MIRACUM-Pipe: An Adaptable Pipeline for Next-Generation Sequencing Analysis, Reporting, and Visualization for Clinical Decision Making
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
- WES analysis, i.e., a patient-matched tumor–normal pair of sequencing samples (DNA only),
- tNGS analysis, i.e., tumor-only sequencing on a hybrid capture-based gene panel (DNA and optional RNA),
- Tumor-only analysis, i.e., a single tumor sample from WES (DNA only).
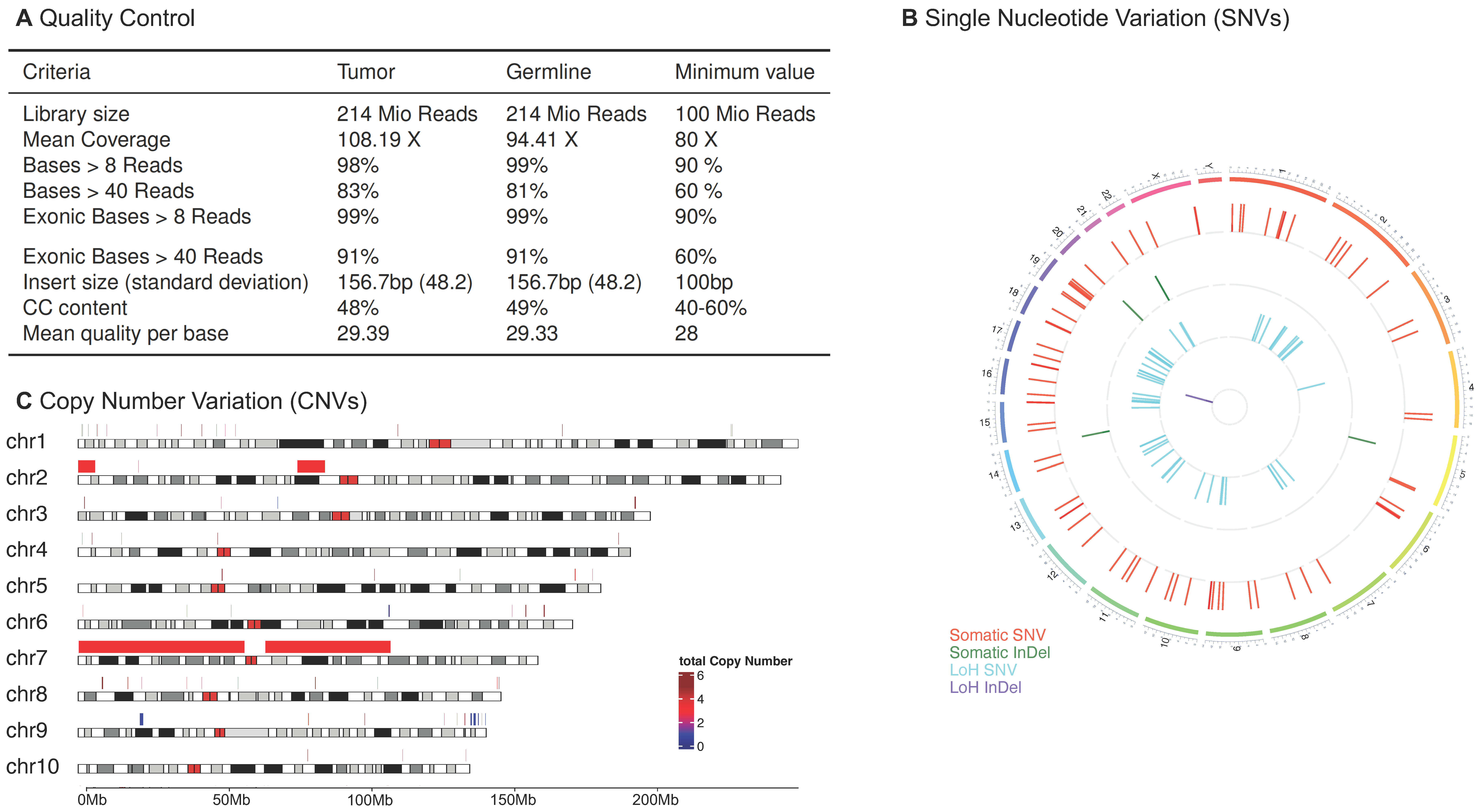
2.1. Quality Control
- mean base quality,
- mean coverage over (exonic) target region,
- library size,
- insert size length.
2.2. Alignment
2.3. Variant Calling
2.4. Copy Number Variation Calling
2.5. RNA Fusion Calling
2.6. Annotation
- tumor mutational burden (TMB),
- homologous recombination deficiency (HRD) score,
- microsatellite instability (MSI),
- bioinformatic tumor purity, and
- ploidy.
2.7. Functional Enrichment Analysis
2.8. Mutational Signature Analysis
2.9. Reporting and Visualization
3. Results
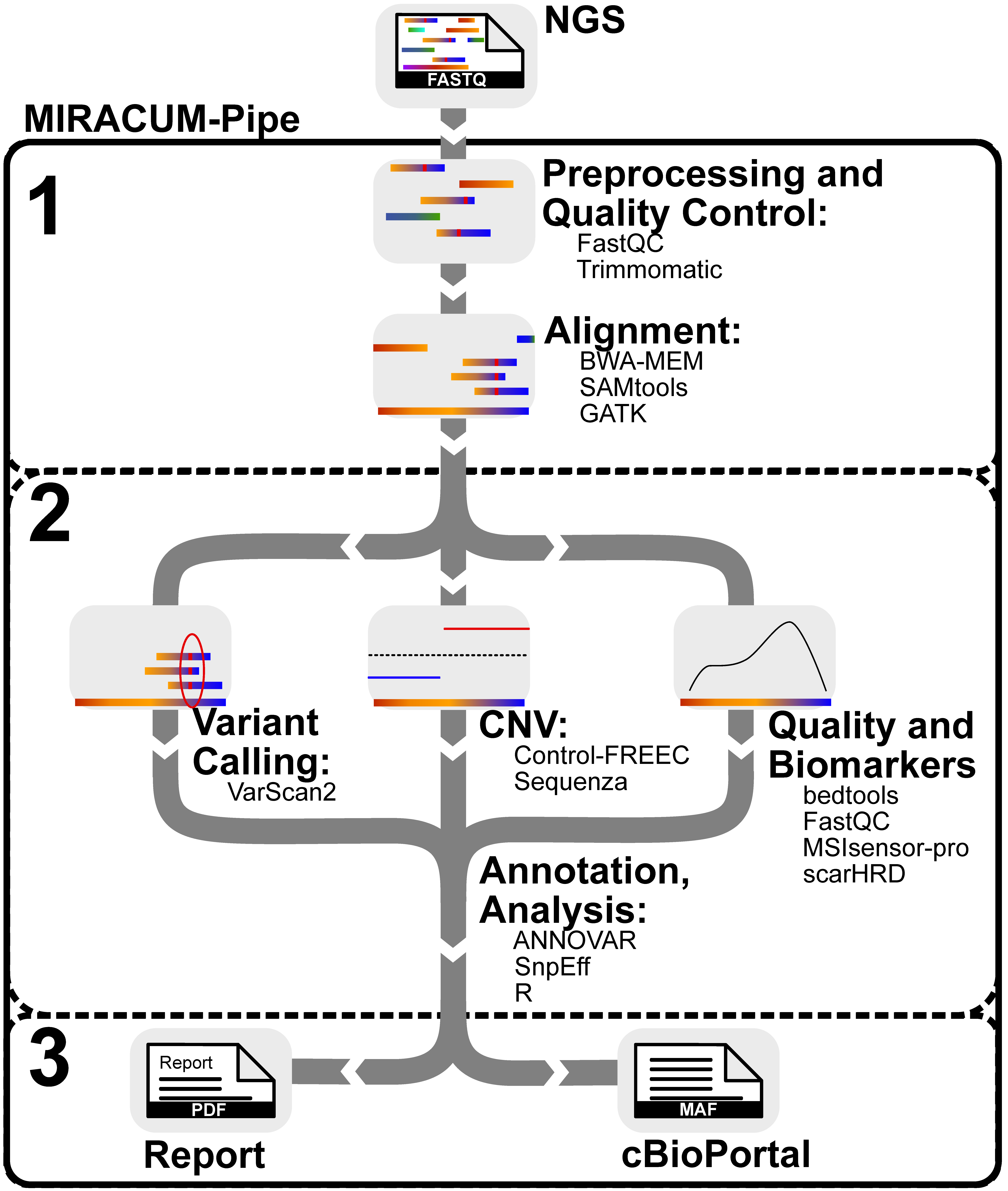
3.1. MIRACUM-Pipe
Performance, Usability, and Configuration
3.2. MIRACUM-Pipe Results
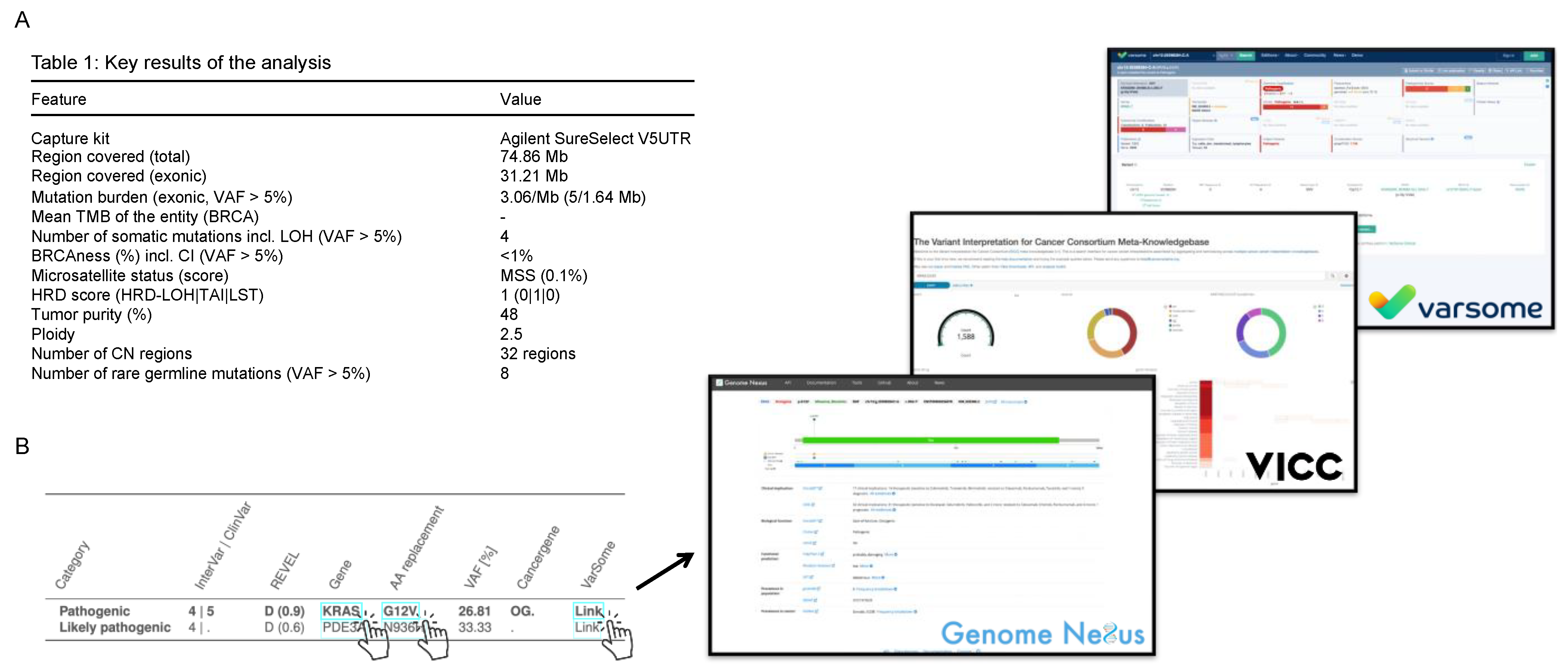
3.2.1. Interactive PDF Report
3.2.2. Visualization in cBioPortal
4. Discussion
4.1. Use and Insights of MIRACUM-Pipe in the Context of an MTB
4.2. Limitations and Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maemondo, M.; Inoue, A.; Kobayashi, K.; Sugawara, S.; Oizumi, S.; Isobe, H.; Gemma, A.; Harada, M.; Yoshizawa, H.; Kinoshita, I.; et al. Gefitinib or Chemotherapy for Non–Small-Cell Lung Cancer with Mutated EGFR. N. Engl. J. Med. 2010, 362, 2380–2388. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Karaszewska, B.; Schachter, J.; Rutkowski, P.; Mackiewicz, A.; Stroiakovski, D.; Lichinitser, M.; Dummer, R.; Grange, F.; Mortier, L.; et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015, 372, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Solomon, B.J.; Mok, T.; Kim, D.-W.; Wu, Y.-L.; Nakagawa, K.; Mekhail, T.; Felip, E.; Cappuzzo, F.; Paolini, J.; Usari, T.; et al. First-Line Crizotinib versus Chemotherapy in ALK-Positive Lung Cancer. N. Engl. J. Med. 2014, 371, 2167–2177. [Google Scholar] [CrossRef] [PubMed]
- Abou-Alfa, G.K.; Sahai, V.; Hollebecque, A.; Vaccaro, G.; Melisi, D.; Al-Rajabi, R.; Paulson, A.S.; Borad, M.J.; Gallinson, D.; Murphy, A.G.; et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: A multicentre, open-label, phase 2 study. Lancet Oncol. 2020, 21, 671–684. [Google Scholar] [CrossRef]
- Tsimberidou, A.M.; Fountzilas, E.; Nikanjam, M.; Kurzrock, R. Review of precision cancer medicine: Evolution of the treatment paradigm. Cancer Treat. Rev. 2020, 86, 102019. [Google Scholar] [CrossRef]
- Tamborero, D.; Dienstmann, R.; Rachid, M.H.; Boekel, J.; Baird, R.; Braña, I.; De Petris, L.; Yachnin, J.; Massard, C.; Opdam, F.L.; et al. Support systems to guide clinical decision-making in precision oncology: The Cancer Core Europe Molecular Tumor Board Portal. Nat. Med. 2020, 26, 992–994. [Google Scholar] [CrossRef]
- Horak, P.; Heining, C.; Kreutzfeldt, S.; Hutter, B.; Mock, A.; Hüllein, J.; Fröhlich, M.; Uhrig, S.; Jahn, A.; Rump, A.; et al. Comprehensive Genomic and Transcriptomic Analysis for Guiding Therapeutic Decisions in Patients with Rare Cancers. Cancer Discov. 2021, 11, 2780–2795. [Google Scholar] [CrossRef]
- Stenzinger, A.; Edsjö, A.; Ploeger, C.; Friedman, M.; Fröhling, S.; Wirta, V.; Seufferlein, T.; Botling, J.; Duyster, J.; Akhras, M.; et al. Trailblazing precision medicine in Europe: A joint view by Genomic Medicine Sweden and the Centers for Personalized Medicine, ZPM, in Germany. Semin. Cancer Biol. 2022, 84, 242–254. [Google Scholar] [CrossRef]
- Westphalen, C.B.; Bokemeyer, C.; Büttner, R.; Fröhling, S.; Gaidzik, V.I.; Glimm, H.; Hacker, U.T.; Heinemann, V.; Illert, A.L.; Keilholz, U.; et al. Conceptual framework for precision cancer medicine in Germany: Consensus statement of the Deutsche Krebshilfe working group ‘Molecular Diagnostics and Therapy’. Eur. J. Cancer 2020, 135, 1–7. [Google Scholar] [CrossRef]
- Hoefflin, R.; Geißler, A.-L.; Fritsch, R.; Claus, R.; Wehrle, J.; Metzger, P.; Reiser, M.; Mehmed, L.; Fauth, L.; Heiland, D.H.; et al. Personalized Clinical Decision Making Through Implementation of a Molecular Tumor Board: A German Single-Center Experience. JCO Precis. Oncol. 2018, 2, 1–16. [Google Scholar] [CrossRef]
- Hoefflin, R.; Lazarou, A.; Hess, M.E.; Reiser, M.; Wehrle, J.; Metzger, P.; Frey, A.V.; Becker, H.; Aumann, K.; Berner, K.; et al. Transitioning the Molecular Tumor Board from Proof of Concept to Clinical Routine: A German Single-Center Analysis. Cancers 2021, 13, 1151. [Google Scholar] [CrossRef]
- Hinderer, M.; Boerries, M.; Haller, F.; Wagner, S.; Sollfrank, S.; Acker, T.; Prokosch, H.-U.; Christoph, J. Supporting molecular tumor boards in molecular-guided decision-making—The current status of five German university hospitals. Stud. Health Technol. Inform. 2017, 236, 48–54. [Google Scholar] [CrossRef]
- Buechner, P.; Hinderer, M.; Unberath, P.; Metzger, P.; Boeker, M.; Acker, T.; Haller, F.; Mack, E.; Nowak, D.; Paret, C.; et al. Requirements Analysis and Specification for a Molecular Tumor Board Platform Based on cBioPortal. Diagnostics 2020, 10, 93. [Google Scholar] [CrossRef]
- Hinderer, M.; Boerries, M.; Boeker, M.; Neumaier, M.; Loubal, F.-P.; Acker, T.; Brunner, M.; Prokosch, H.-U.; Christoph, J. Implementing Pharmacogenomic Clinical Decision Support into German Hospitals. Stud. Health Technol. Inform. 2018, 247, 870–874. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Bonfield, J.K.; Marshall, J.; Danecek, P.; Li, H.; Ohan, V.; Whitwham, A.; Keane, T.; Davies, R.M. HTSlib: C library for reading/writing high-throughput sequencing data. Gigascience 2021, 10, giab007. [Google Scholar] [CrossRef]
- Quinlan, A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinform. 2014, 47, 11.12.1–11.12.34. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef] [PubMed]
- Boeva, V.; Popova, T.; Bleakley, K.; Chiche, P.; Cappo, J.; Schleiermacher, G.; Janoueix-Lerosey, I.; Delattre, O.; Barillot, E. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 2012, 28, 423–425. [Google Scholar] [CrossRef]
- Boeva, V.; Zinovyev, A.; Bleakley, K.; Vert, J.-P.; Janoueix-Lerosey, I.; Delattre, O.; Barillot, E. Control-free calling of copy number alterations in deep-sequencing data using GC-content normalization. Bioinformatics 2010, 27, 268–269. [Google Scholar] [CrossRef]
- Favero, F.; Joshi, T.; Marquard, A.M.; Birkbak, N.J.; Krzystanek, M.; Li, Q.; Szallasi, Z.; Eklund, A.C. Sequenza: Allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol. 2014, 26, 64–70. [Google Scholar] [CrossRef]
- Sztupinszki, Z.; Diossy, M.; Krzystanek, M.; Reiniger, L.; Csabai, I.; Favero, F.; Birkbak, N.J.; Eklund, A.C.; Syed, A.; Szallasi, Z. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. NPJ Breast Cancer 2018, 4, 16. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 26 May 2023).
- Nicorici, D.; Şatalan, M.; Edgren, H.; Kangaspeska, S.; Murumägi, A.; Kallioniemi, O.; Virtanen, S.; Kilkku, O. FusionCatcher—A tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv 2014, 011650. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Yang, H.; Wang, K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015, 10, 1556–1566. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014, 42, 980–985. [Google Scholar] [CrossRef]
- Li, Q.; Wang, K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am. J. Hum. Genet. 2017, 100, 267–280. [Google Scholar] [CrossRef]
- Liu, X.; Jian, X.; Boerwinkle, E. dbNSFP: A lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 2011, 32, 894–899. [Google Scholar] [CrossRef]
- Liu, X.; Li, C.; Mou, C.; Dong, Y.; Tu, Y. dbNSFP v4: A comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020, 12, 103. [Google Scholar] [CrossRef]
- Ioannidis, N.M.; Rothstein, J.H.; Pejaver, V.; Middha, S.; McDonnell, S.K.; Baheti, S.; Musolf, A.; Li, Q.; Holzinger, E.; Karyadi, D.; et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016, 99, 877–885. [Google Scholar] [CrossRef]
- Chakravarty, D.; Gao, J.; Phillips, S.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 1, 1–16. [Google Scholar] [CrossRef]
- Chang, M.T.; Bhattarai, T.S.; Schram, A.M.; Bielski, C.M.; Donoghue, M.T.; Jonsson, P.; Chakravarty, D.; Phillips, S.; Kandoth, C.; Penson, A.; et al. Accelerating Discovery of Functional Mutant Alleles in Cancer. Cancer Discov. 2018, 8, 174–183. [Google Scholar] [CrossRef]
- Chang, M.T.; Asthana, S.; Gao, S.P.; Lee, B.H.; Chapman, J.S.; Kandoth, C.; Gao, J.; Socci, N.D.; Solit, D.B.; Olshen, A.B.; et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 2015, 34, 155–163. [Google Scholar] [CrossRef]
- Popova, T.; Manié, E.; Rieunier, G.; Caux-Moncoutier, V.; Tirapo, C.; Dubois, T.; Delattre, O.; Sigal-Zafrani, B.; Bollet, M.; Longy, M.; et al. Ploidy and Large-Scale Genomic Instability Consistently Identify Basal-like Breast Carcinomas with BRCA1/2 Inactivation. Cancer Res. 2012, 72, 5454–5462. [Google Scholar] [CrossRef] [PubMed]
- Birkbak, N.J.; Wang, Z.C.; Kim, J.-Y.; Eklund, A.C.; Li, Q.; Tian, R.; Bowman-Colin, C.; Li, Y.; Greene-Colozzi, A.; Iglehart, J.D.; et al. Telomeric Allelic Imbalance Indicates Defective DNA Repair and Sensitivity to DNA-Damaging Agents. Cancer Discov. 2012, 2, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Abkevich, V.; Timms, K.M.; Hennessy, B.T.; Potter, J.; Carey, M.S.; Meyer, L.A.; Smith-McCune, K.; Broaddus, R.; Lu, K.H.; Chen, J.; et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer 2012, 107, 1776–1782. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Yang, X.; Guo, L.; Liu, B.; Lin, J.; Liang, H.; Sun, J.; Zhang, C.; Ye, K. MSIsensor-pro: Fast, Accurate, and Matched-normal-sample-free Detection of Microsatellite Instability. Genom. Proteom. Bioinform. 2020, 18, 65–71. [Google Scholar] [CrossRef]
- Niu, B.; Ye, K.; Zhang, Q.; Lu, C.; Xie, M.; McLellan, M.D.; Wendl, M.C.; Ding, L. MSIsensor: Microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2013, 30, 1015. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Hübschmann, D.; Jopp-Saile, L.; Andresen, C.; Krämer, S.; Gu, Z.; Heilig, C.E.; Kreutzfeldt, S.; Teleanu, V.; Fröhling, S.; Eils, R.; et al. Analysis of mutational signatures with yet another package for signature analysis. Genes Chromosom. Cancer 2020, 60, 314–331. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Campbell, P.J.; Stratton, M.R. Deciphering Signatures of Mutational Processes Operative in Human Cancer. Cell Rep. 2013, 3, 246–259. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.J.R.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.-L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Nik-Zainal, S.; Davies, H.; Staaf, J.; Ramakrishna, M.; Glodzik, D.; Zou, X.; Martincorena, I.; Alexandrov, L.B.; Martin, S.; Wedge, D.C.; et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016, 534, 47–54. [Google Scholar] [CrossRef]
- de Bruijn, I.; Li, X.; Sumer, S.O.; Gross, B.; Sheridan, R.; Ochoa, A.; Wilson, M.; Wang, A.; Zhang, H.; Lisman, A.; et al. Genome Nexus: A Comprehensive Resource for the Annotation and Interpretation of Genomic Variants in Cancer. JCO Clin. Cancer Inform. 2022, 6, e2100144. [Google Scholar] [CrossRef]
- Wagner, A.H.; Variant Interpretation for Cancer Consortium; Walsh, B.; Mayfield, G.; Tamborero, D.; Sonkin, D.; Krysiak, K.; Deu-Pons, J.; Duren, R.P.; Gao, J.; et al. A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nat. Genet. 2020, 52, 448–457. [Google Scholar] [CrossRef]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Aguilera, M.A.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.E.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. gtrellis: An R/Bioconductor package for making genome-level Trellis graphics. BMC Bioinform. 2016, 17, 169. [Google Scholar] [CrossRef]
- Unberath, P.; Mahlmeister, L.; Reimer, N.; Busch, H.; Boerries, M.; Christoph, J. Searching of Clinical Trials Made Easier in cBioPortal Using Patients’ Genetic and Clinical Profiles. Appl. Clin. Inform. 2022, 13, 363–369. [Google Scholar] [CrossRef]
- Reimer, N.; Unberath, P.; Busch, H.; Börries, M.; Metzger, P.; Ustjanzew, A.; Renner, C.; Prokosch, H.-U.; Christoph, J. Challenges and Experiences Extending the cBioPortal for Cancer Genomics to a Molecular Tumor Board Platform. In Studies in Health Technology and Informatics; IOS Press: Amsterdam, The Netherlands, 2021; pp. 139–143. [Google Scholar] [CrossRef]
- Ustjanzew, A.; Desuki, A.; Ritzel, C.; Dolezilek, A.C.; Wagner, D.-C.; Christoph, J.; Unberath, P.; Kindler, T.; Faber, J.; Marini, F.; et al. cbpManager: A web application to streamline the integration of clinical and genomic data in cBioPortal to support the Molecular Tumor Board. BMC Med. Inform. Decis. Mak. 2021, 21, 358. [Google Scholar] [CrossRef]
- Garcia, M.; Juhos, S.; Larsson, M.; Olason, P.I.; Martin, M.; Eisfeldt, J.; DiLorenzo, S.; Sandgren, J.; De Ståhl, T.D.; Ewels, P.; et al. Sarek: A portable workflow for whole-genome sequencing analysis of germline and somatic variants [version 2; peer review: 2 approved]. F1000Research 2020, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- Marriott, H.; Kabiljo, R.; Al Khleifat, A.; Dobson, R.J.; Al-Chalabi, A.; Iacoangeli, A. DNAscan2: A versatile, scalable, and user-friendly analysis pipeline for human next-generation sequencing data. Bioinformatics 2023, 39, btad152. [Google Scholar] [CrossRef] [PubMed]
- Sithara, A.A.; Maripuri, D.P.; Moorthy, K.; Ganesh, S.S.A.; Philip, P.; Banerjee, S.; Sudhakar, M.; Raman, K. iCOMIC: A graphical interface-driven bioinformatics pipeline for analyzing cancer omics data. NAR Genom. Bioinform. 2022, 4, lqac053. [Google Scholar] [CrossRef]
- Ahmed, Z.; Renart, E.G.; Mishra, D.; Zeeshan, S. JWES: A new pipeline for whole genome/exome sequence data processing, management, and gene-variant discovery, annotation, prediction, and genotyping. FEBS Open Bio 2021, 11, 2441–2452. [Google Scholar] [CrossRef]
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef]
- Ma, J.; Fong, S.H.; Luo, Y.; Bakkenist, C.J.; Shen, J.P.; Mourragui, S.; Wessels, L.F.A.; Hafner, M.; Sharan, R.; Peng, J.; et al. Few-shot learning creates predictive models of drug response that translate from high-throughput screens to individual patients. Nat. Cancer 2021, 2, 233–244. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Metzger, P.; Hess, M.E.; Blaumeiser, A.; Pauli, T.; Schipperges, V.; Mertes, R.; Christoph, J.; Unberath, P.; Reimer, N.; Scheible, R.; et al. MIRACUM-Pipe: An Adaptable Pipeline for Next-Generation Sequencing Analysis, Reporting, and Visualization for Clinical Decision Making. Cancers 2023, 15, 3456. https://doi.org/10.3390/cancers15133456
Metzger P, Hess ME, Blaumeiser A, Pauli T, Schipperges V, Mertes R, Christoph J, Unberath P, Reimer N, Scheible R, et al. MIRACUM-Pipe: An Adaptable Pipeline for Next-Generation Sequencing Analysis, Reporting, and Visualization for Clinical Decision Making. Cancers. 2023; 15(13):3456. https://doi.org/10.3390/cancers15133456
Chicago/Turabian StyleMetzger, Patrick, Maria Elena Hess, Andreas Blaumeiser, Thomas Pauli, Vincent Schipperges, Ralf Mertes, Jan Christoph, Philipp Unberath, Niklas Reimer, Raphael Scheible, and et al. 2023. "MIRACUM-Pipe: An Adaptable Pipeline for Next-Generation Sequencing Analysis, Reporting, and Visualization for Clinical Decision Making" Cancers 15, no. 13: 3456. https://doi.org/10.3390/cancers15133456
APA StyleMetzger, P., Hess, M. E., Blaumeiser, A., Pauli, T., Schipperges, V., Mertes, R., Christoph, J., Unberath, P., Reimer, N., Scheible, R., Illert, A. L., Busch, H., Andrieux, G., & Boerries, M. (2023). MIRACUM-Pipe: An Adaptable Pipeline for Next-Generation Sequencing Analysis, Reporting, and Visualization for Clinical Decision Making. Cancers, 15(13), 3456. https://doi.org/10.3390/cancers15133456






