Targeting Transcription Factor YY1 for Cancer Treatment: Current Strategies and Future Directions
Abstract
Simple Summary
Abstract
1. Introduction
2. The Roles of YY1 in Tumor Development and Progression
2.1. YY1 and Hallmarks of Cancer
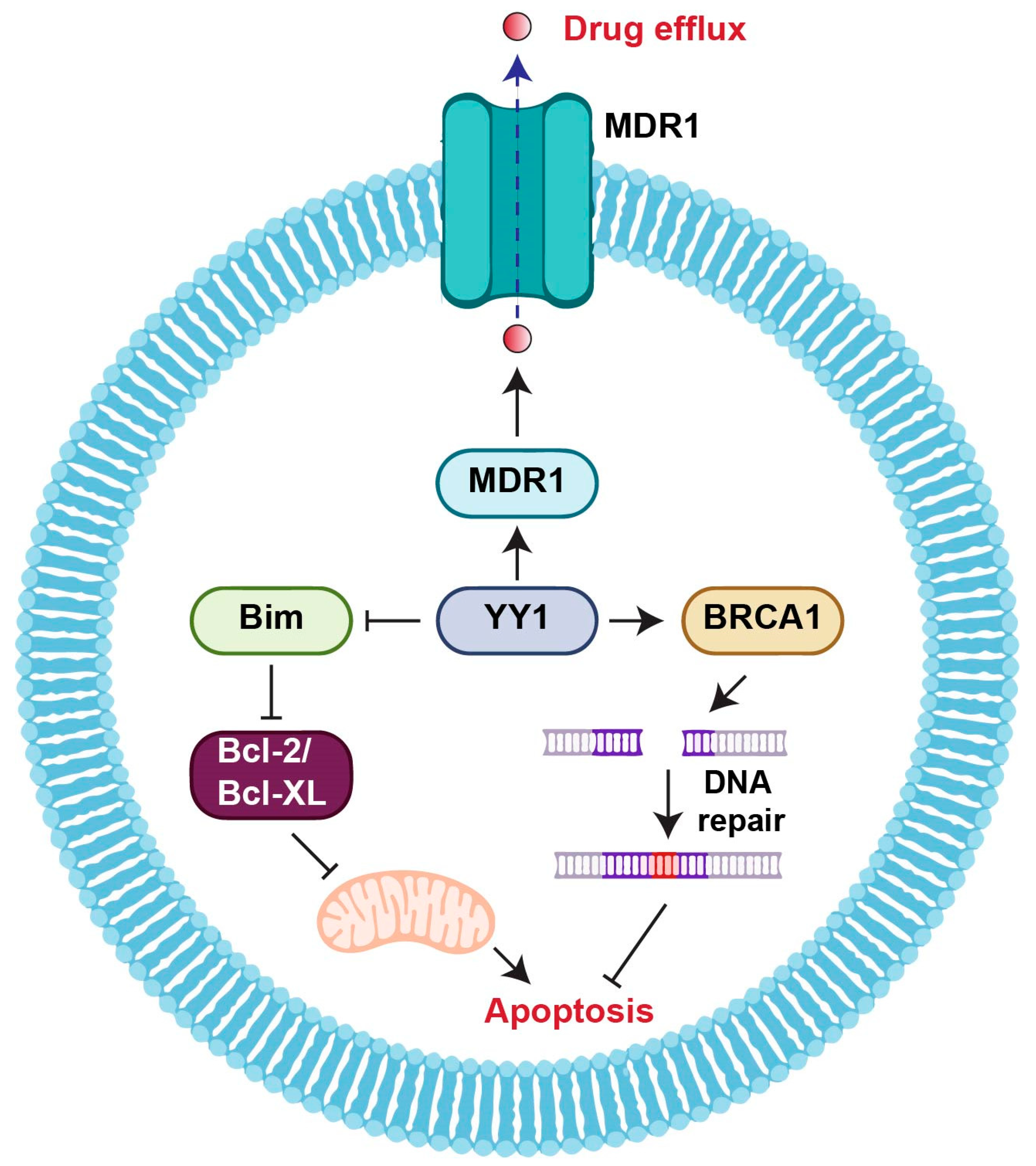
2.2. YY1 and Drug Resistance
2.3. YY1 and Cancer Stem Cells
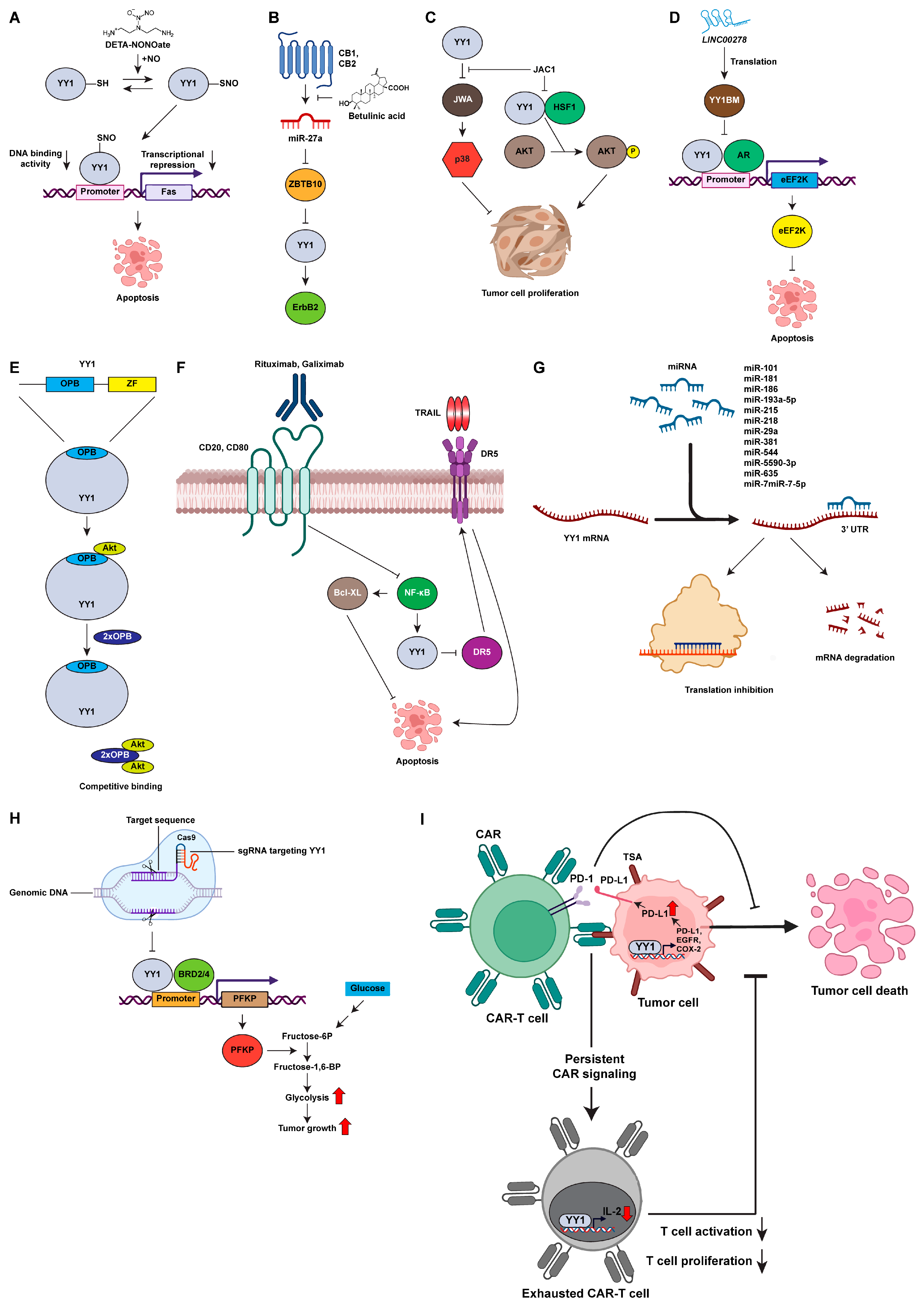
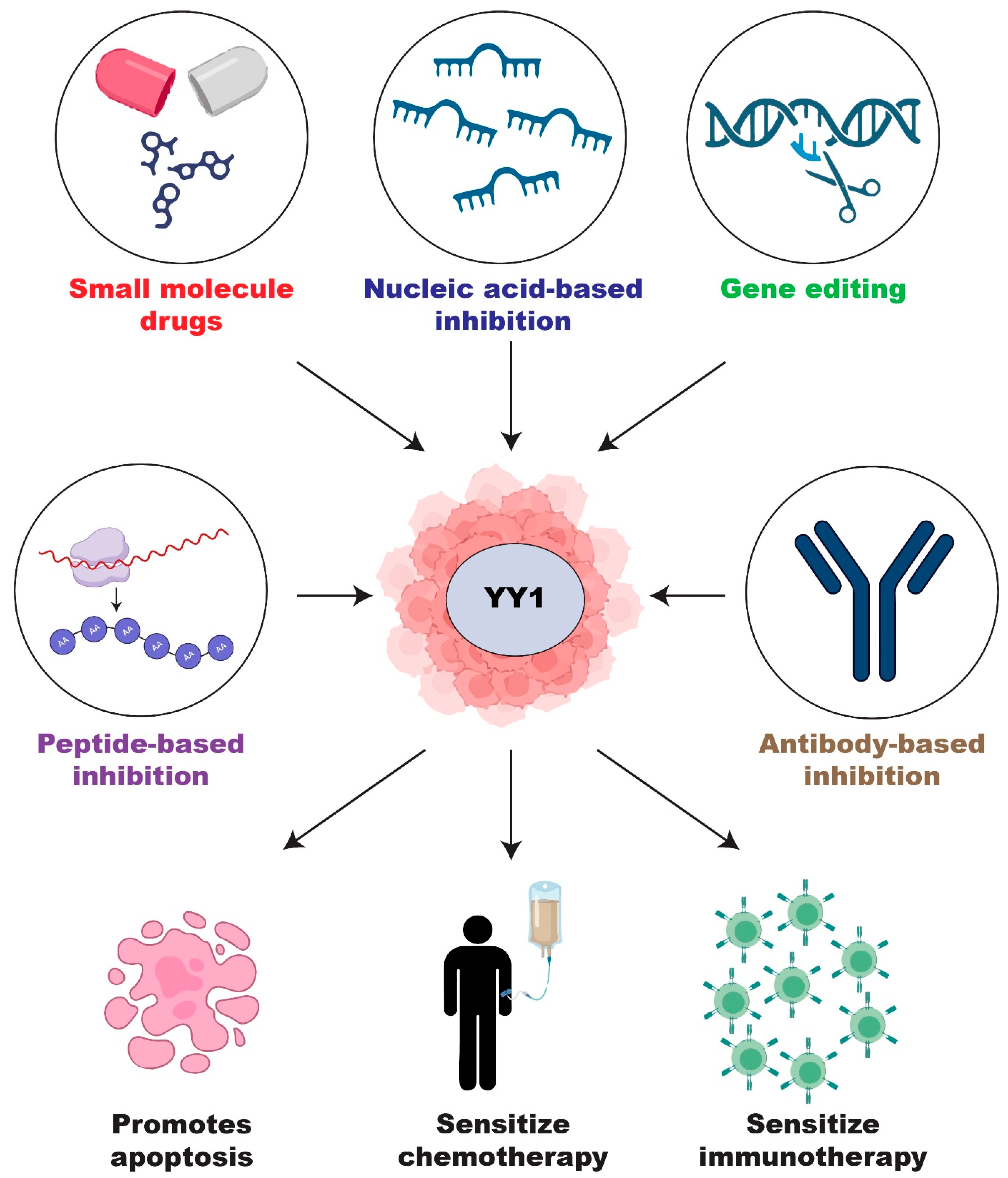
2.4. Current Development of YY1 Inhibitors
2.4.1. Small Molecule Inhibitors of YY1
2.4.2. Diethylenetriamine NONOate (DETA-NONOate)
2.4.3. Betulinic Acid
2.4.4. ADP Ribosylation Factor like GTPase 6 Interacting Protein 5 (ARL6IP5) Gene Activating Compound (JAC1)
2.4.5. Peptide-Based Inhibition
2.4.6. Antibody-Based Inhibition
2.4.7. Nucleic-Acid-Based Inhibition
2.5. CRISPR/Cas9 Genome Editing of YY1
2.6. YY1 and Immunotherapy
2.7. Other Regulatory Functions of YY1
| Result | ||
|---|---|---|
| YY1 Inhibitors | In Vitro | In Vivo |
| DETA-NONOate | Increased Fas-induced apoptosis [150] | Downregulated Bcl-xL expression in mice bearing PC-3 tumor xenograft [76] |
| Sensitized cells to TRAIL-induced apoptosis in prostate cancer cell line (DU145, PC-3, CL-1, and LNCaP) [150] | Inhibited tumor growth [76] | |
| RRx-001 | Enhanced sensitivity to radiotherapy in HT29 and SCCVII cell lines [171] | Enhanced sensitivity to radiotherapy in mouse model [171] |
| RRx-001 already passed phase I clinical trial. RRx-001 was well tolerated, with no notable toxicities nor adverse effects (NCT01359982) RRx-001 is currently in a phase 2 clinical trial [171,172,173] | ||
| Inhibited Ikβ kinase complex [172,173] | Inhibited Iκβ kinase complex [172,173] | |
| Betulinic acid | Downregulated YY1 in MDA-MB-453 cell line [75] | Downregulated YY1 in BT474 xenografted nude mice [75] |
| Downregulated YY1-dependent HER2 expression in the MDA-MB-453 cell line [75] | Decreased tumor growth [75] | |
| Induced cell cycle arrest in G2/M phase [181] | Decreased β2-microglobulin mRNA [181] | |
| Decreased cell proliferation [180,181] | Inhibited tumor growth and metastases [181,182] | |
| JAC1 | Upregulated expression of ARL6IP5 [186,188] | Inhibited formation of neo-vessels in gastric-cancer-bearing nude mice [188] |
| Downregulated HER2 expression [186] | Inhibited angiogenesis of melanoma [188] | |
| Reduced cell migration [186] | ||
| YY1BM (LINC00278) | Downregulated eEF2K; induced apoptosis of ESCC cells [194] | Increased apoptosis [194] |
| Synthetic peptides (YPB and OPB) | Disrupted YY1-EZH2 [197] | Inhibited tumor growth in xenograft of MDA-MB-231 cells [197] |
| Reduced H3K27me3 [197] | ||
| Upregulated PTENP1 and PTEN expression [197] | ||
| Inhibited cell proliferation of TNBC cell lines (MDA-MB-231 and MDA-MB-453) [197] | ||
| Reduced viability, reduced cell migration, in MDA-MB-231 [197] | ||
| Rituximab | Inhibited NF-κB and Bcl-xL activity [108,203] | Increased tumor regression [108] |
| Increased chemotherapy drug sensitivity [108,203] | ||
| Sensitized cells to immune-mediated killing [108,204] | ||
| Galiximab | Inhibited NF-κB activity [207] | Chemosensitized malignant B cells [207] |
| Reduced proliferation of B-NHL cell lines [207] | Galiximab already passed phase I/II clinical trial, result indicates that galiximab can be safely used. Galiximab is currently in phase III clinical trials [207] | |
| Sensitized resistant B cells to chemotherapy and immunotherapy [207] | ||
| Induced malignant B cell apoptosis [207] | ||
| miR-29a | Downregulated DNMT 3A and 3B in A549 cells [211] | |
| Suppressed cell proliferation and migration in A549 cells [211] | ||
| Inhibited IL-13-induced YY1 in A549 cells [211] | ||
| Inhibited tumorigenicity in A549 cells [211] | ||
| Decreased cell migration and invasion of A549 cells [211] | ||
| miR-186 | Inhibited proliferation, invasion, and migration of A549 and HCC827 cells [212] | |
| Induced apoptosis of A549 and HCC827 cells [212] | ||
| miR-181 | Reduced cell proliferation of HeLa, HeLa-229, SiHa, and C33 cells [213] | Suppressed tumor growth in nude mice with HeLa cells [213] |
| Increased cell apoptosis of HeLa, HeLa-229, SiHa, and C33 cells [213] | ||
| miR-193a-5p | Decreased cell proliferation and migration of HEC-1-A, HEC-1-B, AN3CA, RL95-2, and KLE [95] | Inhibited development and progression of primary endometrioid endometrial adenocarcinoma [95] |
| miR-215 | Suppressed cell proliferation, cell migration, and invasion in LS174T, LoVo, HT29, HCT116, SW480, and SW620 cells [214] | |
| miR-218 | Inhibited cell proliferation of U251MG and 293T cells [220] | |
| miR-381 | Inhibited cell proliferation, cell migration, and invasion of OVCAR3, Caov-3, OVCA429, SKOV3, A2780, and COV644 cells [217] | |
| miR-544 | Decreased cell viability, proliferation, and migration of SW173 and 8350C [74] | Suppressed tumorigenicity of ATC cells [74] |
| miR-5590-3p | Inhibited cell proliferation and migration of MDA-MB-436, MDA-MB-468, BT549, and MDA-MB-231 [219] | Suppressed tumor growth xenograft mice model with BT549 cell [219] |
| miR-635 | Inhibited invasion of H522 and H1299 cells [218] | Inhibited tumor growth in null mice with H522 cells [218] |
| miR-7 | Suppressed cell proliferation of HCT116, LoVo, and DLD-1 cells [40] | Suppressed tumor growth in xenograft mice model [40] |
| Induced apoptosis of HCT116, LoVo, and DLD-1 cells [40] | ||
| miR-7-5p | Sensitized LN229 cells to temozolomide [222] | Sensitized LN229 cells to chemotherapy drug temozolomide in nude mice [222] |
| Suppressed cell stemness of LN229 [222] | ||
| TargomiRs | Already passed phase I Clinical trial against malignant pleural mesothelioma and NSCLC (NCT02369198) [230]; the result indicated that TargomiRs were well tolerated in the first 5 patients and associated with transient cytokine-mediated reactions. | |
| MRG106 (Cobomarsen) | Cobomarsen already passed phase I clinical trial against lymphoma and leukemia (NCT02580552) [226]; the result indicated that cobomarsen was well tolerated, has potential clinical activity, and has the potential to improve the life quality of myelofibrosis patients. | |
| CRISPR/Cas9 | Reduced glycolysis of HEK293 and HEK293T cells [79] | Reduced cell proliferation in tumor xenograft of NOD/SCID/gamma null mice with 22Rv1 cells [79] |
| Increased apoptosis of HEK293 and HEK293T cells [79] | ||
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Mellman, I.; Coukos, G.; Dranoff, G. Cancer immunotherapy comes of age. Nature 2011, 480, 480–489. [Google Scholar] [CrossRef]
- Soerjomataram, I.; Bray, F. Planning for tomorrow: Global cancer incidence and the role of prevention 2020–2070. Nat. Rev. Clin. Oncol. 2021, 18, 663–672. [Google Scholar] [CrossRef]
- Lee, T.I.; Young, R.A. Transcriptional regulation and its misregulation in disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef]
- Darnell, J.E., Jr. Transcription factors as targets for cancer therapy. Nat. Rev. Cancer 2002, 2, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Bushweller, J.H. Targeting transcription factors in cancer—From undruggable to reality. Nat. Rev. Cancer 2019, 19, 611–624. [Google Scholar] [CrossRef]
- Henley, M.J.; Koehler, A.N. Advances in targeting ‘undruggable’transcription factors with small molecules. Nat. Rev. Drug Discov. 2021, 20, 669–688. [Google Scholar] [CrossRef]
- Wu, S.; Shi, Y.; Mulligan, P.; Gay, F.; Landry, J.; Liu, H.; Lu, J.; Qi, H.H.; Wang, W.; Nickoloff, J.A.; et al. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat. Struct. Mol. Biol. 2007, 14, 1165–1172. [Google Scholar] [CrossRef]
- Nicholson, S.; Whitehouse, H.; Naidoo, K.; Byers, R. Yin Yang 1 in Human Cancer. Crit. Rev. Oncog. 2011, 16, 245–260. [Google Scholar] [CrossRef]
- Deng, Z.; Cao, P.; Wan, M.M.; Sui, G. Yin Yang 1: A multifaceted protein beyond a transcription factor. Transcription 2010, 1, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Kwan, K.M. Yin Yang 1 is critical for mid-hindbrain neuroepithelium development and involved in cerebellar agenesis. Mol. Brain 2020, 13, 104. [Google Scholar] [CrossRef] [PubMed]
- Varum, S.; Baggiolini, A.; Zurkirchen, L.; Atak, Z.K.; Cantù, C.; Marzorati, E.; Bossart, R.; Wouters, J.; Häusel, J.; Tuncer, E. Yin Yang 1 orchestrates a metabolic program required for both neural crest development and melanoma formation. Cell Stem Cell 2019, 24, 637–653.e9. [Google Scholar] [CrossRef]
- Zurkirchen, L.; Varum, S.; Giger, S.; Klug, A.; Häusel, J.; Bossart, R.; Zemke, M.; Cantù, C.; Atak, Z.K.; Zamboni, N. Yin Yang 1 sustains biosynthetic demands during brain development in a stage-specific manner. Nat. Commun. 2019, 10, 2192. [Google Scholar] [CrossRef]
- Shi, Y.; Seto, E.; Chang, L.-S.; Shenk, T. Transcriptional repression by YY1, a human GLI-Krüippel-related protein, and relief of repression by adenovirus E1A protein. Cell 1991, 67, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Atchison, M.; Basu, A.; Zaprazna, K.; Papasani, M. Mechanisms of Yin Yang 1 in oncogenesis: The importance of indirect effects. Crit. Rev. Oncog. 2011, 16, 143–161. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, A.S.; Li, C.H.; Zamudio, A.V.; Sigova, A.A.; Hannett, N.M.; Day, D.S.; Abraham, B.J.; Cohen, M.A.; Nabet, B.; Buckley, D.L.; et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 2017, 171, 1573–1588.e28. [Google Scholar] [CrossRef] [PubMed]
- Sui, G. The regulation of YY1 in tumorigenesis and its targeting potential in cancer therapy. Mol. Cell. Pharmacol. 2009, 1, 157–176. [Google Scholar] [CrossRef]
- Gordon, S.; Akopyan, G.; Garban, H.; Bonavida, B. Transcription factor YY1: Structure, function, and therapeutic implications in cancer biology. Oncogene 2005, 25, 1125–1142. [Google Scholar] [CrossRef]
- Meliala, I.T.S.; Hosea, R.; Kasim, V.; Wu, S. The biological implications of Yin Yang 1 in the hallmarks of cancer. Theranostics 2020, 10, 4183–4200. [Google Scholar] [CrossRef]
- Cho, A.A.; Bonavida, B. Targeting the Overexpressed YY1 in Cancer Inhibits EMT and Metastasis. Crit. Rev. Oncog. 2017, 22, 49–61. [Google Scholar] [CrossRef]
- Hays, E.; Bonavida, B. YY1 regulates cancer cell immune resistance by modulating PD-L1 expression. Drug Resist. Updates 2019, 43, 10–28. [Google Scholar] [CrossRef]
- Matsumura, N.; Huang, Z.; Baba, T.; Lee, P.S.; Barnett, J.C.; Mori, S.; Chang, J.T.; Kuo, W.-L.; Gusberg, A.H.; Whitaker, R.S.; et al. Yin yang 1 modulates taxane response in epithelial ovarian cancer. Mol. Cancer Res. 2009, 7, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowska, D.; Mazur, E.; Reich, A. YY1 is a key player in melanoma immunotherapy/targeted treatment resistance. Front. Oncol. 2022, 12, 856963. [Google Scholar] [CrossRef]
- Fu, X.; Ji, F.; He, Q.; Qiu, X. A Systematic Pan-Cancer Analysis of YY1 Aberrations and their Relationship with Clinical Outcome, Tumor Microenvironment, and Therapeutic Targets. J. Immunol. Res. 2022, 2022, 5826741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Stovall, D.B.; Inoue, K.; Sui, G. The oncogenic role of Yin Yang 1. Crit. Rev. Oncog. 2011, 16, 163–197. [Google Scholar] [CrossRef]
- Bonavida, B.; Kaufhold, S. Prognostic significance of YY1 protein expression and mRNA levels by bioinformatics analysis in human cancers: A therapeutic target. Pharmacol. Ther. 2015, 150, 149–168. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Steer, C.J.; Song, G. MicroRNA-206 enhances antitumor immunity by disrupting the communication between malignant hepatocytes and regulatory T cells in c-Myc mice. Hepatology 2022, 76, 32–47. [Google Scholar] [CrossRef]
- Bonavida, B.; Huerta-Yepez, S.; Baritaki, S.; Vega, M.I.; Liu, H.; Chen, H.; Berenson, J.R. Overexpression of Yin Yang 1 in the pathogenesis of human hematopoietic malignancies. Crit. Rev. Oncog. 2011, 16, 261–267. [Google Scholar] [CrossRef]
- Tsang, D.P.F.; Wu, W.K.K.; Kang, W.; Lee, Y.-Y.; Wu, F.; Yu, Z.; Xiong, L.; Chan, A.W.; Tong, J.H.; Yang, W.; et al. Yin Yang 1-mediated epigenetic silencing of tumour-suppressive microRNAs activates nuclear factor-κB in hepatocellular carcinoma. J. Pathol. 2016, 238, 651–664. [Google Scholar] [CrossRef]
- Wan, M.; Huang, W.; Kute, T.E.; Miller, L.D.; Zhang, Q.; Hatcher, H.; Wang, J.; Stovall, D.B.; Russell, G.B.; Cao, P.D. Yin Yang 1 plays an essential role in breast cancer and negatively regulates p27. Am. J. Pathol. 2012, 180, 2120–2133. [Google Scholar] [CrossRef]
- de Nigris, F.; Zanella, L.; Cacciatore, F.; De Chiara, A.; Fazioli, F.; Chiappetta, G.; Apice, G.; Infante, T.; Monaco, M.; Rossiello, R.; et al. YY1 overexpression is associated with poor prognosis and metastasis-free survival in patients suffering osteosarcoma. BMC Cancer 2011, 11, 472. [Google Scholar] [CrossRef]
- Wang, H.; Garzon, R.; Sun, H.; Ladner, K.J.; Singh, R.; Dahlman, J.; Cheng, A.; Hall, B.M.; Qualman, S.J.; Chandler, D.S.; et al. NF-κB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 2008, 14, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Li, Y.; Wu, Z.; Wang, Y.; Xing, N.; Yang, W.; Wu, S. Transcription factor YY1 mediates epithelial-mesenchymal transition through the TGFβ signaling pathway in bladder cancer. Med. Oncol. 2020, 37, 93. [Google Scholar] [CrossRef] [PubMed]
- Qiao, K.; Ning, S.; Wan, L.; Wu, H.; Wang, Q.; Zhang, X.; Xu, S.; Pang, D. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 418. [Google Scholar] [CrossRef]
- Wang, W.; Yue, Z.; Tian, Z.; Xie, Y.; Zhang, J.; She, Y.; Yang, B.; Ye, Y.; Yang, Y. Expression of Yin Yang 1 in cervical cancer and its correlation with E-cadherin expression and HPV16 E6. PLoS ONE 2018, 13, e0193340. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Wang, Q.; Zhou, Y.; Wu, X.; Wang, L.; Duru, N.; Kong, X.; Zhang, P.; Wan, B.; Sui, L.; et al. YY1 is a novel potential therapeutic target for the treatment of HPV infection-induced cervical cancer by arsenic trioxide. Int. J. Gynecol. Cancer 2011, 21, 1097–1104. [Google Scholar] [CrossRef]
- Chinnappan, D.; Xiao, D.; Ratnasari, A.; Andry, C.; King, T.C.; Weber, C.H. Transcription factor YY1 expression in human gastrointestinal cancer cells. Int. J. Oncol. 2009, 34, 1417–1423. [Google Scholar]
- Pothoulakis, C.; Torre-Rojas, M.; Duran-Padilla, M.A.; Gevorkian, J.; Zoras, O.; Chrysos, E.; Chalkiadakis, G.; Baritaki, S. CRHR2/Ucn2 signaling is a novel regulator of miR-7/YY1/Fas circuitry contributing to reversal of colorectal cancer cell resistance to Fas-mediated apoptosis. Int. J. Cancer 2018, 142, 334–346. [Google Scholar] [CrossRef]
- Zhang, N.; Li, X.; Wu, C.W.; Dong, Y.; Cai, M.; Mok, M.T.S.; Wang, H.; Chen, J.; Ng, S.S.M.; Chen, M.; et al. microRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene 2012, 32, 5078–5088. [Google Scholar] [CrossRef]
- Zhu, G.; Qian, M.; Lu, L.; Chen, Y.; Zhang, X.; Wu, Q.; Liu, Y.; Bian, Z.; Yang, Y.; Guo, S.; et al. O-GlcNAcylation of YY1 stimulates tumorigenesis in colorectal cancer cells by targeting SLC22A15 and AANAT. Carcinogenesis 2019, 40, 1121–1131. [Google Scholar] [CrossRef]
- Luo, J.; Jiang, X.; Cao, L.; Dai, K.; Zhang, S.; Ge, X.; Zhou, X.; Lu, X. Expression of YY1 correlates with progression and metastasis in esophageal squamous cell carcinomas. OncoTargets Ther. 2014, 7, 1753–1759. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Chen, Y.; Ye, L.; Jiao, W.; Song, H.; Mei, H.; Li, D.; Yang, F.; Li, H.; Huang, K.; et al. miRNA-584-3p inhibits gastric cancer progression by repressing Yin Yang 1- facilitated MMP-14 expression. Sci. Rep. 2017, 7, 8967. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.; Tong, J.H.M.; Chan, A.W.H.; Zhao, J.; Dong, Y.; Wang, S.; Yang, W.; Sin, F.M.C.; Ng, S.S.M.; Yu, J.; et al. Yin Yang 1 contributes to gastric carcinogenesis and its nuclear expression correlates with shorter survival in patients with early stage gastric adenocarcinoma. J. Transl. Med. 2014, 12, 80. [Google Scholar] [CrossRef]
- Baritaki, S.; Chatzinikola, A.M.; Vakis, A.F.; Soulitzis, N.; Karabetsos, D.A.; Neonakis, I.; Bonavida, B.; Spandidos, D.A. YY1 Over-Expression in Human Brain Gliomas and Meningiomas Correlates with TGF-β1, IGF-1 and FGF-2 mRNA Levels. Cancer Investig. 2009, 27, 184–192. [Google Scholar] [CrossRef]
- Waters, M.R.; Gupta, A.S.; Mockenhaupt, K.; Brown, L.N.; Biswas, D.D.; Kordula, T. RelB acts as a molecular switch driving chronic inflammation in glioblastoma multiforme. Oncogenesis 2019, 8, 37. [Google Scholar] [CrossRef]
- Li, J.; Song, J.; Guo, F. miR-186 reverses cisplatin resistance and inhibits the formation of the glioblastoma-initiating cell phenotype by degrading Yin Yang 1 in glioblastoma. Int. J. Mol. Med. 2018, 43, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Dukers, D.F.; van Galen, J.C.; Giroth, C.; Jansen, P.; Sewalt, R.G.A.B.; Otte, A.P.; Kluin-Nelemans, H.C.; Meijer, C.J.L.M.; Raaphorst, F.M. Unique polycomb gene expression pattern in Hodgkin’s lymphoma and Hodgkin’s lymphoma-derived cell lines. Am. J. Pathol. 2004, 164, 873–881. [Google Scholar] [CrossRef]
- Li, Z.-J.; Cheng, J.; Song, Y.; Li, H.-H.; Zheng, J.-F. LncRNA SNHG5 upregulation induced by YY1 contributes to angiogenesis via miR-26b/CTGF/VEGFA axis in acute myelogenous leukemia. Lab. Investig. 2021, 101, 341–352. [Google Scholar] [CrossRef]
- Antonio-Andres, G.; Martinez-Ruiz, G.U.; Morales-Martinez, M.; Jimenez-Hernandez, E.; Martinez-Torres, E.; Lopez-Perez, T.V.; Estrada-Abreo, L.A.; Patino-Lopez, G.; Juarez-Mendez, S.; Davila-Borja, V.M.; et al. Transcriptional Regulation of Yin-Yang 1 Expression through the Hypoxia Inducible Factor-1 in Pediatric Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2022, 23, 1728. [Google Scholar] [CrossRef]
- Antonio-Andrés, G.; Rangel-Santiago, J.; Tirado-Rodríguez, B.; Martinez-Ruiz, G.U.; Klunder-Klunder, M.; Vega, M.I.; Lopez-Martinez, B.; Jiménez-Hernández, E.; Torres Nava, J.; Medina-Sanson, A.; et al. Role of Yin Yang-1 (YY1) in the transcription regulation of the multi-drug resistance (MDR1) gene. Leuk. Lymphoma 2018, 59, 2628–2638. [Google Scholar] [CrossRef]
- Kim, J.S.; Son, S.H.; Kim, M.Y.; Choi, D.; Jang, I.-S.; Paik, S.S.; Chae, J.H.; Uversky, V.N.; Kim, C.G. Diagnostic and prognostic relevance of CP2c and YY1 expression in hepatocellular carcinoma. Oncotarget 2017, 8, 24389–24400. [Google Scholar] [CrossRef]
- Gao, D.; Wang, L.; Zhang, H.; Yan, X.; Yang, J.; Zhou, R.; Chang, X.; Sun, Y.; Tian, S.; Yao, Z.; et al. Spleen tyrosine kinase SYK(L) interacts with YY1 and coordinately suppresses SNAI2 transcription in lung cancer cells. FEBS J. 2018, 285, 4229–4245. [Google Scholar] [CrossRef]
- Zhao, G.; Li, Q.; Wang, A.; Jiao, J. YY1 regulates melanoma tumorigenesis through a miR-9 ~ RYBP axis. J. Exp. Clin. Cancer Res. 2015, 34, 66. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.B.; Majumder, P.; Cooper, J.C.; Yoon, H.; Wade, P.A.; Boss, J.M. Yin yang 1 regulates the expression of snail through a distal enhancer. Mol. Cancer Res. 2009, 7, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Ren, W.; Yao, F.; Wang, H.; Zhang, K.; Luo, M.; Shang, Y.; O’Connell, D.; Bei, Z.; Wang, H.; et al. YY1 cooperates with TFEB to regulate autophagy and lysosomal biogenesis in melanoma. Mol. Carcinog. 2019, 58, 2149–2160. [Google Scholar] [CrossRef]
- Huerta-Yepez, S.; Liu, H.; Baritaki, S.; Del Lourdes Cebrera-MuÑOz, M.; Rivera-Pazos, C.; Maldonado-Valenzuela, A.; Valencia-Hipolito, A.; Vega, M.I.; Chen, H.; Berenson, J.R.; et al. Overexpression of Yin Yang 1 in bone marrow-derived human multiple myeloma and its clinical significance. Int. J. Oncol. 2014, 45, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, Y.; Wei, Y.; Wu, C.; Meng, H.; Niu, W.; Zhou, Y.; Wang, H.; Wen, Q.; Fan, S.; et al. Zinc-finger protein YY1 suppresses tumor growth of human nasopharyngeal carcinoma by inactivating c-Myc-mediated microRNA-141 transcription. J. Biol. Chem. 2019, 294, 6172–6187. [Google Scholar] [CrossRef]
- Hafsi, S.; Candido, S.; Maestro, R.; Falzone, L.; Soua, Z.; Bonavida, B.; Spandidos, D.A.; Libra, M. Correlation between the overexpression of Yin Yang 1 and the expression levels of miRNAs in Burkitt’s lymphoma: A computational study. Oncol. Lett. 2016, 11, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Castellano, G.; Torrisi, E.; Ligresti, G.; Nicoletti, F.; Malaponte, G.; Traval, S.; McCubrey, J.A.; Canevari, S.; Libra, M. Yin Yang 1 overexpression in diffuse large B-cell lymphoma is associated with B-cell transformation and tumor progression. Cell Cycle 2010, 9, 557–563. [Google Scholar] [CrossRef]
- Vivarelli, S.; Falzone, L.; Ligresti, G.; Candido, S.; Garozzo, A.; Magro, G.G.; Bonavida, B.; Libra, M. Role of the transcription factor Yin Yang 1 and its selectively identified target Survivin in high-grade B-cells non-Hodgkin lymphomas: Potential diagnostic and therapeutic targets. Int. J. Mol. Sci. 2020, 21, 6446. [Google Scholar] [CrossRef] [PubMed]
- Sakhinia, E.; Glennie, C.; Hoyland, J.A.; Menasce, L.P.; Brady, G.; Miller, C.; Radford, J.A.; Byers, R.J. Clinical quantitation of diagnostic and predictive gene expression levels in follicular and diffuse large B-cell lymphoma by RT-PCR gene expression profiling. Blood 2007, 109, 3922–3928. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, K.; Clay, V.; Hoyland, J.A.; Swindell, R.; Linton, K.; Illidge, T.; Radford, J.A.; Byers, R.J. YY1 expression predicts favourable outcome in follicular lymphoma. J. Clin. Pathol. 2010, 64, 125–129. [Google Scholar] [CrossRef]
- Qian, S.; Wang, W.; Li, M. Transcriptional factor Yin Yang 1 facilitates the stemness of ovarian cancer via suppressing miR-99a activity through enhancing its deacetylation level. Biomed. Pharmacother. 2020, 126, 110085. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, X.; Zhu, L.; Yang, Y.; Yin, X. YY1-Induced lncRNA PART1 Enhanced Resistance of Ovarian Cancer Cells to Cisplatin by Regulating miR-512-3p/CHRAC1 Axis. DNA Cell Biol. 2021, 40, 821–832. [Google Scholar] [CrossRef]
- Jiang, W.; Zhao, S.; Shen, J.; Guo, L.; Sun, Y.; Zhu, Y.; Ma, Z.; Zhang, X.; Hu, Y.; Xiao, W.; et al. The MiR-135b-BMAL1-YY1 loop disturbs pancreatic clockwork to promote tumourigenesis and chemoresistance. Cell Death Dis. 2018, 9, 149. [Google Scholar] [CrossRef]
- Huang, Y.; Tao, T.; Liu, C.; Guan, H.; Zhang, G.; Ling, Z.; Zhang, L.; Lu, K.; Chen, S.; Xu, B.; et al. Upregulation of miR-146a by YY1 depletion correlates with delayed progression of prostate cancer. Int. J. Oncol. 2017, 50, 421–431. [Google Scholar] [CrossRef]
- Zapata-Tarres, M.; Juarez-Villegas, L.E.; Maldonado-Valenzuela, A.; Baay-Guzman, G.J.; Lopez-Perez, T.V.; Cabrera-Muñoz, L.; Sadowinski-Pine, S.; Huerta-Yepez, S. Expression of YY1 in Wilms tumors with favorable histology is a risk factor for adverse outcomes. Future Oncol. 2019, 15, 1231–1241. [Google Scholar] [CrossRef]
- de Nigris, F.; Botti, C.; de Chiara, A.; Rossiello, R.; Apice, G.; Fazioli, F.; Fiorito, C.; Sica, V.; Napoli, C. Expression of transcription factor Yin Yang 1 in human osteosarcomas. Eur. J. Cancer 2006, 42, 2420–2424. [Google Scholar] [CrossRef]
- Gashaw, I.; Grümmer, R.; Klein-Hitpass, L.; Dushaj, O.; Bergmann, M.; Brehm, R.; Grobholz, R.; Kliesch, S.; Neuvians, T.P.; Schmid, K.W.; et al. Gene signatures of testicular seminoma with emphasis on expression of ets variant gene 4. Cell. Mol. Life Sci. 2005, 62, 2359–2368. [Google Scholar] [CrossRef]
- Zaravinos, A.; Spandidos, D.A. Yin yang 1 expression in human tumors. Cell Cycle 2010, 9, 512–522. [Google Scholar] [CrossRef]
- Arribas, J.; Castellví, J.; Marcos, R.; Zafón, C.; Velázquez, A. Expression of YY1 in Differentiated Thyroid Cancer. Endocr. Pathol. 2015, 26, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Huang, W.; Wu, X.; Gao, Y.; Ou, J.; Zhang, X.; Li, Y. MiR-141-3p Suppresses Tumor Growth and Metastasis in Papillary Thyroid Cancer via Targeting Yin Yang 1. Anat. Rec. 2019, 302, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, Z.; Sun, B. miR-544 inhibits the migration and invasion of anaplastic thyroid cancer by targeting Yin Yang-1. Oncol. Lett. 2019, 17, 2983–2992. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jutooru, I.; Lei, P.; Kim, K.; Lee, S.-o.; Brents, L.K.; Prather, P.L.; Safe, S. Betulinic Acid Targets YY1 and ErbB2 through Cannabinoid Receptor-Dependent Disruption of MicroRNA-27a: ZBTB10 in Breast CancerBetulinic Acid Downregulates ErbB2 in Breast Cancer Cells. Mol. Cancer Ther. 2012, 11, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Yepez, S.; Baritaki, S.; Baay-Guzman, G.; Hernandez-Luna, M.A.; Hernandez-Cueto, A.; Vega, M.I.; Bonavida, B. Contribution of either YY1 or BclXL-induced inhibition by the NO-donor DETANONOate in the reversal of drug resistance, both in vitro and in vivo. YY1 and BclXL are overexpressed in prostate cancer. Nitric Oxide 2013, 29, 17–24. [Google Scholar] [CrossRef]
- Rafii, S.; Tashkandi, E.; Bukhari, N.; Al-Shamsi, H.O. Current status of CRISPR/Cas9 application in clinical cancer research: Opportunities and challenges. Cancers 2022, 14, 947. [Google Scholar] [CrossRef]
- Breier, D.; Peer, D. Genome editing in cancer: Challenges and potential opportunities. Bioact. Mater. 2023, 21, 394–402. [Google Scholar] [CrossRef]
- Xu, C.; Tsai, Y.-H.; Galbo Jr, P.M.; Gong, W.; Storey, A.J.; Xu, Y.; Byrum, S.D.; Xu, L.; Whang, Y.E.; Parker, J.S. Cistrome analysis of YY1 uncovers a regulatory axis of YY1: BRD2/4-PFKP during tumorigenesis of advanced prostate cancer. Nucleic Acids Res. 2021, 49, 4971–4988. [Google Scholar] [CrossRef]
- Sarvagalla, S.; Kolapalli, S.P.; Vallabhapurapu, S. The Two Sides of YY1 in Cancer: A Friend and a Foe. Front. Oncol. 2019, 9, 1230. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of cancer: New dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Kasim, V.; Kano, M.R.; Tanaka, S.; Ohba, S.; Miura, Y.; Miyata, K.; Liu, X.; Matsuhashi, A.; Chung, U.-i.; et al. Transcription Factor YY1 Contributes to Tumor Growth by Stabilizing Hypoxia Factor HIF-1α in a p53-Independent Manner. Cancer Res. 2013, 73, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, H.; Li, Y.; Xie, Y.; Huang, C.; Zhao, H.; Miyagishi, M.; Kasim, V. Transcription Factor YY1 Promotes Cell Proliferation by Directly Activating the Pentose Phosphate Pathway. Cancer Res. 2018, 78, 4549–4562. [Google Scholar] [CrossRef] [PubMed]
- Santiago, F.; Ishii, H.; Shafi, S.; Khurana, R.; Kanellakis, P.; Bhindi, R.; Ramirez, M.; Bobik, A.; Martin, J.; Chesterman, C.; et al. Yin Yang-1 inhibits vascular smooth muscle cell growth and intimal thickening by repressing p21WAF1/Cip1 transcription and p21WAF1/Cip1-Cdk4-cyclin D1 assembly. Circ. Res. 2007, 101, 146–155. [Google Scholar] [CrossRef]
- Riggs, K.J.; Saleque, S.; Wong, K.K.; Merrell, K.T.; Lee, J.S.; Shi, Y.; Calame, K. Yin-yang 1 activates the c-myc promoter. Mol. Cell. Biol. 1993, 13, 7487–7495. [Google Scholar] [CrossRef] [PubMed]
- Sui, G.; El Bachir, A.; Shi, Y.; Brignone, C.; Wall, N.R.; Yin, P.; Donohoe, M.; Luke, M.P.; Calvo, D.; Grossman, S.R.; et al. Yin Yang 1 Is a Negative Regulator of p53. Cell 2004, 117, 859–872. [Google Scholar] [CrossRef]
- Grönroos, E.; Terentiev, A.A.; Punga, T.; Ericsson, J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc. Natl. Acad. Sci. USA 2004, 101, 12165–12170. [Google Scholar] [CrossRef] [PubMed]
- Sui, Y.; Wu, T.; Li, F.; Wang, F.; Cai, Y.; Jin, J. YY1/BCCIP Coordinately Regulates P53-Responsive Element (p53RE)-Mediated Transactivation of p21(Waf1/Cip1). Int. J. Mol. Sci. 2019, 20, 2095. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, S.; Huang, C.; Li, Y.; Zhao, H.; Kasim, V. Yin Yang 1 promotes the Warburg effect and tumorigenesis via glucose transporter GLUT3. Cancer Sci. 2018, 109, 2423–2434. [Google Scholar] [CrossRef]
- Li, Y.; Kasim, V.; Yan, X.; Li, L.; Meliala, I.T.S.; Huang, C.; Li, Z.; Lei, K.; Song, G.; Zheng, X.; et al. Yin Yang 1 facilitates hepatocellular carcinoma cell lipid metabolism and tumor progression by inhibiting PGC-1β-induced fatty acid oxidation. Theranostics 2019, 9, 7599–7615. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Li, Z.; Wei, M.; Zhao, H.; Miyagishi, M.; Wu, S.; Kasim, V. Homeostasis Imbalance of YY2 and YY1 Promotes Tumor Growth by Manipulating Ferroptosis. Adv. Sci. 2022, 9, e2104836. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, Y.; Zhou, C.; Mei, W.; Zeng, C. PI3K/Akt/mTOR Pathway and Its Role in Cancer Therapeutics: Are We Making Headway? Front. Oncol. 2022, 12, 819128. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, L.; Lu, L.; Wang, L.; Li, X.; Jiang, P.; Chan, L.K.Y.; Zhang, T.; Yu, J.; Kwong, J.; et al. A novel miR-193a-5p-YY1-APC regulatory axis in human endometrioid endometrial adenocarcinoma. Oncogene 2012, 32, 3432–3442. [Google Scholar] [CrossRef]
- Potluri, V.; Noothi, S.K.; Vallabhapurapu, S.D.; Yoon, S.-O.; Driscoll, J.J.; Lawrie, C.H.; Vallabhapurapu, S. Transcriptional repression of Bim by a novel YY1-RelA complex is essential for the survival and growth of Multiple Myeloma. PLoS ONE 2013, 8, e66121. [Google Scholar] [CrossRef]
- Wang, X.; Feng, Y.; Xu, L.; Chen, Y.; Zhang, Y.; Su, D.; Ren, G.; Lu, J.; Huang, B. YY1 restrained cell senescence through repressing the transcription of p16. Biochim. Biophys. Acta 2008, 1783, 1876–1883. [Google Scholar] [CrossRef]
- Shalapour, S.; Karin, M. Immunity, inflammation, and cancer: An eternal fight between good and evil. J. Clin. Investig. 2015, 125, 3347–3355. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, J.; Wu, Y.; Shi, G.; Yuan, H.; Lu, Z.; Zhu, Q.; Wu, P.; Lu, C.; Guo, F.; et al. YY1 suppresses proliferation and migration of pancreatic ductal adenocarcinoma by regulating the CDKN3/MdM2/P53/P21 signaling pathway. Int. J. Cancer. 2017, 142, 1392–1404. [Google Scholar] [CrossRef]
- Miller, D.M.; Thomas, S.D.; Islam, A.; Muench, D.; Sedoris, K. c-Myc and cancer metabolism. Clin. Cancer Res. 2012, 18, 5546–5553. [Google Scholar] [CrossRef]
- Joo, M.; Wright, J.G.; Hu, N.N.; Sadikot, R.T.; Park, G.Y.; Blackwell, T.S.; Christman, J.W. Yin Yang 1 enhances cyclooxygenase-2 gene expression in macrophages. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007, 292, L1219–L1226. [Google Scholar] [CrossRef]
- Pu, D.; Yin, L.; Huang, L.; Qin, C.; Zhou, Y.; Wu, Q.; Li, Y.; Zhou, Q.; Li, L. Cyclooxygenase-2 Inhibitor: A Potential Combination Strategy With Immunotherapy in Cancer. Front. Oncol. 2021, 11, 637504. [Google Scholar] [CrossRef]
- de Nigris, F.; Crudele, V.; Giovane, A.; Casamassimi, A.; Giordano, A.; Garban, H.J.; Cacciatore, F.; Pentimalli, F.; Marquez-Garban, D.C.; Petrillo, A.; et al. CXCR4/YY1 inhibition impairs VEGF network and angiogenesis during malignancy. Proc. Natl. Acad. Sci. USA 2010, 107, 14484–14489. [Google Scholar] [CrossRef]
- Chen, Y.; Jacamo, R.; Konopleva, M.; Garzon, R.; Croce, C.; Andreeff, M. CXCR4 downregulation of let-7a drives chemoresistance in acute myeloid leukemia. J. Clin. Investig. 2013, 123, 2395–2407. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, J.; Wang, S.; Luo, X.; Li, Y.; Lv, Z.; Zhu, J.; Lin, J.; Ding, L.; Ye, Q. The DEK oncogene activates VEGF expression and promotes tumor angiogenesis and growth in HIF-1α-dependent and -independent manners. Oncotarget 2016, 7, 23740–23756. [Google Scholar] [CrossRef]
- Sitwala, K.V.; Adams, K.; Markovitz, D.M. YY1 and NF-Y binding sites regulate the transcriptional activity of the dek and dek-can promoter. Oncogene 2002, 21, 8862–8870. [Google Scholar] [CrossRef]
- Baritaki, S.; Huerta-Yepez, S.; Sakai, T.; Spandidos, D.A.; Bonavida, B. Chemotherapeutic drugs sensitize cancer cells to TRAIL-mediated apoptosis: Up-regulation of DR5 and inhibition of Yin Yang 1. Mol. Cancer Ther. 2007, 6, 1387–1399. [Google Scholar] [CrossRef]
- Bonavida, B. Rituximab-induced inhibition of antiapoptotic cell survival pathways: Implications in chemo/immunoresistance, rituximab unresponsiveness, prognostic and novel therapeutic interventions. Oncogene 2007, 26, 3629–3636. [Google Scholar] [CrossRef]
- Huang, H.-C.; Chao, C.-C.; Wu, P.-H.; Chung, H.-Y.; Lee, H.-Y.; Suen, C.-S.; Hwang, M.-J.; Cai, B.-H.; Kannagi, R. Epigenetic silencing of the synthesis of immunosuppressive Siglec ligand glycans by NF-κB/EZH2/YY1 axis in early-stage colon cancers. Biochim. Biophys. Acta 2019, 1862, 173–183. [Google Scholar] [CrossRef]
- Yao, R.; Jiang, H.; Ma, Y.; Wang, L.; Wang, L.; Du, J.; Hou, P.; Gao, Y.; Zhao, L.; Wang, G.; et al. PRMT7 Induces Epithelial-to-Mesenchymal Transition and Promotes Metastasis in Breast Cancer. Cancer Res. 2014, 74, 5656–5667. [Google Scholar] [CrossRef]
- Yin, D.; Ogawa, S.; Kawamata, N.; Leiter, A.; Ham, M.; Li, D.; Doan, N.B.; Said, J.W.; Black, K.L.; Phillip Koeffler, H. miR-34a functions as a tumor suppressor modulating EGFR in glioblastoma multiforme. Oncogene 2013, 32, 1155–1163. [Google Scholar] [CrossRef]
- Tseng, H.Y.; Chen, Y.A.; Jen, J.; Shen, P.C.; Chen, L.M.; Lin, T.D.; Wang, Y.C.; Hsu, H.L. Oncogenic MCT-1 activation promotes YY1-EGFR-MnSOD signaling and tumor progression. Oncogenesis 2017, 6, e313. [Google Scholar] [CrossRef]
- Allouche, A.; Nolens, G.; Tancredi, A.; Delacroix, L.; Mardaga, J.; Fridman, V.; Winkler, R.; Boniver, J.; Delvenne, P.; Begon, D.Y. The combined immunodetection of AP-2alpha and YY1 transcription factors is associated with ERBB2 gene overexpression in primary breast tumors. Breast Cancer Res. 2008, 10, R9. [Google Scholar] [CrossRef]
- Macheda, M.L.; Rogers, S.; Best, J.D. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J. Cell. Physiol. 2004, 202, 654–662. [Google Scholar] [CrossRef]
- Hyuga, S.; Wada, H.; Eguchi, H.; Otsuru, T.; Iwgami, Y.; Yamada, D.; Noda, T.; Asaoka, T.; Kawamoto, K.; Gotoh, K.; et al. Expression of carbonic anhydrase IX is associated with poor prognosis through regulation of the epithelial-mesenchymal transition in hepatocellular carcinoma. Int. J. Oncol. 2017, 51, 1179–1190. [Google Scholar] [CrossRef]
- Chafe, S.C.; McDonald, P.C.; Saberi, S.; Nemirovsky, O.; Venkateswaran, G.; Burugu, S.; Gao, D.; Delaidelli, A.; Kyle, A.H.; Baker, J.H.E.; et al. Targeting Hypoxia-Induced Carbonic Anhydrase IX Enhances Immune-Checkpoint Blockade Locally and Systemically. Cancer Immunol. Res. 2019, 7, 1064–1078. [Google Scholar] [CrossRef]
- Fu, Q.; Yu, Z. Phosphoglycerate kinase 1 (PGK1) in cancer: A promising target for diagnosis and therapy. Life Sci. 2020, 256, 117863. [Google Scholar] [CrossRef]
- Yang, T.; An, Z.; Zhang, C.; Wang, Z.; Wang, X.; Liu, Y.; Du, E.; Liu, R.; Zhang, Z.; Xu, Y. HnRNPM is a potential mediator of YY1 which promotes EMT in prostate cancer cells. Prostate 2019, 79, 1199–1210. [Google Scholar] [CrossRef]
- Pentland, I.; Campos-León, K.; Cotic, M.; Davies, K.-J.; Wood, C.D.; Groves, I.J.; Burley, M.; Coleman, N.; Stockton, J.D.; Noyvert, B.; et al. Disruption of CTCF-YY1-dependent looping of the human papillomavirus genome activates differentiation-induced viral oncogene transcription. PLoS Biol. 2018, 16, e2005752. [Google Scholar] [CrossRef]
- Morales-Martinez, M.; Valencia-Hipolito, A.; Vega, G.G.; Neri, N.; Nambo, M.J.; Alvarado, I.; Cuadra, I.; Duran-Padilla, M.A.; Martinez-Maza, O.; Huerta-Yepez, S.; et al. Regulation of Krüppel-Like Factor 4 (KLF4) expression through the transcription factor Yin-Yang 1 (YY1) in non-Hodgkin B-cell lymphoma. Oncotarget 2019, 10, 2173–2188. [Google Scholar] [CrossRef]
- Zhao, J.-L.; Huang, F.; He, F.; Gao, C.-C.; Liang, S.-Q.; Ma, P.-F.; Dong, G.-Y.; Han, H.; Qin, H.-Y. Forced Activation of Notch in Macrophages Represses Tumor Growth by Upregulating miR-125a and Disabling Tumor-Associated Macrophages. Cancer Res. 2016, 76, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.H.; Lai, M.C.; Zheng, Y.Y.; Sun, Y.W.; Qiu, J.J.; Gui, F.; Zhang, Q.; Liu, F. MiR-195 inhibits the ubiquitination and degradation of YY1 by Smurf2, and induces EMT and cell permeability of retinal pigment epithelial cells. Cell Death Dis. 2021, 12, 708. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, J.-J.; Peng, Y.-P.; Zhu, Y.; Yin, L.-D.; Wei, J.-S.; Gao, W.-T.; Jiang, K.-R.; Miao, Y. A Yin-Yang 1/miR-30a regulatory circuit modulates autophagy in pancreatic cancer cells. J. Transl. Med. 2017, 15, 211. [Google Scholar] [CrossRef]
- Feng, L.; Ma, Y.; Sun, J.; Shen, Q.; Liu, L.; Lu, H.; Wang, F.; Yue, Y.; Li, J.; Zhang, S.; et al. YY1-MIR372-SQSTM1 regulatory axis in autophagy. Autophagy 2014, 10, 1442–1453. [Google Scholar] [CrossRef]
- Bensaad, K.; Tsuruta, A.; Selak, M.A.; Vidal, M.N.C.; Nakano, K.; Bartrons, R.; Gottlieb, E.; Vousden, K.H. TIGAR, a p53-Inducible Regulator of Glycolysis and Apoptosis. Cell 2006, 126, 107–120. [Google Scholar] [CrossRef]
- Ramkumar, C.; Cui, H.; Kong, Y.; Jones, S.N.; Gerstein, R.M.; Zhang, H. Smurf2 suppresses B-cell proliferation and lymphomagenesis by mediating ubiquitination and degradation of YY1. Nat. Commun. 2013, 4, 2598. [Google Scholar] [CrossRef]
- Jeong, H.M.; Lee, S.H.; Yum, J.; Yeo, C.Y.; Lee, K.Y. Smurf2 regulates the degradation of YY1. Biochim. Biophys. Acta 2014, 1843, 2005–2011. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Murai, S.; Kataoka, K.; Miyagishi, M. Yin Yang 1 induces transcriptional activity of p73 through cooperation with E2F1. Biochem. Biophys. Res. Commun. 2008, 365, 75–81. [Google Scholar] [CrossRef]
- Zhan, S.; Wang, T.; Ge, W.; Li, J. Multiple roles of Ring 1 and YY1 binding protein in physiology and disease. J. Cell. Mol. Med. 2018, 22, 2046–2054. [Google Scholar] [CrossRef]
- Bajusz, I.; Henry, S.; Sutus, E.; Kovács, G.; Pirity, M.K. Evolving Role of RING1 and YY1 Binding Protein in the Regulation of Germ-Cell-Specific Transcription. Genes 2019, 10, 941. [Google Scholar] [CrossRef]
- Chen, Q.; Yang, C.; Chen, L.; Zhang, J.-J.; Ge, W.-L.; Yuan, H.; Meng, L.-D.; Huang, X.-M.; Shen, P.; Miao, Y.; et al. YY1 targets tubulin polymerisation-promoting protein to inhibit migration, invasion and angiogenesis in pancreatic cancer via p38/MAPK and PI3K/AKT pathways. Br. J. Cancer 2019, 121, 912–921. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Li, Z.; Qin, R.; Wang, X.; An, H.; Wang, Y.; Zhu, Y.; Liu, Y.; Cai, S.; Chen, S.; et al. YY1 Promotes Endothelial Cell-Dependent Tumor Angiogenesis in Hepatocellular Carcinoma by Transcriptionally Activating VEGFA. Front. Oncol. 2019, 9, 1187. [Google Scholar] [CrossRef] [PubMed]
- Kojima, T.; Nakahama, K.-I.; Yamamoto, K.; Uematsu, H.; Morita, I. Age-and cell cycle-dependent changes in EPC-1/PEDF promoter activity in human diploid fibroblast-like (HDF) cells. Mol. Cell. Biochem. 2006, 293, 63–69. [Google Scholar] [CrossRef]
- Zhang, J.-J.; Zhu, Y.; Xie, K.-L.; Peng, Y.-P.; Tao, J.-Q.; Tang, J.; Li, Z.; Xu, Z.-K.; Dai, C.-C.; Qian, Z.-Y.; et al. Yin Yang-1 suppresses invasion and metastasis of pancreatic ductal adenocarcinoma by downregulating MMP10 in a MUC4/ErbB2/p38/MEF2C-dependent mechanism. Mol. Cancer 2014, 13, 130. [Google Scholar] [CrossRef]
- Wang, W.; Li, D.; Sui, G. YY1 Is an Inducer of Cancer Metastasis. Crit. Rev. Oncog. 2017, 22, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.-J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Bracken, C.P.; Goodall, G.J. The many regulators of epithelial− mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2022, 23, 89–90. [Google Scholar] [CrossRef]
- Han, J.; Meng, J.; Chen, S.; Wang, X.; Yin, S.; Zhang, Q.; Liu, H.; Qin, R.; Li, Z.; Zhong, W.; et al. YY1 Complex Promotes Quaking Expression via Super-Enhancer Binding during EMT of Hepatocellular Carcinoma. Cancer Res. 2019, 79, 1451–1464. [Google Scholar] [CrossRef]
- Hwang, S.S.; Kim, Y.U.; Lee, S.; Jang, S.W.; Kim, M.K.; Koh, B.H.; Lee, W.; Kim, J.; Souabni, A.; Busslinger, M. Transcription factor YY1 is essential for regulation of the Th2 cytokine locus and for Th2 cell differentiation. Proc. Natl. Acad. Sci. USA 2013, 110, 276–281. [Google Scholar] [CrossRef]
- Hwang, S.S.; Jang, S.W.; Kim, M.K.; Kim, L.K.; Kim, B.-S.; Kim, H.S.; Kim, K.; Lee, W.; Flavell, R.A.; Lee, G.R. YY1 inhibits differentiation and function of regulatory T cells by blocking Foxp3 expression and activity. Nat. Commun. 2016, 7, 10789. [Google Scholar] [CrossRef]
- Banerjee, A.; Sindhava, V.; Vuyyuru, R.; Jha, V.; Hodewadekar, S.; Manser, T.; Atchison, M.L. YY1 is required for germinal center B cell development. PLoS ONE 2016, 11, e0155311. [Google Scholar] [CrossRef]
- Elinav, E.; Nowarski, R.; Thaiss, C.A.; Hu, B.; Jin, C.; Flavell, R.A. Inflammation-induced cancer: Crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer 2013, 13, 759–771. [Google Scholar] [CrossRef]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.A.; Fawell, S.; Floc’h, N.; Flemington, V.; McKerrecher, D.; Smith, P.D. Challenges and opportunities in cancer drug resistance. Chem. Rev. 2020, 121, 3297–3351. [Google Scholar] [CrossRef] [PubMed]
- Garbán, H.J.; Bonavida, B. Nitric Oxide Inhibits the Transcription Repressor Yin-Yang 1 Binding Activity at the Silencer Region of the Fas Promoter: A Pivotal Role for Nitric Oxide in the Up-Regulation of Fas Gene Expression in Human Tumor Cells. J. Immunol. 2001, 167, 75–81. [Google Scholar] [CrossRef]
- Van Nguyen, T.; Puebla-Osorio, N.; Pang, H.; Dujka, M.E.; Zhu, C. DNA damage-induced cellular senescence is sufficient to suppress tumorigenesis: A mouse model. J. Exp. Med. 2007, 204, 1453–1461. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Lahusen, T.; Wang, R.; Xiao, C.; Xu, X.; Hwang, Y.; He, W.; Shi, Y.; Deng, C. Yin Yang 1 positively regulates BRCA1 and inhibits mammary cancer formation. Oncogene 2012, 31, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Hays, E.; Bonavida, B. Nitric Oxide-Mediated Enhancement and Reversal of Resistance of Anticancer Therapies. Antioxidants 2019, 8, 407. [Google Scholar] [CrossRef]
- Bonavida, B. Sensitizing activities of nitric oxide donors for cancer resistance to anticancer therapeutic drugs. Biochem. Pharmacol. 2020, 176, 113913. [Google Scholar] [CrossRef]
- Huerta-Yepez, S.; Vega, M.; Jazirehi, A.; Garban, H.; Hongo, F.; Cheng, G.; Bonavida, B. Nitric oxide sensitizes prostate carcinoma cell lines to TRAIL-mediated apoptosis via inactivation of NF-κB and inhibition of Bcl-xL expression. Oncogene 2004, 23, 4993–5003. [Google Scholar] [CrossRef]
- Stewart, G.D.; Nanda, J.; Katz, E.; Bowman, K.J.; Christie, J.G.; Brown, D.G.; McLaren, D.B.; Riddick, A.C.; Ross, J.A.; Jones, G.D. DNA strand breaks and hypoxia response inhibition mediate the radiosensitisation effect of nitric oxide donors on prostate cancer under varying oxygen conditions. Biochem. Pharmacol. 2011, 81, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Bonavida, B.; Garban, H. Nitric oxide-mediated sensitization of resistant tumor cells to apoptosis by chemo-immunotherapeutics. Redox Biol. 2015, 6, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Bjerkvig, R.; Tysnes, B.B.; Aboody, K.S.; Najbauer, J.; Terzis, A. The origin of the cancer stem cell: Current controversies and new insights. Nat. Rev. Cancer 2005, 5, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Lobo, N.A.; Shimono, Y.; Qian, D.; Clarke, M.F. The Biology of Cancer Stem Cells. Annu. Rev. Cell Dev. Biol. 2007, 23, 675–699. [Google Scholar] [CrossRef]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target Ther. 2020, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Kaufhold, S.; Garbán, H.; Bonavida, B. Yin Yang 1 is associated with cancer stem cell transcription factors (SOX2, OCT4, BMI1) and clinical implication. J. Exp. Clin. Cancer Res. 2016, 35, 84. [Google Scholar] [CrossRef]
- Guo, Q.; Wang, T.; Yang, Y.; Gao, L.; Zhao, Q.; Zhang, W.; Xi, T.; Zheng, L. Transcriptional factor Yin Yang 1 promotes the stemness of breast cancer cells by suppressing miR-873-5p transcriptional activity. Mol. Ther. Nucleic Acids 2020, 21, 527–541. [Google Scholar] [CrossRef]
- You, J.; Tao, B.; Peng, L.; Peng, T.; He, H.; Zeng, S.; Han, J.; Chen, L.; Xia, X.; Yang, X. Transcription factor YY1 mediates self-renewal of glioblastoma stem cells through regulation of the SENP1/METTL3/MYC axis. Cancer Gene Ther. 2022, 30, 683–693. [Google Scholar] [CrossRef]
- Bonavida, B. Overexpression of YY1 Regulates the Resistance of Cancer Stem Cells: Targeting YY1. In Cancer Stem Cell Resistance to Targeted Therapy; Springer: Cham, Switzerland, 2019; pp. 93–113. [Google Scholar]
- Li, H.; Li, T.; Huang, D.; Zhang, P. Long noncoding RNA SNHG17 induced by YY1 facilitates the glioma progression through targeting miR-506-3p/CTNNB1 axis to activate Wnt/β-catenin signaling pathway. Cancer Cell Int. 2020, 20, 29. [Google Scholar] [CrossRef]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T. Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal Transduct. Target Ther. 2021, 6, 201. [Google Scholar] [CrossRef]
- Bedard, P.L.; Hyman, D.M.; Davids, M.S.; Siu, L.L. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet 2020, 395, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Bonavida, B. Therapeutic YY1 Inhibitors in Cancer: ALL in ONE. Crit. Rev. Oncog. 2017, 22, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Hongo, F.; Garban, H.; Huerta-Yepez, S.; Vega, M.; Jazirehi, A.R.; Mizutani, Y.; Miki, T.; Bonavida, B. Inhibition of the transcription factor Yin Yang 1 activity by S-nitrosation. Biochem. Biophys. Res. Commun. 2005, 336, 692–701. [Google Scholar] [CrossRef]
- Sato, Y.; Yoshino, H.; Tsuruga, E.; Kashiwakura, I. Fas ligand enhances apoptosis of human lung cancer cells cotreated with RIG-I-like receptor agonist and radiation. Curr. Cancer Drug Targets 2020, 20, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, B.A.; El-Deiry, W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef]
- Yasuda, H. Solid tumor physiology and hypoxia-induced chemo/radio-resistance: Novel strategy for cancer therapy: Nitric oxide donor as a therapeutic enhancer. Nitric Oxide 2008, 19, 205–216. [Google Scholar] [CrossRef]
- Ribeiro, E.; Costa, B.; Vasques-Nóvoa, F.; Vale, N. In Vitro Drug Repurposing: Focus on Vasodilators. Cells 2023, 12, 671. [Google Scholar] [CrossRef]
- Reynaert, N.L.; Ckless, K.; Korn, S.H.; Vos, N.; Guala, A.S.; Wouters, E.F.; van der Vliet, A.; Janssen-Heininger, Y.M. Nitric oxide represses inhibitory κB kinase through S-nitrosylation. Proc. Natl. Acad. Sci. USA 2004, 101, 8945–8950. [Google Scholar] [CrossRef]
- Kelleher, Z.T.; Matsumoto, A.; Stamler, J.S.; Marshall, H.E. NOS2 regulation of NF-κB by S-nitrosylation of p65. J. Biol. Chem. 2007, 282, 30667–30672. [Google Scholar] [CrossRef]
- Scicinski, J.; Oronsky, B.; Ning, S.; Knox, S.; Peehl, D.; Kim, M.M.; Langecker, P.; Fanger, G. NO to cancer: The complex and multifaceted role of nitric oxide and the epigenetic nitric oxide donor, RRx-001. Redox Biol. 2015, 6, 1–8. [Google Scholar] [CrossRef]
- Oronsky, B.; Scicinski, J.; Ning, S.; Peehl, D.; Oronsky, A.; Cabrales, P.; Bednarski, M.; Knox, S. RRx-001, A novel dinitroazetidine radiosensitizer. Investig. New Drugs 2016, 34, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Oronsky, B.; Caroen, S.; Abrouk, N.; Reid, T.R. RRx-001 and the “Right stuff”: Protection and treatment in outer space. Life Sci. Space Res. 2022, 35, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Alakurtti, S.; Mäkelä, T.; Koskimies, S.; Yli-Kauhaluoma, J. Pharmacological properties of the ubiquitous natural product betulin. Eur. J. Pharm. Sci. 2006, 29, 1–13. [Google Scholar] [CrossRef]
- Fulda, S. Betulinic Acid for cancer treatment and prevention. Int. J. Mol. Sci. 2008, 9, 1096–1107. [Google Scholar] [CrossRef]
- Pisha, E.; Chai, H.; Lee, I.-S.; Chagwedera, T.E.; Farnsworth, N.R.; Cordell, G.A.; Beecher, C.W.W.; Fong, H.H.S.; Kinghorn, A.D.; Brown, D.M.; et al. Discovery of betulinic acid as a selective inhibitor of human melanoma that functions by induction of apoptosis. Nat. Med. 1995, 1, 1046–1051. [Google Scholar] [CrossRef]
- Fulda, S.; Kroemer, G. Targeting mitochondrial apoptosis by betulinic acid in human cancers. Drug Discov. Today 2009, 14, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Fulda, S.; Scaffidi, C.; Susin, S.A.; Krammer, P.H.; Kroemer, G.; Peter, M.E.; Debatin, K.-M. Activation of Mitochondria and Release of Mitochondrial Apoptogenic Factors by Betulinic Acid. J. Biol. Chem. 1998, 273, 33942–33948. [Google Scholar] [CrossRef]
- Chintharlapalli, S.; Papineni, S.; Lei, P.; Pathi, S.; Safe, S. Betulinic acid inhibits colon cancer cell and tumor growth and induces proteasome-dependent and -independent downregulation of specificity proteins (Sp) transcription factors. BMC Cancer 2011, 11, 371. [Google Scholar] [CrossRef]
- Begon, D.Y.; Delacroix, L.; Vernimmen, D.; Jackers, P.; Winkler, R. Yin Yang 1 cooperates with activator protein 2 to stimulate ERBB2 gene expression in mammary cancer cells. J. Biol. Chem. 2005, 280, 24428–24434. [Google Scholar] [CrossRef]
- Mertens-Talcott, S.U.; Noratto, G.D.; Li, X.; Angel-Morales, G.; Bertoldi, M.C.; Safe, S. Betulinic acid decreases ER-negative breast cancer cell growth in vitro and in vivo: Role of Sp transcription factors and microRNA-27a: ZBTB10. Mol. Carcinog. 2013, 52, 591–602. [Google Scholar] [CrossRef]
- Sawada, N.; Kataoka, K.; Kondo, K.; Arimochi, H.; Fujino, H.; Takahashi, Y.; Miyoshi, T.; Kuwahara, T.; Monden, Y.; Ohnishi, Y. Betulinic acid augments the inhibitory effects of vincristine on growth and lung metastasis of B16F10 melanoma cells in mice. Br. J. Cancer 2004, 90, 1672–1678. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Li, X.; Dong, S.; Zhou, W. Betulinic acid in the treatment of tumour diseases: Application and research progress. Biomed. Pharmacother. 2021, 142, 111990. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Ren, Y.; Shu, C.; Chen, D.; Liu, X.; Liang, Y.; Li, A.; Zhou, J. JAC1 targets YY1 mediated JWA/p38 MAPK signaling to inhibit proliferation and induce apoptosis in TNBC. Cell Death Discov. 2022, 8, 169. [Google Scholar] [CrossRef]
- Ren, Y.; Chen, D.; Zhai, Z.; Chen, J.; Li, A.; Liang, Y.; Zhou, J. JAC1 suppresses proliferation of breast cancer through the JWA/p38/SMURF1/HER2 signaling. Cell Death Discov. 2021, 7, 85. [Google Scholar] [CrossRef]
- Qian, J.; Zhu, W.; Wang, K.; Ma, L.; Xu, J.; Xu, T.; Røe, O.D.; Li, A.; Zhou, J.; Shu, Y. JWA loss promotes cell migration and cytoskeletal rearrangement by affecting HER2 expression and identifies a high-risk subgroup of HER2-positive gastric carcinoma patients. Oncotarget 2016, 7, 36865. [Google Scholar] [CrossRef]
- Wang, S.; Gong, Z.; Chen, R.; Liu, Y.; Li, A.; Li, G.; Zhou, J. JWA regulates XRCC1 and functions as a novel base excision repair protein in oxidative-stress-induced DNA single-strand breaks. Nucleic Acids Res. 2009, 37, 1936–1950. [Google Scholar] [CrossRef]
- Ding, K.; Liu, X.; Wang, L.; Zou, L.; Jiang, X.; Li, A.; Zhou, J. Targeting JWA for Cancer Therapy: Functions, Mechanisms and Drug Discovery. Cancers 2022, 14, 4655. [Google Scholar] [CrossRef] [PubMed]
- Setrerrahmane, S.; Li, M.; Zoghbi, A.; Lv, X.; Zhang, S.; Zhao, W.; Lu, J.; Craik, D.J.; Xu, H. Cancer-related micropeptides encoded by ncRNAs: Promising drug targets and prognostic biomarkers. Cancer Lett. 2022, 547, 215723. [Google Scholar] [CrossRef]
- Xing, J.; Liu, H.; Jiang, W.; Wang, L. LncRNA-encoded peptide: Functions and predicting methods. Front. Oncol. 2021, 10, 622294. [Google Scholar] [CrossRef]
- Pueyo, J.I.; Magny, E.G.; Couso, J.P. New peptides under the s (ORF) ace of the genome. Trends Biochem. Sci. 2016, 41, 665–678. [Google Scholar] [CrossRef]
- Pauli, A.; Valen, E.; Schier, A.F. Identifying (non-) coding RNAs and small peptides: Challenges and opportunities. Bioessays 2015, 37, 103–112. [Google Scholar] [CrossRef]
- Chen, Y.; Ho, L.; Tergaonkar, V. sORF-Encoded MicroPeptides: New players in inflammation, metabolism, and precision medicine. Cancer Lett. 2021, 500, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, L.; Deng, J.; Guo, B.; Li, F.; Wang, Y.; Wu, R.; Zhang, S.; Lu, J.; Zhou, Y. A Novel Micropeptide Encoded by Y-Linked LINC00278 Links Cigarette Smoking and AR Signaling in Male Esophageal Squamous Cell CarcinomaRole of Micropeptide Encoded by lncRNA in Male ESCC. Cancer Res. 2020, 80, 2790–2803. [Google Scholar] [CrossRef] [PubMed]
- Qiao, S.; Wang, W.; Yi, C.; Xu, Q.; Wang, W.; Shi, J.; Stovall, D.B.; Li, D.; Sui, G. YY1 Oligomerization Is Regulated by Its OPB Domain and Competes with Its Regulation of Oncoproteins. Cancers 2022, 14, 1611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wan, M.; Shi, J.; Horita, D.A.; Miller, L.D.; Kute, T.E.; Kridel, S.J.; Kulik, G.; Sui, G. Yin Yang 1 promotes mTORC2-mediated AKT phosphorylation. J. Mol. Cell Biol. 2016, 8, 232–243. [Google Scholar] [CrossRef]
- Yi, C.; Li, G.; Wang, W.; Sun, Y.; Zhang, Y.; Zhong, C.; Stovall, D.B.; Li, D.; Shi, J.; Sui, G. Disruption of YY1-EZH2 interaction using synthetic peptides inhibits breast cancer development. Cancers 2021, 13, 2402. [Google Scholar] [CrossRef]
- Qi, Y.; Yan, T.; Chen, L.; Zhang, Q.; Wang, W.; Han, X.; Li, D.; Shi, J.; Sui, G. Characterization of YY1 OPB peptide for its anticancer activity. Curr. Cancer Drug Targets 2019, 19, 504–511. [Google Scholar] [CrossRef]
- Wang, H.; Hertlein, E.; Bakkar, N.; Sun, H.; Acharyya, S.; Wang, J.; Carathers, M.; Davuluri, R.; Guttridge, D.C. NF-κB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol. Cell. Biol. 2007, 27, 4374–4387. [Google Scholar] [CrossRef]
- Zinatizadeh, M.R.; Schock, B.; Chalbatani, G.M.; Zarandi, P.K.; Jalali, S.A.; Miri, S.R. The Nuclear Factor Kappa B (NF-κB) signaling in cancer development and immune diseases. Genes Dis. 2021, 8, 287–297. [Google Scholar] [CrossRef]
- Focosi, D.; Tuccori, M.; Maggi, F. Progressive multifocal leukoencephalopathy and anti-CD20 monoclonal antibodies: What do we know after 20 years of rituximab. Rev. Med. Virol. 2019, 29, e2077. [Google Scholar] [CrossRef]
- Byrd, J.C.; Kitada, S.; Flinn, I.W.; Aron, J.L.; Pearson, M.; Lucas, D.; Reed, J.C. The mechanism of tumor cell clearance by rituximab in vivo in patients with B-cell chronic lymphocytic leukemia: Evidence of caspase activation and apoptosis induction. Blood 2002, 99, 1038–1043. [Google Scholar] [CrossRef]
- Jazirehi, A.R.; Huerta-Yepez, S.; Cheng, G.; Bonavida, B. Rituximab (chimeric anti-CD20 monoclonal antibody) inhibits the constitutive nuclear factor-κB signaling pathway in non-Hodgkin’s lymphoma B-cell lines: Role in sensitization to chemotherapeutic drug-induced apoptosis. Cancer Res. 2005, 65, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Vega, M.I.; Jazirehi, A.R.; Huerta-Yepez, S.; Bonavida, B. Rituximab-induced inhibition of YY1 and Bcl-xL expression in Ramos non-Hodgkin’s lymphoma cell line via inhibition of NF-κB activity: Role of YY1 and Bcl-xL in Fas resistance and chemoresistance, respectively. J. Immunol. 2005, 175, 2174–2183. [Google Scholar] [CrossRef] [PubMed]
- Czuczman, M.; Leonard, J.; Jung, S.; Johnson, J.; Hsi, E.; Byrd, J.; Cheson, B. Phase II trial of galiximab (anti-CD80 monoclonal antibody) plus rituximab (CALGB 50402): Follicular Lymphoma International Prognostic Index (FLIPI) score is predictive of upfront immunotherapy responsiveness. Ann. Oncol. 2012, 23, 2356–2362. [Google Scholar] [CrossRef] [PubMed]
- Baritaki, S.; Suzuki, E.; Vega, M.; Ho, S.; Hariharan, K.; Bonavida, B. Galiximab Sensitizes Malignant Human B Cell Lines to Apoptosis by Chemotherapeutic Drugs. Blood 2007, 110, 3591. [Google Scholar] [CrossRef]
- Martinez-Paniagua, M.A.; Vega, M.I.; Huerta-Yepez, S.; Baritaki, S.; Vega, G.G.; Hariharan, K.; Bonavida, B. Galiximab Signals B-NHL Cells and Inhibits the Activities of NF-κB–Induced YY1-and Snail-Resistant Factors: Mechanism of Sensitization to Apoptosis by Chemoimmunotherapeutic Drugs. Mol. Cancer Ther. 2012, 11, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Entezari, M.; Taheriazam, A.; Orouei, S.; Fallah, S.; Sanaei, A.; Hejazi, E.S.; Kakavand, A.; Rezaei, S.; Heidari, H.; Behroozaghdam, M.; et al. LncRNA-miRNA axis in tumor progression and therapy response: An emphasis on molecular interactions and therapeutic interventions. Biomed. Pharmacother. 2022, 154, 113609. [Google Scholar] [CrossRef] [PubMed]
- Reda El Sayed, S.; Cristante, J.; Guyon, L.; Denis, J.; Chabre, O.; Cherradi, N. MicroRNA therapeutics in cancer: Current advances and challenges. Cancers 2021, 13, 2680. [Google Scholar] [CrossRef]
- Zhang, Y.; He, S.; Mei, R.; Kang, Y.; Duan, J.; Wei, R.; Xiang, C.; Wu, Y.; Lu, X.; Cai, Z.; et al. miR-29a suppresses IL-13-induced cell invasion by inhibiting YY1 in the AKT pathway in lung adenocarcinoma A549 cells. Oncol. Rep. 2018, 39, 2613–2623. [Google Scholar] [CrossRef]
- Huang, T.; Wang, G.; Yang, L.; Peng, B.; Wen, Y.; Ding, G.; Wang, Z. MiR-186 inhibits proliferation, migration, and invasion of non-small cell lung cancer cells by downregulating Yin Yang 1. Cancer Biomark. 2017, 21, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.-Y.; Chen, J.-C.; Jiao, T.-T.; Hui, N.; Qi, X. MicroRNA-181 targets Yin Yang 1 expression and inhibits cervical cancer progression. Mol. Med. Rep. 2015, 11, 4541–4546. [Google Scholar] [CrossRef]
- Chen, Z.; Han, S.; Huang, W.; Wu, J.; Liu, Y.; Cai, S.; He, Y.; Wu, S.; Song, W. MicroRNA-215 suppresses cell proliferation, migration and invasion of colon cancer by repressing Yin-Yang 1. Biochem. Biophys. Res. Commun. 2016, 479, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Ge, X.I.N.; Geng, Y.; Cao, H.A.N.; Zhu, W.E.I.; Jiao, Y.; Wu, J.; Zhou, J.; Cao, J. miR-34a inhibits the migration and invasion of esophageal squamous cell carcinoma by targeting Yin Yang-1. Oncol. Rep. 2015, 34, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.-M.; Huang, T.-T.; Hsu, K.-W.; Huang, K.-H.; Fang, W.-L.; Yang, M.-H.; Lo, S.-S.; Chi, C.-W.; Lin, J.-J.; Yeh, T.-S. Yin Yang 1 is a target of microRNA-34 family and contributes to gastric carcinogenesis. Oncotarget 2014, 5, 5002–5016. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Li, H.; Yang, S.; Liu, T.; Lou, G. MiR-381 inhibits epithelial ovarian cancer malignancy via YY1 suppression. Tumor Biol. 2016, 37, 9157–9167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, Z.; Zhang, Y.; Fu, T.; Liu, C.; Liu, Y.; Lin, Y. The microRNA-635 suppresses tumorigenesis in non-small cell lung cancer. Biomed. Pharmacother. 2016, 84, 1274–1281. [Google Scholar] [CrossRef]
- Liang, F.; Fu, X.; Wang, L. miR-5590-3p-YY1 feedback loop promotes the proliferation and migration of triple-negative breast cancer cells. J. Cell. Biochem. 2019, 120, 18415–18424. [Google Scholar] [CrossRef]
- Gao, Y.; Sun, L.; Wu, Z.; Xuan, C.; Zhang, J.; You, Y.; Chen, X. miR-218 inhibits the proliferation of human glioma cells through downregulation of Yin Yang 1. Mol. Med. Rep. 2018, 17, 1926–1932. [Google Scholar] [CrossRef]
- Zhou, G.; Han, F.; Shi, Z.; Yu, L.; Li, X.; Yu, C.; Shen, C.; Wan, D.; Zhu, X.; Li, R. DNMT3A-mediated down-regulation of microRNA-105 promotes gastric cancer cell proliferation. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3377–3383. [Google Scholar]
- Jia, B.; Liu, W.; Gu, J.; Wang, J.; Lv, W.; Zhang, W.; Hao, Q.; Pang, Z.; Mu, N.; Zhang, W.; et al. MiR-7-5p suppresses stemness and enhances temozolomide sensitivity of drug-resistant glioblastoma cells by targeting Yin Yang 1. Exp. Cell Res. 2019, 375, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Liu, L.; Zhang, Y.; Wang, J.; Zhao, Y. Long noncoding RNA NPCCAT1 promotes nasopharyngeal carcinoma progression via upregulating YY1. Biochimie 2019, 157, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Rong, D.; Dong, Q.; Qu, H.; Deng, X.; Gao, F.; Li, Q.; Sun, P. m(6)A-induced LINC00958 promotes breast cancer tumorigenesis via the miR-378a-3p/YY1 axis. Cell Death Discov. 2021, 7, 27. [Google Scholar] [CrossRef]
- Cheng, Z.; Liu, G.; Huang, C.; Zhao, X. KLF5 activates lncRNA DANCR and inhibits cancer cell autophagy accelerating gastric cancer progression. NPJ Genom. Med. 2021, 6, 75. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, Y.; Lin, M.; Wu, D.; Zhao, M. LncRNA HOTAIR promotes proliferation and inhibits apoptosis by sponging miR-214-3p in HPV16 positive cervical cancer cells. Cancer Cell Int. 2021, 21, 400. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Thomson, S.B.; Chen, S.; Leavitt, B.R.; Cullis, P.R.; van der Meel, R. The current landscape of nucleic acid therapeutics. Nat. Nanotechnol. 2021, 16, 630–643. [Google Scholar] [CrossRef]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.-J. Therapeutic siRNA: State of the art. Signal Transduct. Target Ther. 2020, 5, 101. [Google Scholar] [CrossRef]
- Cuciniello, R.; Filosa, S.; Crispi, S. Novel approaches in cancer treatment: Preclinical and clinical development of small non-coding RNA therapeutics. J. Exp. Clin. Cancer Res. 2021, 40, 383. [Google Scholar] [CrossRef]
- Kao, S.C.; Fulham, M.; Wong, K.; Cooper, W.; Brahmbhatt, H.; MacDiarmid, J.; Pattison, S.; Sagong, J.O.; Huynh, Y.; Leslie, F.; et al. A significant metabolic and radiological response after a novel targeted microRNA-based treatment approach in malignant pleural mesothelioma. Am. J. Respir. Crit. Care Med. 2015, 191, 1467–1469. [Google Scholar] [CrossRef]
- Querfeld, C.; Pacheco, T.; Foss, F.M.; Halwani, A.S.; Porcu, P.; Seto, A.G.; Ruckman, J.; Landry, M.L.; Jackson, A.L.; Pestano, L.A.; et al. Preliminary Results of a Phase 1 Trial Evaluating MRG-106, a Synthetic microRNA Antagonist (LNA antimiR) of microRNA-155, in Patients with CTCL. Blood 2016, 128, 1829. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, D.-Y.; Huang, L. In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Adv. Drug Deliv. Rev. 2015, 81, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, A.; Miyata, K.; Kataoka, K. Recent progress in development of siRNA delivery vehicles for cancer therapy. Adv. Drug Deliv. Rev. 2016, 104, 61–77. [Google Scholar] [CrossRef]
- Yao, Y.; Zhou, Y.; Liu, L.; Xu, Y.; Chen, Q.; Wang, Y.; Wu, S.; Deng, Y.; Zhang, J.; Shao, A. Nanoparticle-based drug delivery in cancer therapy and its role in overcoming drug resistance. Front. Mol. Biosci. 2020, 7, 193. [Google Scholar] [CrossRef] [PubMed]
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. miRNA: A promising therapeutic target in cancer. Int. J. Mol. Sci. 2022, 23, 11502. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Meliala, I.T.S.; Hosea, R.; Kasim, V.; Wu, S. Yin and Yang of YY1 regulation on tumor metabolic reprogramming. In YY1 in the Control of the Pathogenesis and Drug Resistance of Cancer; Elsevier: Amsterdam, The Netherlands, 2021; pp. 79–99. [Google Scholar]
- Yap, T.A.; Parkes, E.E.; Peng, W.; Moyers, J.T.; Curran, M.A.; Tawbi, H.A. Development of Immunotherapy Combination Strategies in CancerImmunotherapy Combination Strategies in Cancer. Cancer Discov. 2021, 11, 1368–1397. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, Y.; Nakatsuji, M.; Seno, H.; Ishizu, S.; Akitake-Kawano, R.; Kanda, K.; Ueo, T.; Komekado, H.; Kawada, M.; Minami, M.; et al. COX-2 inhibition alters the phenotype of tumor-associated macrophages from M2 to M1 in ApcMin/+ mouse polyps. Carcinogenesis 2011, 32, 1333–1339. [Google Scholar] [CrossRef]
- Balkhi, M.Y.; Wittmann, G.; Xiong, F.; Junghans, R.P. YY1 Upregulates Checkpoint Receptors and Downregulates Type I Cytokines in Exhausted, Chronically Stimulated Human T Cells. iScience 2018, 2, 105–122. [Google Scholar] [CrossRef]
- Robert, C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 2020, 11, 3801. [Google Scholar] [CrossRef]
- Karasarides, M.; Cogdill, A.P.; Robbins, P.B.; Bowden, M.; Burton, E.M.; Butterfield, L.H.; Cesano, A.; Hammer, C.; Haymaker, C.L.; Horak, C.E. Hallmarks of Resistance to Immune-Checkpoint Inhibitors. Cancer Immunol. Res. 2022, 10, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Liu, Y.; Wang, C.; Zheng, H.; Chen, Y.; Liu, W.; Chen, X.; Zhang, J.; Chen, H.; Yang, Y. Inhibition of COX-2 and EGFR by melafolone improves anti-PD-1 therapy through vascular normalization and PD-L1 downregulation in lung cancer. J. Pharmacol. Exp. Ther. 2019, 368, 401–413. [Google Scholar] [CrossRef]
- Lemoine, J.; Ruella, M.; Houot, R. Born to survive: How cancer cells resist CAR T cell therapy. J. Hematol. Oncol. 2021, 14, 199. [Google Scholar] [CrossRef]
- Hirayama, A.V.; Gauthier, J.; Hay, K.A.; Sheih, A.; Cherian, S.; Chen, X.; Pender, B.S.; Hawkins, R.M.; Vakil, A.; Steinmetz, R.N. Efficacy and toxicity of JCAR014 in combination with durvalumab for the treatment of patients with relapsed/refractory aggressive B-cell non-Hodgkin lymphoma. Blood 2018, 132, 1680. [Google Scholar] [CrossRef]
- Wang, C.; Shi, F.; Liu, Y.; Zhang, Y.; Dong, L.; Li, X.; Tong, C.; Wang, Y.; Su, L.; Nie, J. Anti-PD-1 antibodies as a salvage therapy for patients with diffuse large B cell lymphoma who progressed/relapsed after CART19/20 therapy. J. Hematol. Oncol. 2021, 14, 106. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, N.; Feng, K.; Chen, M.; Zhang, Y.; Liu, Y.; Yang, Q.; Nie, J.; Tang, N.; Zhang, X. Phase I study of CAR-T cells with PD-1 and TCR disruption in mesothelin-positive solid tumors. Cell. Mol. Immunol. 2021, 18, 2188–2198. [Google Scholar] [CrossRef]
- Rafiq, S.; Yeku, O.O.; Jackson, H.J.; Purdon, T.J.; Van Leeuwen, D.G.; Drakes, D.J.; Song, M.; Miele, M.M.; Li, Z.; Wang, P. Targeted delivery of a PD-1-blocking scFv by CAR-T cells enhances anti-tumor efficacy in vivo. Nat. Biotechnol. 2018, 36, 847–856. [Google Scholar] [CrossRef]
- Nebbioso, A.; Tambaro, F.P.; Dell’Aversana, C.; Altucci, L. Cancer epigenetics: Moving forward. PLoS Genet. 2018, 14, e1007362. [Google Scholar] [CrossRef]
- Atchison, L.; Ghias, A.; Wilkinson, F.; Bonini, N.; Atchison, M.L. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 2003, 22, 1347–1358. [Google Scholar] [CrossRef]
- Wilkinson, F.H.; Park, K.; Atchison, M.L. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc. Natl. Acad. Sci. USA 2006, 103, 19296–19301. [Google Scholar] [CrossRef]
- Yang, W.-M.; Yao, Y.-L.; Sun, J.-M.; Davie, J.R.; Seto, E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Biol. Chem. 1997, 272, 28001–28007. [Google Scholar] [CrossRef]
- Lee, J.-S.; Galvin, K.M.; See, R.H.; Eckner, R.; Livingston, D.; Moran, E.; Shi, Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995, 9, 1188–1198. [Google Scholar] [CrossRef]
- Rezai-Zadeh, N.; Zhang, X.; Namour, F.; Fejer, G.; Wen, Y.-D.; Yao, Y.-L.; Gyory, I.; Wright, K.; Seto, E. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 2003, 17, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Jin, J.; Yao, T.; Gottschalk, A.J.; Swanson, S.K.; Wu, S.; Shi, Y.; Washburn, M.P.; Florens, L.; Conaway, R.C. YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 2007, 14, 872–874. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, X.; Wei, C.; Huang, X.; Ma, Q.; Huang, X.; Faiola, F.; Guallar, D.; Fidalgo, M.; Huang, T. YY1 positively regulates transcription by targeting promoters and super-enhancers through the BAF complex in embryonic stem cells. Stem Cell Rep. 2018, 10, 1324–1339. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Feng, Y.; Pauklin, S. 3D chromatin architecture and transcription regulation in cancer. J. Hematol. Oncol. 2022, 15, 49. [Google Scholar] [CrossRef]
- Schwalie, P.C.; Ward, M.C.; Cain, C.E.; Faure, A.J.; Gilad, Y.; Odom, D.T.; Flicek, P. Co-binding by YY1 identifies the transcriptionally active, highly conserved set of CTCF-bound regions in primate genomes. Genome Biol. 2013, 14, R148. [Google Scholar] [CrossRef]
- Pan, X.; Papasani, M.; Hao, Y.; Calamito, M.; Wei, F.; Quinn, W.J.; Basu, A.; Wang, J.; Hodawadekar, S.; Zaprazna, K. YY1 controls Igκ repertoire and B-cell development, and localizes with condensin on the Igκ locus. EMBO J. 2013, 32, 1168–1182. [Google Scholar] [CrossRef]
- Morris, E.C.; Neelapu, S.S.; Giavridis, T.; Sadelain, M. Cytokine release syndrome and associated neurotoxicity in cancer immunotherapy. Nat. Rev. Immunol. 2022, 22, 85–96. [Google Scholar] [CrossRef]
- Drago, J.Z.; Modi, S.; Chandarlapaty, S. Unlocking the potential of antibody–drug conjugates for cancer therapy. Nat. Rev. Clin. Oncol. 2021, 18, 327–344. [Google Scholar] [CrossRef]
- Li, L.; Li, Y.; Timothy Sembiring Meliala, I.; Kasim, V.; Wu, S. Biological roles of Yin Yang 2: Its implications in physiological and pathological events. J. Cell. Mol. Med. 2020, 24, 12886–12899. [Google Scholar] [CrossRef]
- Nguyen, N.; Zhang, X.; Olashaw, N.; Seto, E. Molecular Cloning and Functional Characterization of the Transcription Factor YY2. J. Biol. Chem. 2004, 279, 25927–25934. [Google Scholar] [CrossRef]
- Kasim, V.; Xie, Y.-D.; Wang, H.-M.; Huang, C.; Yan, X.-S.; Nian, W.-Q.; Zheng, X.-D.; Miyagishi, M.; Wu, S.-R. Transcription factor Yin Yang 2 is a novel regulator of the p53/p21 axis. Oncotarget 2017, 8, 54694–54707. [Google Scholar] [CrossRef]
- Chen, L.; Shioda, T.; Coser, K.R.; Lynch, M.C.; Yang, C.; Schmidt, E.V. Genome-wide analysis of YY2 versus YY1 target genes. Nucleic Acids Res. 2010, 38, 4011–4026. [Google Scholar] [CrossRef]
- Figiel, M.; Łakomska, J.; Miłek, P.; Dziedzicka-Wasylewska, M.; Górecki, A. The transcription factor YY2 has less momentous properties of an intrinsically disordered protein than its paralog YY1. FEBS Lett. 2019, 593, 1787–1798. [Google Scholar] [CrossRef]
- Kaufhold, S.; Aziz, N.; Bonavida, B. The forgotten YY2 in reported YY1 expression levels in human cancers. Crit. Rev. Oncog. 2017, 22, 63–73. [Google Scholar] [CrossRef]
- Jin, H.; Wang, L.; Bernards, R. Rational combinations of targeted cancer therapies: Background, advances and challenges. Nat. Rev. Drug Discov. 2023, 22, 213–234. [Google Scholar] [CrossRef]
- Hinshaw, D.C.; Shevde, L.A. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef]
- Turley, S.J.; Cremasco, V.; Astarita, J.L. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat. Rev. Immunol. 2015, 15, 669–682. [Google Scholar] [CrossRef]
- Liu, X.; Cao, Z.; Liu, N.; Gao, G.; Du, M.; Wang, Y.; Cheng, B.; Zhu, M.; Jia, B.; Pan, L. Kill two birds with one stone: Engineered exosome-mediated delivery of cholesterol modified YY1-siRNA enhances chemoradiotherapy sensitivity of glioblastoma. Front. Pharmacol. 2022, 13, 975291. [Google Scholar] [CrossRef]
- Gambardella, V.; Tarazona, N.; Cejalvo, J.M.; Lombardi, P.; Huerta, M.; Roselló, S.; Fleitas, T.; Roda, D.; Cervantes, A. Personalized medicine: Recent progress in cancer therapy. Cancers 2020, 12, 1009. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Zhang, G.; Han, F.; Fu, S.; Cao, Y.; Zhang, F.; Zhang, Q.; Meslamani, J.; Xu, Y.; Ji, D.; et al. Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth. Proc. Natl. Acad. Sci. USA 2018, 115, 7949–7954. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shen, L.; Zhu, Y.; Xu, R.; Deng, Z.; Liu, X.; Ding, Y.; Wang, C.; Shi, Y.; Bei, L.; et al. KDM6A promotes imatinib resistance through YY1-mediated transcriptional upregulation of TRKA independently of its demethylase activity in chronic myelogenous leukemia. Theranostics 2021, 11, 2691–2705. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Han, J.; Wang, X.; Wu, T.; Zhang, H.; An, H.; Qin, L.; Sun, Y.; Zhong, W.; Yang, C. Twist1-YY1-p300 complex promotes the malignant progression of HCC through activation of miR-9 by forming phase-separated condensates at super-enhancers and relieved by Metformin. Pharmacol. Res. 2023, 188, 106661. [Google Scholar] [CrossRef] [PubMed]
| Cancer Types | YY1 Expression Level | Prognosis | Refs. |
|---|---|---|---|
| Bladder | Upregulated | Poor | [33,34] |
| Breast | Upregulated | Poor | [35] |
| Cervical | Upregulated | Poor | [36,37] |
| Colon | Upregulated | Poor | [38,39,40,41] |
| Esophageal | Upregulated | Poor | [42] |
| Gastric | Upregulated | Poor | [43,44] |
| Glioma | Upregulated | Poor | [45,46,47] |
| Hodgkin lymphoma | Upregulated | n/a | [48] |
| Leukemia | Upregulated | Poor | [49,50,51] |
| Liver | Upregulated | Poor | [52] |
| Lung | Upregulated | Poor | [27,53] |
| Melanoma | Upregulated | Poor | [51,54,55,56] |
| Multiple myeloma | Upregulated | Poor | [57] |
| Nasopharynx | Downregulated | Good | [58] |
| Non-Hodgkin lymphoma | Upregulated | Poor | [59,60,61,62] |
| Downregulated | Good | [63] | |
| Osteosarcoma | Upregulated | Poor | [32] |
| Ovarian | Upregulated | Poor | [23,64,65] |
| Pancreatic | Upregulated | Poor | [66] |
| Upregulated | Poor | [67] | |
| Renal | Upregulated | Poor | [68] |
| Sarcoma | Upregulated | n/a | [69] |
| Upregulated | Poor | [32] | |
| Testicular seminoma | Upregulated | Poor | [70,71] |
| Thyroid | Upregulated | Poor | [72,73,74] |
| Target | Pathway | Hallmarks | Refs. |
|---|---|---|---|
| AKT | YY1/mTORC2/AKT | Evading apoptosis; limitless replicative potential; sustained angiogenesis; tissue invasion and metastasis | [74,94] |
| APC | miR-193a-5p/YY1/APC | Limitless replicative potential | [95] |
| Atg5 | YY1/TFEB/Atg5-Atg12-Atg16 | Evading apoptosis (by evading autophagy) | [56] |
| Beclin1 | YY1/TFEB/Beclin1 | Evading apoptosis (by evading autophagy) | [56] |
| Bim | YY1/RelA/Bim | Evading apoptosis; limitless replicative potential | [96] |
| CDKN2A | YY1/HDACs/CDKN2A | Evading apoptosis | [97,98] |
| CDKN3 | YY1/CDKN3/MdM2/p53/p21 | Limitless replicative potential; tissue invasion and metastasis | [99] |
| c-Myc | YY1/c-Myc | Deregulated metabolism; evading apoptosis; genome instability; limitless replicative potential; tissue invasion and metastasis | [87,100] |
| COX2 | YY1/COX2/PG | Evading immune system | [101,102] |
| CXCR4 | CXCR4/YY1/VEGF | Sustained angiogenesis; tissue invasion and metastasis | [103] |
| CXCR4 | SDF-1α/CXCR4/YY1/let-7a | Evading apoptosis; evading immune system | [104] |
| DEK | YY1/DEK/HIF-1α/VEGF | Sustained angiogenesis | [105] |
| DEK | YY1/NF-Y/DEK | Limitless replicative potential | [106] |
| DR5 | YY1/DR5/TRAIL/NF-κB | Evading apoptosis; evading immune system | [107,108] |
| DTDST | NF-κB/YY1/PRC2-EZH2/ DTDST | Evading immune system; limitless replicative potential; tissue invasion and metastasis | [109] |
| CDH1 | YY1-PRMT7-HDAC3/H3K4me3/CDH1 | Tissue invasion and metastasis | [110] |
| EGFR | mir-34a/YY1/EGFR | Limitless replicative potential | [111] |
| EGFR | MCT1/YY1/EGFR/MnSOD | Deregulate metabolism; evading apoptosis | [112] |
| ERBB2 | YY1/AP-2α/ERBB2 | Sustained angiogenesis; tissue invasion and metastasis | [113] |
| Fas | miR27a/ZBTB10/Sp/YY1/ ERBB2 | Limitless proliferative potential | [75] |
| G6PD | YY1/G6PD/PPP/r5p | Deregulated metabolism | [85] |
| GLUT3 | YY1/GLUT3 | Deregulated metabolism; limitless replicative potential | [91] |
| HIF-1α | YY1/HIF-1α/GLUT1-GLUT3 | Deregulated metabolism; evading apoptosis | [114] |
| HIF-1α | YY1/HIF-1α/VEGF & TGF-α | Sustained angiogenesis; tissue invasion and metastasis | [84,103] |
| HIF-1α | YY1/HIF-1α/CA9 | Evading immune system; tissue invasion and metastasis | [84,115,116] |
| HIF-1α | YY1/HIF-1α/PGK | Evading apoptosis; deregulated metabolism; sustaining proliferative signaling | [84,117] |
| hnRNPM | YY1/hnRNPM/CD44 | Tissue invasion and metastasis | [118] |
| HPV18 | YY1-CTCF/HPV18 | Insensitivity to anti-growth signals; limitless replicative potential | [119] |
| IL6 | YY1/IL6/STAT3/PD-L1 | Evading immune system | [22] |
| KLF4 | YY1/KLF4/p53 | Evading apoptosis | [120] |
| KLF5 | YY1/KLF4/p21 | Limitless replicative potential | [120] |
| KLF6 | YY1/KLF4/c-Myc | Deregulated metabolism; evading apoptosis; genome instability; limitless replicative potential; tissue invasion and metastasis | [87,120] |
| KLF7 | YY1/KLF4/cyclin D2 | Limitless replicative potential | [120] |
| MAP1LC3B | YY1/TFEB/MAP1LC3B | Evading apoptosis | [56] |
| miR-125a | RYBP/YY1/pri-miR-125a | Evading apoptosis, evading immune system | [121] |
| miR-195 | miR-195/Smurf2YY1/VEGFA/ Snail1 | Tissue invasion and metastasis | [122] |
| miR-30a | YY1/miR-30a/ATG5 & Beclin1 | Evading apoptosis | [123] |
| miR-372 | YY1/mIR-372/SQSTM1 | Evading apoptosis | [124] |
| miR-9 | YY1/EZH2/H3K27me3/miR9/ NF-κB1 | Evading apoptosis; tissue invasion and metastasis | [30] |
| p21 | YY1/BCCIP/p53re/p21 | Evading apoptosis; limitless replicative potential | [90] |
| p53 | YY1/BCCIP/p53re/p21 | Evading apoptosis; limitless replicative potential | [90] |
| p53 | YY1/MDM2/p53 | Evading apoptosis | [89] |
| p53 | p14ARF/YY1/Hdm2/p53 | Evading apoptosis; insensitivity to anti-growth signals | [88] |
| p53 | YY1/TIGAR/PDK2/PFK-1 | Deregulated metabolism; evading apoptosis | [125] |
| p53 | YY1/p300/MDM2/p53 | Evading apoptosis | [88] |
| p53 | Smurf2/YY1/p53 | Evading apoptosis; evading immune system | [126,127] |
| p73 | YY1/E2F1/p73 | Evading apoptosis; insensitivity to anti-growth signals | [128] |
| PGC-1β | YY1/PGC-1β/MCAD & LCAD | Deregulated metabolism | [92] |
| RelB | YY1/RelB/p65 & p50 | Evading apoptosis; evading immune system | [46] |
| RYBP | YY1/miR-9/RYBP/SP1 | Evading apoptosis; insensitivity to anti-growth signals; tissue invasion and metastasis | [129] |
| RYBP | RYBP/YY1/E2F6/Mae1 or Staq3 or Smc1β | Insensitivity to anti-growth signals; limitless replicative potential | [130] |
| RYBP | RYBP/YY1/E2F2 or E2F3/CDC7 | Genome instability; insensitivity to anti-growth signals; limitless replicative potential | [130] |
| ST6GalNAc6 | YY1/PRC2/EZH2/H3K27me3/ DTDST/ST6GalNAc6 | Evading immune system; genome instability | [97,109] |
| TPPP | YY1/TPPP/p38/MAPK | Evading apoptosis; sustained angiogenesis; tissue invasion and metastasis | [131] |
| TPPP | YY1/TPPP/PI3K/AKT | Sustained angiogenesis; tissue invasion and metastasis | [131] |
| VEGF | CXCR4/YY1/VEGF | Sustained angiogenesis; tissue invasion and metastasis | [103] |
| VEGF | YY1/VEGFA/VEGFR2 | Evading apoptosis; sustained angiogenesis | [132] |
| VEGFB | CXCR4/YY1/VEGFB | Sustained angiogenesis; tissue invasion and metastasis | [103] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hosea, R.; Hillary, S.; Wu, S.; Kasim, V. Targeting Transcription Factor YY1 for Cancer Treatment: Current Strategies and Future Directions. Cancers 2023, 15, 3506. https://doi.org/10.3390/cancers15133506
Hosea R, Hillary S, Wu S, Kasim V. Targeting Transcription Factor YY1 for Cancer Treatment: Current Strategies and Future Directions. Cancers. 2023; 15(13):3506. https://doi.org/10.3390/cancers15133506
Chicago/Turabian StyleHosea, Rendy, Sharon Hillary, Shourong Wu, and Vivi Kasim. 2023. "Targeting Transcription Factor YY1 for Cancer Treatment: Current Strategies and Future Directions" Cancers 15, no. 13: 3506. https://doi.org/10.3390/cancers15133506
APA StyleHosea, R., Hillary, S., Wu, S., & Kasim, V. (2023). Targeting Transcription Factor YY1 for Cancer Treatment: Current Strategies and Future Directions. Cancers, 15(13), 3506. https://doi.org/10.3390/cancers15133506







