RETRACTED: Prediction of Ovarian Cancer Response to Therapy Based on Deep Learning Analysis of Histopathology Images
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. TCGA Ovarian Cancer Whole-Slide Image Dataset
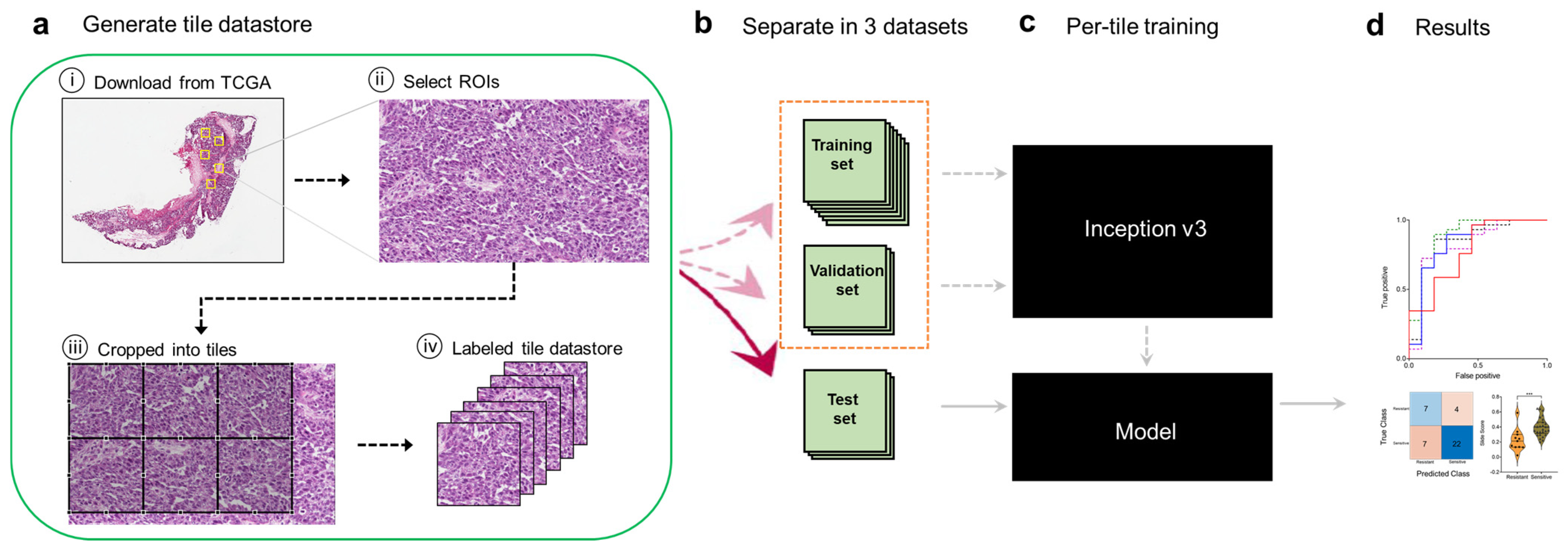
2.2. Tile Datastore Generation via Image Preprocessing
2.3. Deep Learning with Convolutional Neural Network
2.4. Training the Inception V3 Network
2.5. Statistical Analysis
2.6. Identification of Histopathologic Features Associated with Chemotherapy Response
3. Results
3.1. A Deep Learning Framework for Digital Analysis of Histopathology Images
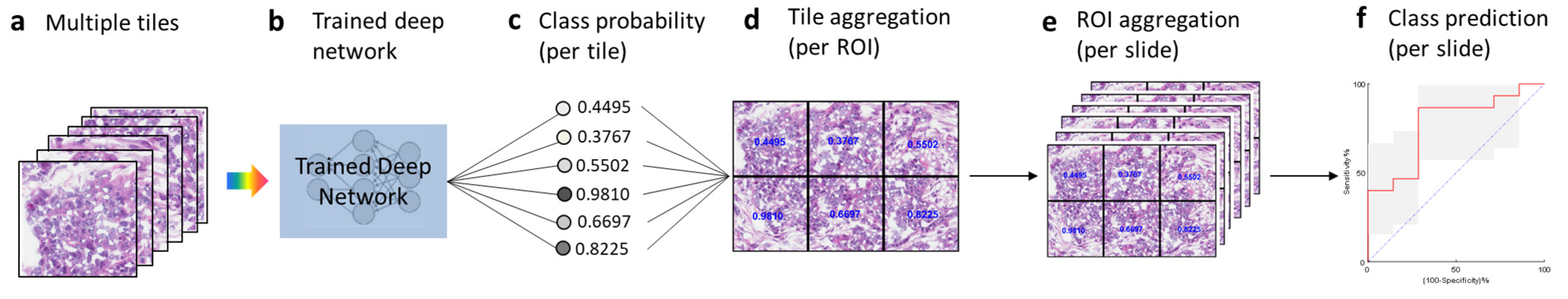
3.2. Testing and Tile Aggregation Pipeline
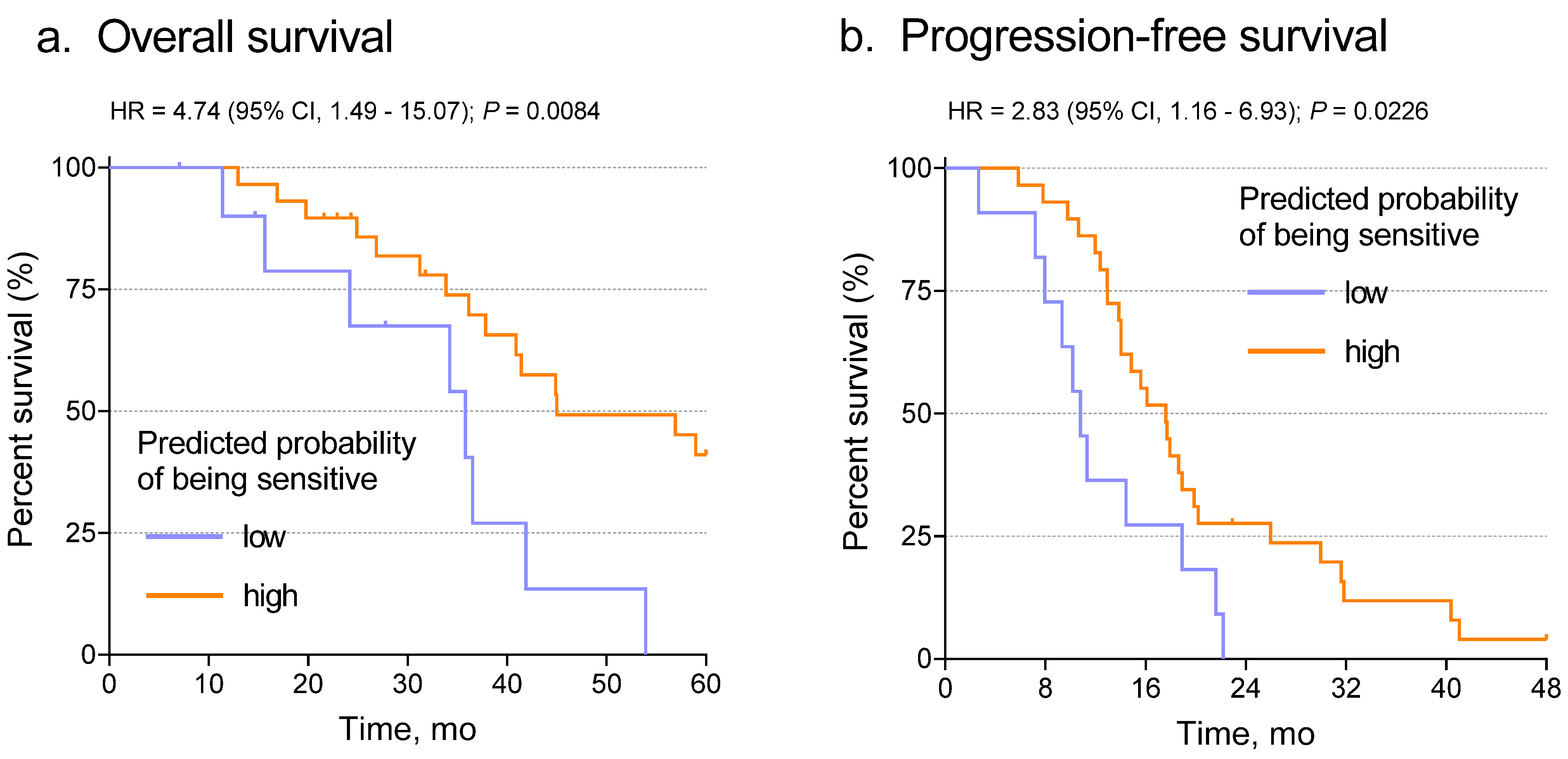
3.3. The Deep Learning Model Predicts Chemotherapy Response from Ovarian Histopathology Images
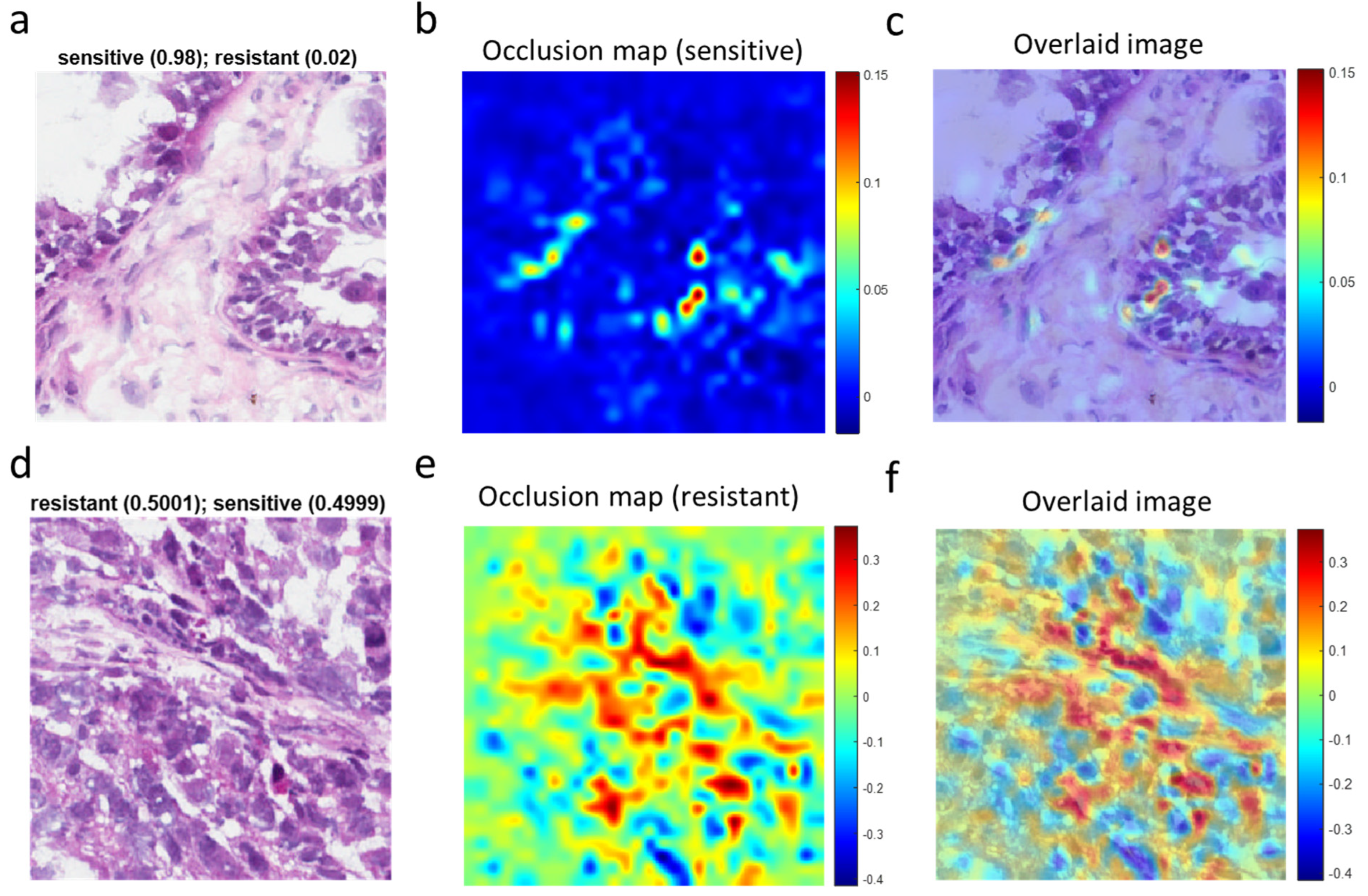
3.4. Visualization of Chemotherapy Response-Associated Features Identified by the Deep Learning Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; MIller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Cannistra, S.A. Cancer of the ovary. N. Engl. J Med. 2004, 351, 2519–2529. [Google Scholar] [CrossRef]
- Selvanayagam, Z.E.; Cheung, T.H.; Wei, N.; Chin, K.V. Prediction of chemotherapeutic response in ovarian cancer with DNA microarray expression profiling. Cancer Genet. Cytogenet. 2004, 154, 63–66. [Google Scholar] [CrossRef]
- Kulkarni, P.M.; Robinson, E.J.; Pradhan, J.S.; Gartrell-Corrado, R.D.; Rohr, B.R.; Trager, M.H.; Geskin, L.J.; Kluger, H.M.; Saenger, Y.M. Deep learning based on standard H&E images of primary melanoma tumors identifes patients at risk of visceral recurrence and death. Clin Cancer Res. 2020, 26, 1126–1134. [Google Scholar] [PubMed]
- Chetrit, A.; Hirsh-Yechezkel, G.; Ben-David, Y. Effect of BRCA1/2 mutations on long-term survival of patients with invasive ovarian cancer: The national Israeli study of ovarian cancer. J. Clin. Oncol. 2008, 26, 20–25. [Google Scholar] [CrossRef]
- Liu, Y.; Yasukawa, M.; Chen, K.; Hu, L.; Broaddus, R.R.; Ding, L.; Mardis, E.R.; Spellman, P.; Levine, D.A.; Mills, G.B.; et al. Association of Somatic Mutations of ADAMTS Genes With Chemotherapy Sensitivity and Survival in High-Grade Serous Ovarian Carcinoma. JAMA Oncol. 2015, 1, 486–494. [Google Scholar] [CrossRef]
- Yasukawa, M.; Liu, Y.; Hu, L.; Cogdell, D.; Gharpure, K.M.; Pradeep, S.; Nagaraja, A.S.; Sood, A.K.; Zhang, W. ADAMTS16 mutations sensitize ovarian cancer cells to platinum-based chemotherapy. Oncotarget 2016, 8, 88410–88420. [Google Scholar] [CrossRef]
- Huang, S.; Ingber, D.E. The structural and mechanical complexity of cell-growth control. Nat. Cell Biol. 1999, 1, E131–E138. [Google Scholar] [CrossRef] [PubMed]
- Capo-chichi, C.D.; Cai, K.Q.; Smedberg, J.; Ganjei-Azar, P.; Godwin, A.K.; Xu, X.X. Loss of A-type lamin expression compromises nuclear envelope integrity in breast cancer. Chin. J. Cancer 2011, 30, 415–425. [Google Scholar] [PubMed]
- Kilian, K.A.; Bugarija, B.; Lahn, B.T.; Mrksich, M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. USA 2010, 107, 4872–4877. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Szegedy, C. Going Deeper with Convolutions. In Proceedings of the the IEEE Conference on Computer Vision and Pattern Recogniztion, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef]
- Xing, F.; Xie, Y.; Yang, L. An automatic learning-based framework for robust nucleus segmentation. IEEE Trans. Med. Imaging 2016, 35, 550–566. [Google Scholar] [CrossRef] [PubMed]
- Simon, O.; Yacoub, R.; Jain, S.; Tomaszewski, J.E.; Sarder, P. Multi-radial LBP features as a tool for rapid glomerular detection and assessment in whole slide histopathology images. Sci. Rep. 2018, 8, 2032. [Google Scholar] [CrossRef]
- Cruz-Roa, A. Accurate and reproducible invasive breast cancer detection in whole-slide images: A deep learning approach for quantifying tumor extent. Sci. Rep. 2017, 7, 46450. [Google Scholar] [CrossRef] [PubMed]
- Sirinukunwattana, K. Locality sensitive deep learning for detection and classificaiton of nuclei in routine colon cancer histology images. IEEE Trans. Med. Imaging 2016, 35, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Linder, N.; Taylor, J.C.; Colling, R.; Pell, R.; Alveyn, E.; Joseph, J. Deep learning for detecting tumour-infiltraing lymphocytes in testicular germ cell tumours. J. Clin. Pathol. 2019, 72, 157–164. [Google Scholar] [CrossRef]
- Saltz, J.; Gupta, R.; Hou, L.; Kurc, T.; Singh, P.; Nguyen, V. Spatial organization and molecular correlaton of tumor-infiltrating lymphocytes usng deep learning on pathology images. Cell Rep. 2018, 23, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Xia, D.; Casanova, R.; Machiraju, D.; McKee, T.D.; Weder, W.; Beck, A.H. Computatoinally-guided development of a stromal inflammation histologic biomarker in lung squamous cell carcinoma. Sci. Rep. 2018, 8, 3941. [Google Scholar] [CrossRef] [PubMed]
- Ehteshami, B.B.; Mullooly, M.; Pfeiffer, R.M.; FAn, S.; Vacek, P.M.; Weaver, D.L. Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod. Pathol. 2018, 31, 1502–1512. [Google Scholar] [CrossRef]
- Arvaniti, E.; Fricker, K.S.; Moret, M.; Rupp, N.; Hermanns, T.; Frankhauser, C. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci. Rep. 2018, 8, 12054. [Google Scholar] [CrossRef] [PubMed]
- Casanova, R.; Xia, D.; Rulle, U.; Nanni, P.; Grossmann, J.; Vrugt, B. Morphoproteomic characterization of lung squamous cell carcinoma fragmentation, a histological marker of increased tumor invasiveness. Cancer Res. 2017, 77, 2585–2593. [Google Scholar] [CrossRef] [PubMed]
- Coudray, N.; Sakellaropoulos, P.S.; Sakellaropoulos, T.; Narula, N.; Snuder, M.; Fenyo, D. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, B.; Topatana, W.; Cao, J.; Zhu, H.; Juengpanich, S.; Mao, Q.; Yu, H.; Cai, X. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. npj Precis. Oncol. 2020, 4, 14. [Google Scholar]
- Wu, M.; Yan, C.; Liu, H.; Liu, Q. Automatic classification of ovarian cancer types from cytological images using deep convolutional neural networks. Biosci. Rep. 2018, 38, 29. [Google Scholar] [CrossRef]
- Shin, S.J.; You, S.C.; Jeon, H.; Jung, J.W.; An, M.H.; Park, R.W.; Roh, J. Style transfer strategy for developing a generalizable deep learning application in digital pathology. Comput. Methods Programs Biomed. 2021, 198, 105815. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.W.; Chang, C.C.; Lee, Y.C.; Lin, Y.J.; Lo, S.C.; Hsu, P.C.; Liou, Y.A.; Wang, C.H.; Chao, T.K. Weakly supervised deep learning for prediciton of treatment effectiveness on ovarian cancer from histopathology images. Comput. Medican Imaging Graph. 2022, 99, 102093. [Google Scholar] [CrossRef]
- Laury, A.R.; Blom, S.; Ropponen, T.; Virtanen, A.; Carpen, O.M. Artificial intelligence-based image analysis can predict outcome in high-grade serous carcinoma via histology alone. Sci. Rep. 2021, 11, 19165. [Google Scholar] [CrossRef]
- Yu, K.H.; Hu, V.; Wang, F.; Matulonis, U.A.; Mutter, G.I.; Golden, J.A.; Kohane, I.S. Deciphering serous ovarian carcinoma histopathology and platinum-response by convolutional neural networks. BMC Med. 2020, 18, 236. [Google Scholar] [CrossRef]
- Akazawa, M.; Hashimoto, K. Artificial intelligence in gynecologic cancers: Current status and future challenges—A systematic review. Artif. Intell. Med. 2021, 120, 102164. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, Y.; Broaddus, R.; Liu, J.; Sood, A.K.; Shmulevich, J.; Zhang, W. Integrated analysis of gene expression and tumor nuclear image profiles associated with chemotherapy response in serous ovarian carcinoma. PLoS ONE 2012, 7, e36383. [Google Scholar] [CrossRef] [PubMed]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Slens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 2818–2826. [Google Scholar]
- The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef]
- Ruopp, M.D.; Perkins, N.J.; Whitcomb, B.W.; Schisterman, E.F. Youden index and optimal cut-point estimated from observations affected by a lower limit of detection. Biom. J. 2008, 50, 419–430. [Google Scholar] [CrossRef]
- Kaplan, E.L.; Meier, P. Nonparametric estimation from incomplete observations. J. Amer. Statist. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Fergus, R.; Fleet, D. Visualizing and Understanding Convolutional Networks; Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Springer: New York, NY, USA, 2014. [Google Scholar]
- Laak, J.; Litjens, G.; Ciompi, F. Deep learning in histopathology: The path to the clinic. Nat. Med. 2021, 27, 775–784. [Google Scholar] [CrossRef]
- Weinstein, J.N. Fishing expeditions. Science 1998, 282, 628–629. [Google Scholar] [CrossRef]

| No. of Patients | 248 | ||
|---|---|---|---|
| Chemotherapy response ξ | |||
| Resistant | 74 | ||
| Sensitive | 174 | ||
| Age | |||
| Mean, years [SD] | 60.0 [11.4] | ||
| Range | 30.5–87.5 | ||
| FIGO Stage ¶ | |||
| II | 13 | ||
| III | 196 | ||
| IV | 36 | ||
| Unknown | 3 | ||
| WHO Grade | |||
| 2 | 37 | ||
| 3 | 204 | ||
| Unknown | 7 | ||
| Vital status | |||
| Alive | 94 | ||
| Dead | 150 | ||
| Unknown | 4 | ||
| Recurrent disease ζ | |||
| Yes | 216 | ||
| No | 29 | ||
| Unknown | 3 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Lawson, B.C.; Huang, X.; Broom, B.M.; Weinstein, J.N. RETRACTED: Prediction of Ovarian Cancer Response to Therapy Based on Deep Learning Analysis of Histopathology Images. Cancers 2023, 15, 4044. https://doi.org/10.3390/cancers15164044
Liu Y, Lawson BC, Huang X, Broom BM, Weinstein JN. RETRACTED: Prediction of Ovarian Cancer Response to Therapy Based on Deep Learning Analysis of Histopathology Images. Cancers. 2023; 15(16):4044. https://doi.org/10.3390/cancers15164044
Chicago/Turabian StyleLiu, Yuexin, Barrett C. Lawson, Xuelin Huang, Bradley M. Broom, and John N. Weinstein. 2023. "RETRACTED: Prediction of Ovarian Cancer Response to Therapy Based on Deep Learning Analysis of Histopathology Images" Cancers 15, no. 16: 4044. https://doi.org/10.3390/cancers15164044
APA StyleLiu, Y., Lawson, B. C., Huang, X., Broom, B. M., & Weinstein, J. N. (2023). RETRACTED: Prediction of Ovarian Cancer Response to Therapy Based on Deep Learning Analysis of Histopathology Images. Cancers, 15(16), 4044. https://doi.org/10.3390/cancers15164044






