150-5p and miR-99b-5p as Diagnostic Biomarkers for Colorectal Cancer. Front. Oncol. 2019, 9, 1129. [Google Scholar] [CrossRef]
- He, Z.; Dang, J.; Song, A.; Cui, X.; Ma, Z.; Zhang, Y. The involvement of miR-150/β-catenin axis in colorectal cancer progression. Biomed. Pharmacother. 2020, 121, 109495. [Google Scholar] [CrossRef]
- Paunescu, I.A.; Bardan, R.; Marcu, A.; Nitusca, D.; Dema, A.; Negru, S.; Balacescu, O.; Balacescu, L.; Cumpanas, A.; Sirbu, I.O.; et al. Biomarker Potential of Plasma MicroRNA-150-5p in Prostate Cancer. Medicina 2019, 55, 564. [Google Scholar] [CrossRef]
- Yu, J.; Feng, Y.; Wang, Y.; An, R. Aryl hydrocarbon receptor enhances the expression of miR-150-5p to suppress in prostate cancer progression by regulating MAP3K12. Arch. Biochem. Biophys. 2018, 654, 47–54. [Google Scholar] [CrossRef]
- Wang, X.; Luo, G.; Zhang, K.; Cao, J.; Huang, C.; Jiang, T.; Liu, B.; Su, L.; Qiu, Z. Hypoxic Tumor-Derived Exosomal miR-301a Mediates M2 Macrophage Polarization via PTEN/PI3Kγ to Promote Pancreatic Cancer Metastasis. Cancer Res. 2018, 78, 4586–4598. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Qian, T.; Bao, S.; Zhao, H.; Chen, H.; Xing, Z.; Li, Y.; Zhang, M.; Meng, X.; Wang, C.; et al. Circulating exosomal miR-363-5p inhibits lymph node metastasis by downregulating PDGFB and serves as a potential noninvasive biomarker for breast cancer. Mol. Oncol. 2021, 15, 2466–2479. [Google Scholar] [CrossRef] [PubMed]

| Total of Samples | Metastatic Lymph Node | Non-Metastatic Lymph Node | Tumor | |||
|---|---|---|---|---|---|---|
| n = 7 | n = 8 | n = 3 | ||||
| Age | ||||||
| Mean | 63.71 | 65.125 | 67.67 | |||
| Range | 57–71 | 52–71 | 62–71 | |||
| PSA (ng/mL) | ||||||
| Mean | 15.64 | 13.06 | 18.18 | |||
| Range | 8.35–27 | 8.13–18.11 | 9.43–27 | |||
| Gleason Score | n | % | n | % | n | % |
| 3 + 3 | 3 | 42.86 | 3 | 37.50 | 2 | 66.67 |
| 3 + 4 | 1 | 14.29 | 3 | 37.50 | ||
| 4 + 3 | 1 | 12.50 | ||||
| 4 + 4 | 2 | 28.57 | 1 | 12.50 | 1 | 33.33 |
| 5 + 5 | 1 | 14.29 | ||||
| Positive Nodes | ||||||
| 1–3 | 5 | 71.43 | ||||
| 4–6 | 1 | 14.29 | ||||
| >7 | 1 | 14.29 | ||||
| Total of Samples ** | Prostate Cancer | Healthy Donors | |
|---|---|---|---|
| n = 16 | n = 10 | ||
| Age | |||
| Mean | 75.06 | 28.1 | |
| Range | 55–93 | 22–45 | |
| PSA (ng/mL) | |||
| Mean | 84.72 | - | |
| Range | 2.74–566 | - | |
| Gleason Score * | n | % | |
| 3 + 3 | 4 | 57.14 | |
| 3 + 4 | 2 | 28.57 | |
| 4 + 3 | 1 | 14.29 | |
| 4 + 4 | 3 | 42.86 | |
| 4 + 5 | 1 | 14.29 | |
| 5 + 4 | 1 | 14.29 | |
| 5 + 5 | 2 | 28.57 | |
| TNM Stage | n | % | |
| T1A | 1 | 6.25 | |
| T2A | 9 | 56.25 | |
| T2B | 3 | 18.75 | |
| T2C | 3 | 18.75 | |
| Tissue | miRNA | Fold Change | p-Value |
|---|---|---|---|
| Metastasis vs. Tumor | hsa-miR-150-5p | 30.81 | 0.003 |
| hsa-miR-23b-3p | −2.03 | 0.0472 | |
| hsa-miR-140-3p | 3.87 | 0.0213 | |
| Metastasis vs. Lymph Node | hsa-miR-150-5p | No significant differential expression | |
| hsa-miR-23b-3p | 2.36 | 0.0283 | |
| hsa-miR-140-3p | No significant differential expression | ||
| Tumor vs. Lymph Node | hsa-miR-150-5p | −23.14 | 0.0001 |
| hsa-miR-23b-3p | 4.79 | 0.0009 | |
| hsa-miR-140-3p | −3.8 | 0.0049 | |
| PCa vs. Healthy Plasma | hsa-miR-150-5p | −2.47 | 0.0162 |
| hsa-miR-23b-3p | −2.4 | 0.0308 | |
| hsa-miR-140-3p | −1.63 | 0.0262 | |
| Total of Samples * | Prostate Cancer | Healthy Donors | |
|---|---|---|---|
| n = 31 | n = 12 | ||
| Age | |||
| Mean | 73.35 | 35.25 | |
| Range | 57–86 | 26–45 | |
| PSA (ng/mL) | |||
| Mean | 16.709 | - | |
| Range | 1.99–87.6 | - | |
| Gleason Score | n | % | |
| 3 + 3 | 8 | 25.81 | |
| 3 + 4 | 7 | 22.58 | |
| 4 + 3 | 8 | 25.81 | |
| 4 + 4 | 4 | 12.90 | |
| 4 + 5 | 3 | 9.68 | |
| 5 + 5 | 1 | 3.23 | |
| Primary Tumor Tissue | Metastatic Lymph Node Tissue | Normal Prostate Tissue | ||||
|---|---|---|---|---|---|---|
| Total of samples | n = 15 | n = 5 | n = 20 | |||
| Age | ||||||
| Mean | 75.46 | 70.2 | 25.75 | |||
| Range | 51–90 | 62–78 | 22–30 | |||
| PSA (ng/mL) | ||||||
| Mean | 33.2 | - | - | |||
| Range | 0.7–106 | - | - | |||
| Gleason Score * | n | % | Gleason Score | n | % | |
| 3 + 4 | 4 | 26.67 | 3 + 4 | - | - | |
| 4 + 3 | 3 | 20 | 4 + 3 | - | - | |
| 4 + 4 | - | - | 4 + 4 | 1 | 20 | |
| 4 + 5 | 3 | 20 | 4 + 5 | 2 | 40 | |
| 5 + 4 | 3 | 20 | 5 + 4 | 2 | 40 | |
| 5 + 5 | 1 | 6.67 | 5 + 5 | - | - | |
| Stage ** | n | % | Number of Positive LNs | |||
| IIB | 2 | 13.33 | 2 | 1 | 20 | |
| IIIB | 2 | 13.33 | 4 | 1 | 20 | |
| IIC | 1 | 6.67 | 6 | 2 | 40 | |
| IIIC | 4 | 26.67 | 7 | 1 | 20 | |
| IV | 3 | 20 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz-Burgos, M.; Cortés-Ramírez, S.A.; Losada-García, A.; Morales-Pacheco, M.; Martínez-Martínez, E.; Morales-Montor, J.G.; Servín-Haddad, A.; Izquierdo-Luna, J.S.; Rodríguez-Martínez, G.; Ramos-Godínez, M.d.P.; et al. Unraveling the Role of EV-Derived miR-150-5p in Prostate Cancer Metastasis and Its Association with High-Grade Gleason Scores: Implications for Diagnosis. Cancers 2023, 15, 4148. https://doi.org/10.3390/cancers15164148
Cruz-Burgos M, Cortés-Ramírez SA, Losada-García A, Morales-Pacheco M, Martínez-Martínez E, Morales-Montor JG, Servín-Haddad A, Izquierdo-Luna JS, Rodríguez-Martínez G, Ramos-Godínez MdP, et al. Unraveling the Role of EV-Derived miR-150-5p in Prostate Cancer Metastasis and Its Association with High-Grade Gleason Scores: Implications for Diagnosis. Cancers. 2023; 15(16):4148. https://doi.org/10.3390/cancers15164148
Chicago/Turabian StyleCruz-Burgos, Marian, Sergio A. Cortés-Ramírez, Alberto Losada-García, Miguel Morales-Pacheco, Eduardo Martínez-Martínez, Jorge Gustavo Morales-Montor, Alejandro Servín-Haddad, J. Samuel Izquierdo-Luna, Griselda Rodríguez-Martínez, María del Pilar Ramos-Godínez, and et al. 2023. "Unraveling the Role of EV-Derived miR-150-5p in Prostate Cancer Metastasis and Its Association with High-Grade Gleason Scores: Implications for Diagnosis" Cancers 15, no. 16: 4148. https://doi.org/10.3390/cancers15164148
APA StyleCruz-Burgos, M., Cortés-Ramírez, S. A., Losada-García, A., Morales-Pacheco, M., Martínez-Martínez, E., Morales-Montor, J. G., Servín-Haddad, A., Izquierdo-Luna, J. S., Rodríguez-Martínez, G., Ramos-Godínez, M. d. P., González-Covarrubias, V., Cañavera-Constantino, A., González-Ramírez, I., Su, B., Leong, H. S., & Rodríguez-Dorantes, M. (2023). Unraveling the Role of EV-Derived miR-150-5p in Prostate Cancer Metastasis and Its Association with High-Grade Gleason Scores: Implications for Diagnosis. Cancers, 15(16), 4148. https://doi.org/10.3390/cancers15164148






