Dissecting Microbiome-Derived SCFAs in Prostate Cancer: Analyzing Gut Microbiota, Racial Disparities, and Epigenetic Mechanisms
Abstract
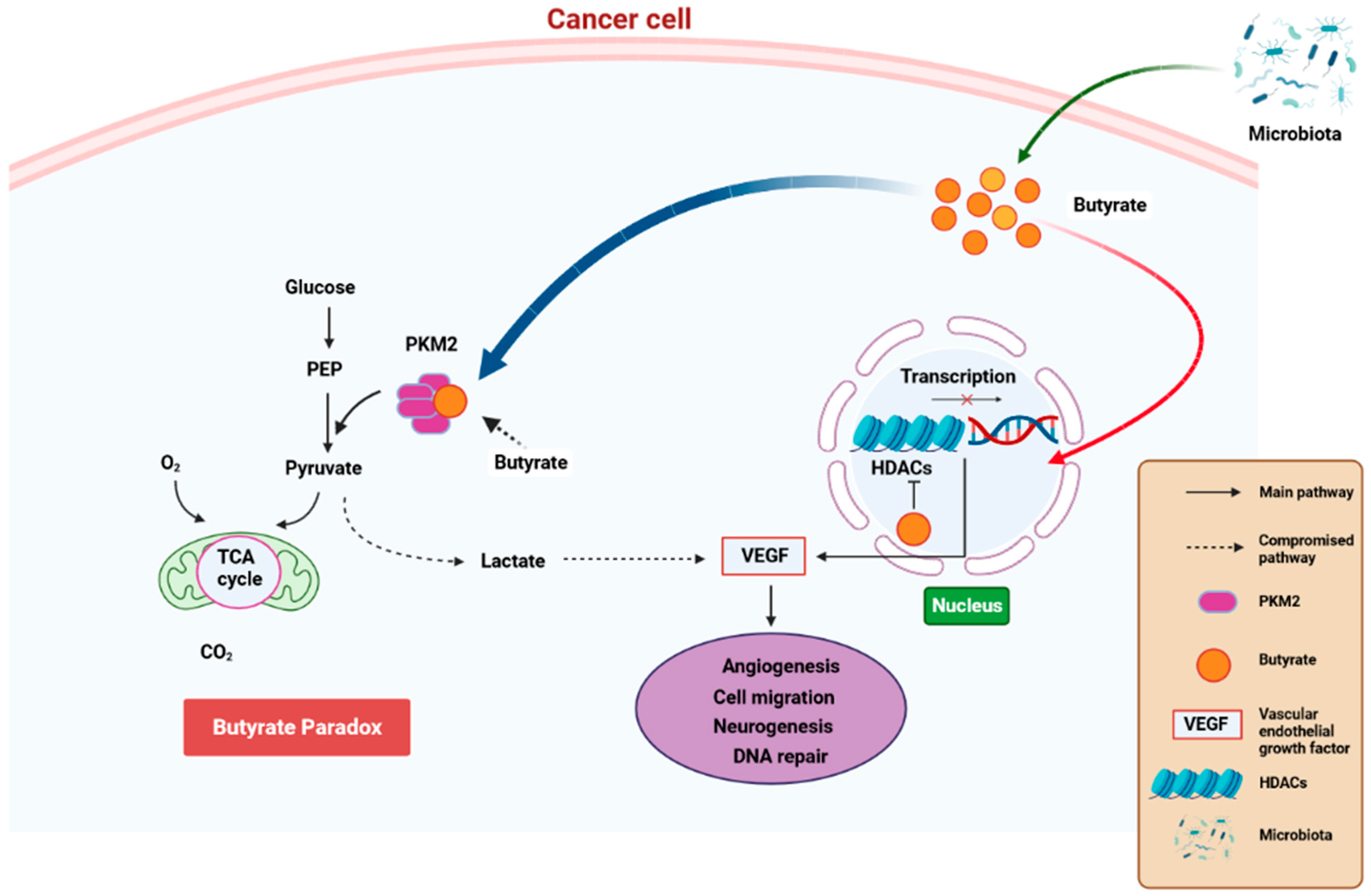
Simple Summary
Abstract
1. Introduction
Association between the Human Microbiome and PCa Pathogenesis
2. The “Prostate Microbiome”
2.1. Intraprostatic Microbiome
2.2. Genitourinary Microbiome
2.3. Gut Microbiome
3. Microbiome-Derived Short-Chain Fatty Acids (SCFAs)
3.1. The Mechanism of SCFAs in Cells
3.2. The Role of SCFAs in PCa Carcinogenesis
3.3. The Effect of Microbiome-Derived SCFAs on Response to Cancer Immunotherapy
3.4. Gut Microbiota and SCFA Profile Disparities among Different Race/Ethnic Groups
3.5. The Effect of Microbiome-Derived SCFAs on PCa Progression via Epigenetics
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Wang, L.; Lu, B.; He, M.; Wang, Y.; Wang, Z.; Du, L. Prostate cancer incidence and mortality: Global status and temporal trends in 89 countries from 2000 to 2019. Front. Public Health 2022, 10, 811044. [Google Scholar] [CrossRef] [PubMed]
- Moul, J.W. The evolving definition of advanced prostate cancer. Rev. Urol. 2004, 6, S10. [Google Scholar] [PubMed]
- Siegel, R.; Miller, K.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. Am. Cancer Soc. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Swami, U.; McFarland, T.R.; Nussenzveig, R.; Agarwal, N. Advanced prostate cancer: Treatment advances and future directions. Trends Cancer 2020, 6, 702–715. [Google Scholar] [CrossRef]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef]
- Porter, C.M.; Shrestha, E.; Peiffer, L.B.; Sfanos, K.S. The microbiome in prostate inflammation and prostate cancer. Prostate Cancer Prostatic Dis. 2018, 21, 345–354. [Google Scholar] [CrossRef]
- Ma, J.; Gnanasekar, A.; Lee, A.; Li, W.T.; Haas, M.; Wang-Rodriguez, J.; Chang, E.Y.; Rajasekaran, M.; Ongkeko, W.M. Influence of intratumor microbiome on clinical outcome and immune processes in prostate cancer. Cancers 2020, 12, 2524. [Google Scholar] [CrossRef]
- Cai, Z.; Lv, H.; Cao, W.; Zhou, C.; Liu, Q.; Li, H.; Zhou, F. Targeting strategies of adenovirus-mediated gene therapy and virotherapy for prostate cancer. Mol. Med. Rep. 2017, 16, 6443–6458. [Google Scholar] [CrossRef][Green Version]
- Rescigno, M. A ‘fit’microbiota to potentiate cancer immunotherapy. Genome Med. 2015, 7, 131. [Google Scholar] [CrossRef]
- Dzutsev, A.; Goldszmid, R.S.; Viaud, S.; Zitvogel, L.; Trinchieri, G. The role of the microbiota in inflammation, carcinogenesis, and cancer therapy. Eur. J. Immunol. 2015, 45, 17–31. [Google Scholar] [CrossRef]
- Bosch, T.C.; McFall-Ngai, M.J. Metaorganisms as the new frontier. Zoology 2011, 114, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Tulstrup, M.V.-L.; Christensen, E.G.; Carvalho, V.; Linninge, C.; Ahrné, S.; Højberg, O.; Licht, T.R.; Bahl, M.I. Antibiotic treatment affects intestinal permeability and gut microbial composition in Wistar rats dependent on antibiotic class. PLoS ONE 2015, 10, e0144854. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L. The long-term stability of the human gut microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Park, M.; Kong, H.H.; Segre, J.A. Temporal stability of the human skin microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef]
- Massari, F.; Mollica, V.; Di Nunno, V.; Gatto, L.; Santoni, M.; Scarpelli, M.; Cimadamore, A.; Lopez-Beltran, A.; Cheng, L.; Battelli, N. The human microbiota and prostate cancer: Friend or foe? Cancers 2019, 11, 459. [Google Scholar] [CrossRef]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, S.A.; Razvi, H.; Dave, S.; Reid, G.; Burton, J.P. The microbiome of the urinary tract—A role beyond infection. Nat. Rev. Urol. 2015, 12, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, H.; Nederbragt, A.J.; Lagesen, K.; Jeansson, S.L.; Jakobsen, K.S. Assessing diversity of the female urine microbiota by high throughput sequencing of 16S rDNA amplicons. BMC Microbiol. 2011, 11, 244. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.A.; Brown, R.; Williams, J.; White, P.; Jacobson, S.K.; Marchesi, J.R.; Drake, M.J. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front. Cell. Infect. Microbiol. 2013, 3, 41. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.M.; Zilliox, M.J.; Rosenfeld, A.B.; Thomas-White, K.J.; Richter, H.E.; Nager, C.W.; Visco, A.G.; Nygaard, I.E.; Barber, M.D.; Schaffer, J. The female urinary microbiome in urgency urinary incontinence. Am. J. Obstet. Gynecol. 2015, 213, 347.e1–347.e11. [Google Scholar] [CrossRef]
- Shrestha, E.; White, J.R.; Yu, S.-H.; Kulac, I.; Ertunc, O.; De Marzo, A.M.; Yegnasubramanian, S.; Mangold, L.A.; Partin, A.W.; Sfanos, K.S. Profiling the urinary microbiome in men with positive versus negative biopsies for prostate cancer. J. Urol. 2018, 199, 161–171. [Google Scholar] [CrossRef]
- Yow, M.A.; Tabrizi, S.N.; Severi, G.; Bolton, D.M.; Pedersen, J.; Giles, G.G.; Southey, M.C. Characterisation of microbial communities within aggressive prostate cancer tissues. Infect. Agents Cancer 2017, 12, 4. [Google Scholar] [CrossRef]
- Feng, Y.; Ramnarine, V.R.; Bell, R.; Volik, S.; Davicioni, E.; Hayes, V.M.; Ren, S.; Collins, C.C. Metagenomic and metatranscriptomic analysis of human prostate microbiota from patients with prostate cancer. BMC Genom. 2019, 20, 146. [Google Scholar] [CrossRef]
- Kustrimovic, N.; Bombelli, R.; Baci, D.; Mortara, L. Microbiome and Prostate Cancer: A Novel Target for Prevention and Treatment. Int. J. Mol. Sci. 2023, 24, 1511. [Google Scholar] [CrossRef]
- Cavarretta, I.; Ferrarese, R.; Cazzaniga, W.; Saita, D.; Lucianò, R.; Ceresola, E.R.; Locatelli, I.; Visconti, L.; Lavorgna, G.; Briganti, A. The microbiome of the prostate tumor microenvironment. Eur. Urol. 2017, 72, 625–631. [Google Scholar] [CrossRef]
- Yu, H.; Meng, H.; Zhou, F.; Ni, X.; Shen, S.; Das, U.N. Urinary microbiota in patients with prostate cancer and benign prostatic hyperplasia. Arch. Med. Sci. 2015, 11, 385–394. [Google Scholar] [CrossRef]
- Hochreiter, W.W.; Duncan, J.L.; Schaeffer, A.J. Evaluation of the bacterial flora of the prostate using a 16S rRNA gene based polymerase chain reaction. J. Urol. 2000, 163, 127–130. [Google Scholar] [CrossRef]
- Sfanos, K.S.; Sauvageot, J.; Fedor, H.L.; Dick, J.D.; De Marzo, A.M.; Isaacs, W.B. A molecular analysis of prokaryotic and viral DNA sequences in prostate tissue from patients with prostate cancer indicates the presence of multiple and diverse microorganisms. Prostate 2008, 68, 306–320. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, D.B.; Vaghasia, A.M.; Yu, S.H.; Mak, T.N.; Brüggemann, H.; Nelson, W.G.; De Marzo, A.M.; Yegnasubramanian, S.; Sfanos, K.S. A mouse model of chronic prostatic inflammation using a human prostate cancer-derived isolate of Propionibacterium acnes. Prostate 2013, 73, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Shannon, B.A.; Garrett, K.L.; Cohen, R.J. Links between Propionibacterium Acnes and Prostate Cancer. Future Oncol. 2006, 2, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Eisenhofer, R.; Minich, J.J.; Marotz, C.; Cooper, A.; Knight, R.; Weyrich, L.S. Contamination in low microbial biomass microbiome studies: Issues and recommendations. Trends Microbiol. 2019, 27, 105–117. [Google Scholar] [CrossRef]
- Banerjee, S.; Alwine, J.C.; Wei, Z.; Tian, T.; Shih, N.; Sperling, C.; Guzzo, T.; Feldman, M.D.; Robertson, E.S. Microbiome signatures in prostate cancer. Carcinogenesis 2019, 40, 749–764. [Google Scholar] [CrossRef]
- André, A.R.; Ferreira, M.V.P.; Mota, R.M.S.; Ferrasi, A.C.; Pardini, M.I.d.M.C.; Rabenhorst, S.H.B. Gastric adenocarcinoma and Helicobacter pylori: Correlation with p53 mutation and p27 immunoexpression. Cancer Epidemiol. 2010, 34, 618–625. [Google Scholar] [CrossRef]
- Miyake, M.; Ohnishi, K.; Hori, S.; Nakano, A.; Nakano, R.; Yano, H.; Ohnishi, S.; Owari, T.; Morizawa, Y.; Itami, Y. Mycoplasma genitalium infection and chronic inflammation in human prostate cancer: Detection using prostatectomy and needle biopsy specimens. Cells 2019, 8, 212. [Google Scholar] [CrossRef]
- De Martel, C.; Franceschi, S. Infections and cancer: Established associations and new hypotheses. Crit. Rev. Oncol. Hematol. 2009, 70, 183–194. [Google Scholar] [CrossRef]
- Wolfe, A.J.; Toh, E.; Shibata, N.; Rong, R.; Kenton, K.; FitzGerald, M.; Mueller, E.R.; Schreckenberger, P.; Dong, Q.; Nelson, D.E. Evidence of uncultivated bacteria in the adult female bladder. J. Clin. Microbiol. 2012, 50, 1376–1383. [Google Scholar] [CrossRef]
- Hilt, E.E.; McKinley, K.; Pearce, M.M.; Rosenfeld, A.B.; Zilliox, M.J.; Mueller, E.R.; Brubaker, L.; Gai, X.; Wolfe, A.J.; Schreckenberger, P.C. Urine is not sterile: Use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J. Clin. Microbiol. 2014, 52, 871–876. [Google Scholar] [CrossRef]
- Kirby, R.; Lowe, D.; Bultitude, M.; Shuttleworth, K. Intra-prostatic urinary reflux: An aetiological factor in abacterial prostatitis. Br. J. Urol. 1982, 54, 729–731. [Google Scholar] [CrossRef]
- Nelson, D.E.; Van Der Pol, B.; Dong, Q.; Revanna, K.V.; Fan, B.; Easwaran, S.; Sodergren, E.; Weinstock, G.M.; Diao, L.; Fortenberry, J.D. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS ONE 2010, 5, e14116. [Google Scholar] [CrossRef]
- Fouts, D.E.; Pieper, R.; Szpakowski, S.; Pohl, H.; Knoblach, S.; Suh, M.-J.; Huang, S.-T.; Ljungberg, I.; Sprague, B.M.; Lucas, S.K. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J. Transl. Med. 2012, 10, 174. [Google Scholar] [CrossRef]
- Dong, Q.; Nelson, D.E.; Toh, E.; Diao, L.; Gao, X.; Fortenberry, J.D.; Van Der Pol, B. The microbial communities in male first catch urine are highly similar to those in paired urethral swab specimens. PLoS ONE 2011, 6, e19709. [Google Scholar] [CrossRef]
- Cohen, R.J.; Shannon, B.A.; McNEAL, J.E.; Shannon, T.; Garrett, K.L. Propionibacterium acnes associated with inflammation in radical prostatectomy specimens: A possible link to cancer evolution? J. Urol. 2005, 173, 1969–1974. [Google Scholar] [CrossRef]
- Mak, T.N.; Yu, S.H.; De Marzo, A.M.; Brüggemann, H.; Sfanos, K.S. Multilocus sequence typing (MLST) analysis of Propionibacterium acnes isolates from radical prostatectomy specimens. Prostate 2013, 73, 770–777. [Google Scholar] [CrossRef]
- Sfanos, K.S.; Isaacs, W.B. An evaluation of PCR primer sets used for detection of Propionibacterium acnes in prostate tissue samples. Prostate 2008, 68, 1492–1495. [Google Scholar] [CrossRef]
- Davidsson, S.; Mölling, P.; Rider, J.R.; Unemo, M.; Karlsson, M.G.; Carlsson, J.; Andersson, S.-O.; Elgh, F.; Söderquist, B.; Andrén, O. Frequency and typing of Propionibacterium acnes in prostate tissue obtained from men with and without prostate cancer. Infect. Agents Cancer 2016, 11, 26. [Google Scholar] [CrossRef]
- Brede, C.M.; Shoskes, D.A. The etiology and management of acute prostatitis. Nat. Rev. Urol. 2011, 8, 207–212. [Google Scholar] [CrossRef]
- Sasaki, M.; Yamaura, C.; Ohara-Nemoto, Y.; Tajika, S.; Kodama, Y.; Ohya, T.; Harada, R.; Kimura, S. Streptococcus anginosus infection in oral cancer and its infection route. Oral Dis. 2005, 11, 151–156. [Google Scholar] [CrossRef]
- Shiga, K.; Tateda, M.; Saijo, S.; Hori, T.; Sato, I.; Tateno, H.; Matsuura, K.; Takasaka, T.; Miyagi, T. Presence of Streptococcus infection in extra-oropharyngeal head and neck squamous cell carcinoma and its implication in carcinogenesis. Oncol. Rep. 2001, 8, 245–248. [Google Scholar] [CrossRef]
- Sfanos, K.S.; Yegnasubramanian, S.; Nelson, W.G.; De Marzo, A.M. The inflammatory microenvironment and microbiome in prostate cancer development. Nat. Rev. Urol. 2018, 15, 11–24. [Google Scholar] [CrossRef]
- Stapleton, A.E. Urinary tract infection pathogenesis: Host factors. Infect. Dis. Clin. 2014, 28, 149–159. [Google Scholar] [CrossRef]
- Ragnarsdóttir, B.; Lutay, N.; Grönberg-Hernandez, J.; Köves, B.; Svanborg, C. Genetics of innate immunity and UTI susceptibility. Nat. Rev. Urol. 2011, 8, 449–468. [Google Scholar] [CrossRef]
- Pearce, M.M.; Hilt, E.E.; Rosenfeld, A.B.; Zilliox, M.J.; Thomas-White, K.; Fok, C.; Kliethermes, S.; Schreckenberger, P.C.; Brubaker, L.; Gai, X. The female urinary microbiome: A comparison of women with and without urgency urinary incontinence. MBio 2014, 5, e01283-14. [Google Scholar] [CrossRef]
- Nienhouse, V.; Gao, X.; Dong, Q.; Nelson, D.E.; Toh, E.; McKinley, K.; Schreckenberger, P.; Shibata, N.; Fok, C.S.; Mueller, E.R. Interplay between bladder microbiota and urinary antimicrobial peptides: Mechanisms for human urinary tract infection risk and symptom severity. PLoS ONE 2014, 9, e114185. [Google Scholar] [CrossRef]
- Sutcliffe, S.; Zenilman, J.M.; Ghanem, K.G.; Jadack, R.A.; Sokoll, L.J.; Elliott, D.J.; Nelson, W.G.; De Marzo, A.M.; Cole, S.R.; Isaacs, W.B. Sexually transmitted infections and prostatic inflammation/cell damage as measured by serum prostate specific antigen concentration. J. Urol. 2006, 175, 1937–1942. [Google Scholar] [CrossRef]
- Huang, W.-Y.; Hayes, R.; Pfeiffer, R.; Viscidi, R.P.; Lee, F.K.; Wang, Y.F.; Reding, D.; Whitby, D.; Papp, J.R.; Rabkin, C.S. Sexually transmissible infections and prostate cancer risk. Cancer Epidemiol. Biomark. Prev. 2008, 17, 2374–2381. [Google Scholar] [CrossRef]
- Hayes, R.; Pottern, L.; Strickler, H.; Rabkin, C.; Pope, V.; Swanson, G.; Greenberg, R.; Schoenberg, J.; Liff, J.; Schwartz, A. Sexual behaviour, STDs and risks for prostate cancer. Br. J. Cancer 2000, 82, 718–725. [Google Scholar] [CrossRef]
- Shoskes, D.A.; Altemus, J.; Polackwich, A.S.; Tucky, B.; Wang, H.; Eng, C. The urinary microbiome differs significantly between patients with chronic prostatitis/chronic pelvic pain syndrome and controls as well as between patients with different clinical phenotypes. Urology 2016, 92, 26–32. [Google Scholar] [CrossRef]
- Ohadian Moghadam, S.; Momeni, S.A. Human microbiome and prostate cancer development: Current insights into the prevention and treatment. Front. Med. 2021, 15, 11–32. [Google Scholar] [CrossRef]
- Fujita, K.; Matsushita, M.; Banno, E.; De Velasco, M.A.; Hatano, K.; Nonomura, N.; Uemura, H. Gut microbiome and prostate cancer. Int. J. Urol. 2022, 29, 793–798. [Google Scholar] [CrossRef]
- Liss, M.A.; White, J.R.; Goros, M.; Gelfond, J.; Leach, R.; Johnson-Pais, T.; Lai, Z.; Rourke, E.; Basler, J.; Ankerst, D. Metabolic biosynthesis pathways identified from fecal microbiome associated with prostate cancer. Eur. Urol. 2018, 74, 575–582. [Google Scholar] [CrossRef]
- Golombos, D.M.; Ayangbesan, A.; O’Malley, P.; Lewicki, P.; Barlow, L.; Barbieri, C.E.; Chan, C.; DuLong, C.; Abu-Ali, G.; Huttenhower, C. The role of gut microbiome in the pathogenesis of prostate cancer: A prospective, pilot study. Urology 2018, 111, 122–128. [Google Scholar] [CrossRef]
- Matsushita, M.; Fujita, K.; Motooka, D.; Hatano, K.; Fukae, S.; Kawamura, N.; Tomiyama, E.; Hayashi, Y.; Banno, E.; Takao, T. The gut microbiota associated with high-Gleason prostate cancer. Cancer Sci. 2021, 112, 3125–3135. [Google Scholar] [CrossRef]
- Wong, S.H.; Kwong, T.N.; Wu, C.-Y.; Yu, J. Clinical Applications of Gut Microbiota in Cancer Biology. In Proceedings of Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2019; pp. 28–36. [Google Scholar]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012, 6, 320–329. [Google Scholar] [CrossRef]
- Picardo, S.L.; Coburn, B.; Hansen, A.R. The microbiome and cancer for clinicians. Crit. Rev. Oncol. Hematol. 2019, 141, 1–12. [Google Scholar] [CrossRef]
- Mirzaei, R.; Afaghi, A.; Babakhani, S.; Sohrabi, M.R.; Hosseini-Fard, S.R.; Babolhavaeji, K.; Akbari, S.K.A.; Yousefimashouf, R.; Karampoor, S. Role of microbiota-derived short-chain fatty acids in cancer development and prevention. Biomed. Pharmacother. 2021, 139, 111619. [Google Scholar] [CrossRef]
- Tsvetikova, S.A.; Koshel, E.I. Microbiota and cancer: Host cellular mechanisms activated by gut microbial metabolites. Int. J. Med. Microbiol. 2020, 310, 151425. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.L.; Wolin, M.J. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl. Environ. Microbiol. 1996, 62, 1589–1592. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, V.; Thangaraju, M.; Prasad, P.D.; Martin, P.M.; Singh, N. Transporters and receptors for short-chain fatty acids as the molecular link between colonic bacteria and the host. Curr. Opin. Pharmacol. 2013, 13, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H. Microbiota or short-chain fatty acids: Which regulates diabetes? Cell. Mol. Immunol. 2018, 15, 88–91. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.; Faber, K.N.; Hermoso, M.A. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Scheppach, W.; Bartram, H.; Richter, F. Role of short-chain fatty acids in the prevention of colorectal cancer. Eur. J. Cancer 1995, 31, 1077–1080. [Google Scholar] [CrossRef]
- Wang, G.; Yu, Y.; Wang, Y.Z.; Wang, J.J.; Guan, R.; Sun, Y.; Shi, F.; Gao, J.; Fu, X.L. Role of SCFAs in gut microbiome and glycolysis for colorectal cancer therapy. J. Cell. Physiol. 2019, 234, 17023–17049. [Google Scholar] [CrossRef]
- Yusuf, F.; Adewiah, S.; Syam, A.F.; Fatchiyah, F. Altered profile of gut microbiota and the level short chain fatty acids in colorectal cancer patients. J. Phys. Conf. Ser. 2019, 1146, 012037. [Google Scholar] [CrossRef]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- O’keefe, S.J. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef]
- Liang, W.; Yang, Y.; Wang, H.; Wang, H.; Yu, X.; Lu, Y.; Shen, S.; Teng, L. Gut microbiota shifts in patients with gastric cancer in perioperative period. Medicine 2019, 98, e16626. [Google Scholar] [CrossRef]
- Yang, Q.; Ouyang, J.; Sun, F.; Yang, J. Short-chain fatty acids: A soldier fighting against inflammation and protecting from tumorigenesis in people with diabetes. Front. Immunol. 2020, 11, 590685. [Google Scholar] [CrossRef]
- Park, S.Y.; Wilkens, L.R.; Kolonel, L.N.; Henderson, B.E.; Le Marchand, L. Inverse associations of dietary fiber and menopausal hormone therapy with colorectal cancer risk in the Multiethnic Cohort Study. Int. J. Cancer 2016, 139, 1241–1250. [Google Scholar] [CrossRef]
- Shaw, E.; Warkentin, M.T.; McGregor, S.E.; Town, S.; Hilsden, R.J.; Brenner, D.R. Intake of dietary fibre and lifetime non-steroidal anti-inflammatory drug (NSAID) use and the incidence of colorectal polyps in a population screened for colorectal cancer. J. Epidemiol. Commun. Health 2017, 71, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Kopp, T.I.; Vogel, U.; Tjonneland, A.; Andersen, V. Meat and fiber intake and interaction with pattern recognition receptors (TLR1, TLR2, TLR4, and TLR10) in relation to colorectal cancer in a Danish prospective, case-cohort study. Am. J. Clin. Nutr. 2018, 107, 465–479. [Google Scholar] [CrossRef]
- Kasubuchi, M.; Hasegawa, S.; Hiramatsu, T.; Ichimura, A.; Kimura, I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 2015, 7, 2839–2849. [Google Scholar] [CrossRef]
- Thangaraju, M.; Cresci, G.A.; Liu, K.; Ananth, S.; Gnanaprakasam, J.P.; Browning, D.D.; Mellinger, J.D.; Smith, S.B.; Digby, G.J.; Lambert, N.A. GPR109A is a G-protein–coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009, 69, 2826–2832. [Google Scholar] [CrossRef]
- Blad, C.C.; Ahmed, K.; IJzerman, A.P.; Offermanns, S. Biological and pharmacological roles of HCA receptors. Adv. Pharmacol. 2011, 62, 219–250. [Google Scholar]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef]
- Yonezawa, T.; Kobayashi, Y.; Obara, Y. Short-chain fatty acids induce acute phosphorylation of the p38 mitogen-activated protein kinase/heat shock protein 27 pathway via GPR43 in the MCF-7 human breast cancer cell line. Cell. Signal. 2007, 19, 185–193. [Google Scholar] [CrossRef]
- Zou, X.; Blank, M. Targeting p38 MAP kinase signaling in cancer through post-translational modifications. Cancer Lett. 2017, 384, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Chen, Y.; Jiang, H.; Robbins, G.T.; Nie, D. G-protein-coupled receptor for short-chain fatty acids suppresses colon cancer. Int. J. Cancer 2011, 128, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Myeroff, L.; Smiraglia, D.; Romero, M.F.; Pretlow, T.P.; Kasturi, L.; Lutterbaugh, J.; Rerko, R.M.; Casey, G.; Issa, J.-P. SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc. Natl. Acad. Sci. USA 2003, 100, 8412–8417. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, P.; Martel, F. Regulation of colonic epithelial butyrate transport: Focus on colorectal cancer. Porto Biomed. J. 2016, 1, 83–91. [Google Scholar] [CrossRef]
- Dietrich, C.G.; Vehr, A.K.; Martin, I.V.; Gaßler, N.; Rath, T.; Roeb, E.; Schmitt, J.; Trautwein, C.; Geier, A. Downregulation of breast cancer resistance protein in colon adenomas reduces cellular xenobiotic resistance and leads to accumulation of a food-derived carcinogen. Int. J. Cancer 2011, 129, 546–552. [Google Scholar] [CrossRef]
- Ropero, S.; Esteller, M. The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol. 2007, 1, 19–25. [Google Scholar] [CrossRef]
- Hinnebusch, B.F.; Meng, S.; Wu, J.T.; Archer, S.Y.; Hodin, R.A. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J. Nutr. 2002, 132, 1012–1017. [Google Scholar] [CrossRef]
- Pesavento, J.J.; Yang, H.; Kelleher, N.L.; Mizzen, C.A. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol. Cell. Biol. 2008, 28, 468–486. [Google Scholar] [CrossRef]
- White, N.R.; Mulligan, P.; King, P.J.; Sanderson, I.R. Sodium butyrate-mediated Sp3 acetylation represses human insulin-like growth factor binding protein-3 expression in intestinal epithelial cells. J. Pediatr. Gastroenterol. Nutr. 2006, 42, 134–141. [Google Scholar] [CrossRef]
- Yang, H.J.; Kim, J.H. Role of microbiome and its metabolite, short chain fatty acid in prostate cancer. Investig. Clin. Urol. 2023, 64, 3–12. [Google Scholar] [CrossRef]
- Walczak, J.; Wood, H.; Wilding, G.; Williams Jr, T.; Bishop, C.W.; Carducci, M. Prostate cancer prevention strategies using antiproliferative or differentiating agents. Urology 2001, 57, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Samid, D.; Shack, S.; Myers, C.E. Selective growth arrest and phenotypic reversion of prostate cancer cells in vitro by nontoxic pharmacological concentrations of phenylacetate. J. Clin. Investig. 1993, 91, 2288–2295. [Google Scholar] [CrossRef] [PubMed]
- Carducci, M.A.; Nelson, J.B.; Chan-Tack, K.M.; Ayyagari, S.R.; Sweatt, W.H.; Campbell, P.A.; Nelson, W.G.; Simons, J.W. Phenylbutyrate induces apoptosis in human prostate cancer and is more potent than phenylacetate. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 1996, 2, 379–387. [Google Scholar]
- Thibault, A.; Cooper, M.R.; Figg, W.D.; Venzon, D.J.; Sartor, A.O.; Tompkins, A.C.; Weinberger, M.S.; Headlee, D.J.; McCall, N.A.; Samid, D. A phase I and pharmacokinetic study of intravenous phenylacetate in patients with cancer. Cancer Res. 1994, 54, 1690–1694. [Google Scholar]
- Carducci, M.A.; Gilbert, J.; Bowling, M.K.; Noe, D.; Eisenberger, M.A.; Sinibaldi, V.; Zabelina, Y.; Chen, T.-L.; Grochow, L.B.; Donehower, R.C. A Phase I clinical and pharmacological evaluation of sodium phenylbutyrate on an 120-h infusion schedule. Clin. Cancer Res. 2001, 7, 3047–3055. [Google Scholar]
- Matsushita, M.; Fujita, K.; Hayashi, T.; Kayama, H.; Motooka, D.; Hase, H.; Jingushi, K.; Yamamichi, G.; Yumiba, S.; Tomiyama, E. Gut Microbiota–Derived Short-Chain Fatty Acids Promote Prostate Cancer Growth via IGF1 SignalingGut Microbiota Regulates Prostate Cancer Growth. Cancer Res. 2021, 81, 4014–4026. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, Q.; Ye, F.; Yang, C.; Jiang, H. Gut Microbiota-Derived Short-Chain Fatty Acids Promote Prostate Cancer Progression through Inducing Cancer Cell Autophagy and M2 Macrophage Polarization. Cancer Res. 2021, 81, 4014–4026. [Google Scholar]
- Zitvogel, L.; Ma, Y.; Raoult, D.; Kroemer, G.; Gajewski, T.F. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science 2018, 359, 1366–1370. [Google Scholar] [CrossRef]
- Zitvogel, L.; Daillère, R.; Roberti, M.P.; Routy, B.; Kroemer, G. Anticancer effects of the microbiome and its products. Nat. Rev. Microbiol. 2017, 15, 465–478. [Google Scholar] [CrossRef]
- Roy, S.; Trinchieri, G. Microbiota: A key orchestrator of cancer therapy. Nat. Rev. Cancer 2017, 17, 271–285. [Google Scholar] [CrossRef]
- Ratajczak, W.; Rył, A.; Mizerski, A.; Walczakiewicz, K.; Sipak, O.; Laszczyńska, M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim. Pol. 2019, 66, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cerf-Bensussan, N.; Gaboriau-Routhiau, V. The immune system and the gut microbiota: Friends or foes? Nat. Rev. Immunol. 2010, 10, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Meijer, K.; de Vos, P.; Priebe, M.G. Butyrate and other short-chain fatty acids as modulators of immunity: What relevance for health? Curr. Opin. Clin. Nutr. Metab. Care 2010, 13, 715–721. [Google Scholar] [CrossRef]
- Canfora, E.E.; Jocken, J.W.; Blaak, E.E. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 2015, 11, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.; Karpinets, T.; Prieto, P.; Vicente, D.; Hoffman, K.; Wei, S.C. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef]
- Dubin, K.; Callahan, M.K.; Ren, B.; Khanin, R.; Viale, A.; Ling, L.; No, D.; Gobourne, A.; Littmann, E.; Huttenhower, C. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 2016, 7, 10391. [Google Scholar] [CrossRef]
- Frankel, A.E.; Coughlin, L.A.; Kim, J.; Froehlich, T.W.; Xie, Y.; Frenkel, E.P.; Koh, A.Y. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia 2017, 19, 848–855. [Google Scholar] [CrossRef]
- Coutzac, C.; Jouniaux, J.-M.; Paci, A.; Schmidt, J.; Mallardo, D.; Seck, A.; Asvatourian, V.; Cassard, L.; Saulnier, P.; Lacroix, L. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 2020, 11, 2168. [Google Scholar] [CrossRef]
- Royston, K.J.; Adedokun, B.; Olopade, O.I. Race, the microbiome and colorectal cancer. World J. Gastrointest. Oncol. 2019, 11, 773. [Google Scholar] [CrossRef]
- Hester, C.M.; Jala, V.R.; Langille, M.G.; Umar, S.; Greiner, K.A.; Haribabu, B. Fecal microbes, short chain fatty acids, and colorectal cancer across racial/ethnic groups. World J. Gastroenterol. WJG 2015, 21, 2759. [Google Scholar] [CrossRef]
- Farhana, L.; Antaki, F.; Murshed, F.; Mahmud, H.; Judd, S.L.; Nangia-Makker, P.; Levi, E.; Yu, Y.; Majumdar, A.P. Gut microbiome profiling and colorectal cancer in African Americans and Caucasian Americans. World J. Gastrointest. Pathophysiol. 2018, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Carson, T.L.; Wang, F.; Cui, X.; Jackson, B.E.; Van Der Pol, W.J.; Lefkowitz, E.J.; Morrow, C.; Baskin, M.L. Associations between race, perceived psychological stress, and the gut microbiota in a sample of generally healthy black and white women: A pilot study on the role of race and perceived psychological stress. Psychosom. Med. 2018, 80, 640. [Google Scholar] [CrossRef] [PubMed]
- Mai, V.; McCrary, Q.M.; Sinha, R.; Glei, M. Associations between dietary habits and body mass index with gut microbiota composition and fecal water genotoxicity: An observational study in African American and Caucasian American volunteers. Nutr. J. 2009, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Yazici, C.; Wolf, P.G.; Kim, H.; Cross, T.-W.L.; Vermillion, K.; Carroll, T.; Augustus, G.J.; Mutlu, E.; Tussing-Humphreys, L.; Braunschweig, C. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut 2017, 66, 1983–1994. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Upadhyay, V.; Poroyko, V.; Kim, T.-j.; Devkota, S.; Fu, S.; Liu, D.; Tumanov, A.V.; Koroleva, E.P.; Deng, L.; Nagler, C. Lymphotoxin regulates commensal responses to enable diet-induced obesity. Nat. Immunol. 2012, 13, 947–953. [Google Scholar] [CrossRef]
- Ou, J.; Carbonero, F.; Zoetendal, E.G.; DeLany, J.P.; Wang, M.; Newton, K.; Gaskins, H.R.; O’Keefe, S.J. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am. J. Clin. Nutr. 2013, 98, 111–120. [Google Scholar] [CrossRef]
- Ou, J.; DeLany, J.P.; Zhang, M.; Sharma, S.; O’Keefe, S.J. Association between low colonic short-chain fatty acids and high bile acids in high colon cancer risk populations. Nutr. Cancer 2012, 64, 34–40. [Google Scholar] [CrossRef]
- Nava, G.M.; Carbonero, F.; Ou, J.; Benefiel, A.C.; O’Keefe, S.J.; Gaskins, H.R. Hydrogenotrophic microbiota distinguish native Africans from African and European Americans. Environ. Microbiol. Rep. 2012, 4, 307–315. [Google Scholar] [CrossRef]
- O’Keefe, S.J.; Ou, J.; Aufreiter, S.; O’Connor, D.; Sharma, S.; Sepulveda, J.; Fukuwatari, T.; Shibata, K.; Mawhinney, T. Products of the colonic microbiota mediate the effects of diet on colon cancer risk. J. Nutr. 2009, 139, 2044–2048. [Google Scholar] [CrossRef]
- O’Keefe, S.J.; Kidd, M.; Espitalier-Noel, G.; Owira, P. Rarity of colon cancer in Africans is associated with low animal product consumption, not fiber. Am. J. Gastroenterol. 1999, 94, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Vikramdeo, K.S.; Anand, S.; Pierce, J.Y.; Singh, A.P.; Singh, S.; Dasgupta, S. Distribution of microbiota in cervical preneoplasia of racially disparate populations. BMC Cancer 2022, 22, 1074. [Google Scholar] [CrossRef]
- Smith, A.; Pierre, J.F.; Makowski, L.; Tolley, E.; Lyn-Cook, B.; Lu, L.; Vidal, G.; Starlard-Davenport, A. Distinct microbial communities that differ by race, stage, or breast-tumor subtype in breast tissues of non-Hispanic Black and non-Hispanic White women. Sci. Rep. 2019, 9, 11940. [Google Scholar] [CrossRef] [PubMed]
- Thyagarajan, S.; Zhang, Y.; Thapa, S.; Allen, M.S.; Phillips, N.; Chaudhary, P.; Kashyap, M.V.; Vishwanatha, J.K. Comparative analysis of racial differences in breast tumor microbiome. Sci. Rep. 2020, 10, 14116. [Google Scholar] [CrossRef] [PubMed]
- Bridges, K.M.; Diaz, F.J.; Wang, Z.; Ahmed, I.; Sullivan, D.K.; Umar, S.; Buckles, D.C.; Greiner, K.A.; Hester, C.M. Relating stool microbial metabolite levels, inflammatory markers and dietary behaviors to screening colonoscopy findings in a racially/ethnically diverse patient population. Genes 2018, 9, 119. [Google Scholar] [CrossRef]
- Hawkins, G.M.; Burkett, W.C.; McCoy, A.N.; Nichols, H.B.; Olshan, A.F.; Broaddus, R.; Merker, J.D.; Weissman, B.; Brewster, W.R.; Roach, J. Differences in the microbial profiles of early stage endometrial cancers between Black and White women. Gynecol. Oncol. 2022, 165, 248–256. [Google Scholar] [CrossRef]
- Carson, T.L.; Byrd, D.A.; Smith, K.S.; Carter, D.; Abaskaron, M.; Little, R.B.; van Der Pol, W.J.; Lefkowitz, E.J.; Morrow, C.D.; Fruge, A.D. A Case-Control Study of the Association between the Gut Microbiota and Colorectal Cancer: Exploring the Roles of Diet, Stress, and Race. Res. Sq. 2023, Preprint. [Google Scholar] [CrossRef]
- Khan, F.H.; Bhat, B.A.; Sheikh, B.A.; Tariq, L.; Padmanabhan, R.; Verma, J.P.; Shukla, A.C.; Dowlati, A.; Abbas, A. Microbiome Dysbiosis and Epigenetic Modulations in Lung Cancer: From Pathogenesis to Therapy. In Proceedings of Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2022; pp. 732–742. [Google Scholar]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef]
- Du, J.; Johnson, L.M.; Jacobsen, S.E.; Patel, D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015, 16, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Gebert, L.F.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef]
- Lindsay, M.A.; Griffiths-Jones, S.; Valadkhan, S.; Gunawardane, L.S. Role of small nuclear RNAs in eukaryotic gene expression. Essays Biochem. 2013, 54, 79–90. [Google Scholar] [CrossRef]
- Sartorelli, V.; Lauberth, S.M. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 2020, 27, 521–528. [Google Scholar] [CrossRef]
- D’Urso, A.; Brickner, J.H. Mechanisms of epigenetic memory. Trends Genet. 2014, 30, 230–236. [Google Scholar] [CrossRef]
- Devaux, C.A.; Raoult, D. The microbiological memory, an epigenetic regulator governing the balance between good health and metabolic disorders. Front. Microbiol. 2018, 9, 1379. [Google Scholar] [CrossRef]
- Krautkramer, K.A.; Rey, F.E.; Denu, J.M. Chemical signaling between gut microbiota and host chromatin: What is your gut really saying? J. Biol. Chem. 2017, 292, 8582–8593. [Google Scholar] [CrossRef]
- Wellen, K.E.; Hatzivassiliou, G.; Sachdeva, U.M.; Bui, T.V.; Cross, J.R.; Thompson, C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 2009, 324, 1076–1080. [Google Scholar] [CrossRef]
- Riggs, M.; Whittaker, R.; Neumann, J.; Ingram, V. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature 1977, 268, 462–464. [Google Scholar] [CrossRef]
- Waldecker, M.; Kautenburger, T.; Daumann, H.; Busch, C.; Schrenk, D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J. Nutr. Biochem. 2008, 19, 587–593. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Holley, D.; Collins, L.B.; Montgomery, S.A.; Whitmore, A.C.; Hillhouse, A.; Curry, K.P.; Renner, S.W.; Greenwalt, A.; Ryan, E.P. A Gnotobiotic Mouse Model Demonstrates That Dietary Fiber Protects against Colorectal Tumorigenesis in a Microbiota-and Butyrate-Dependent MannerFiber–Microbiota–Butyrate Axis in Tumor Suppression. Cancer Discov. 2014, 4, 1387–1397. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef]
- Lukovac, S.; Belzer, C.; Pellis, L.; Keijser, B.J.; de Vos, W.M.; Montijn, R.C.; Roeselers, G. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio 2014, 5, e01438-14. [Google Scholar] [CrossRef]
- Rizzo, A.; Santoni, M.; Mollica, V.; Fiorentino, M.; Brandi, G.; Massari, F. Microbiota and Prostate Cancer. In Proceedings of Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2022; pp. 1058–1065. [Google Scholar]



| Microbiome derived-SCFAs | Metabolite | Effect on Host Epigenetic Changes: Putative or Demonstrated | Model System | Outcome |
| Acetate | Putative | In vitro, mammalian cell culture | Increases histone acetylation [156] | |
| Butyrate | Demonstrated | In vitro, mammalian cell culture, human, mouse | HDAC inhibition [157,158], HAT activation, and protection from colorectal cancer [159,160]; protection from HFD-induced metabolic syndrome, and is linked with deceased HDAC activity [161]; increase in histone acetylation in HT-29 cells [98]; modest increase in HDAC3 and HDAC5 expression in gut organoids [162] | |
| Propionate | Putative | In vitro mammalian cell culture; intestinal organoids | Weak–modest HDC inhibition in vitro (<butyrate) [158]; increase in histone acetylation in HT-29 cells [98]; modest increase in HDAC3 and HDAC5 expression in gut organoids [162] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miya, T.V.; Marima, R.; Damane, B.P.; Ledet, E.M.; Dlamini, Z. Dissecting Microbiome-Derived SCFAs in Prostate Cancer: Analyzing Gut Microbiota, Racial Disparities, and Epigenetic Mechanisms. Cancers 2023, 15, 4086. https://doi.org/10.3390/cancers15164086
Miya TV, Marima R, Damane BP, Ledet EM, Dlamini Z. Dissecting Microbiome-Derived SCFAs in Prostate Cancer: Analyzing Gut Microbiota, Racial Disparities, and Epigenetic Mechanisms. Cancers. 2023; 15(16):4086. https://doi.org/10.3390/cancers15164086
Chicago/Turabian StyleMiya, Thabiso Victor, Rahaba Marima, Botle Precious Damane, Elisa Marie Ledet, and Zodwa Dlamini. 2023. "Dissecting Microbiome-Derived SCFAs in Prostate Cancer: Analyzing Gut Microbiota, Racial Disparities, and Epigenetic Mechanisms" Cancers 15, no. 16: 4086. https://doi.org/10.3390/cancers15164086
APA StyleMiya, T. V., Marima, R., Damane, B. P., Ledet, E. M., & Dlamini, Z. (2023). Dissecting Microbiome-Derived SCFAs in Prostate Cancer: Analyzing Gut Microbiota, Racial Disparities, and Epigenetic Mechanisms. Cancers, 15(16), 4086. https://doi.org/10.3390/cancers15164086






