Higher Plasma Creatinine Is Associated with an Increased Risk of Death in Patients with Non-Metastatic Rectal but Not Colon Cancer: Results from an International Cohort Consortium
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Data Collection
2.3. Sample Analysis
2.4. Study Endpoint
2.5. Statistical Analysis
3. Results
3.1. Study Population Characteristics
3.2. Associations of Metabolites with All-Cause Mortality
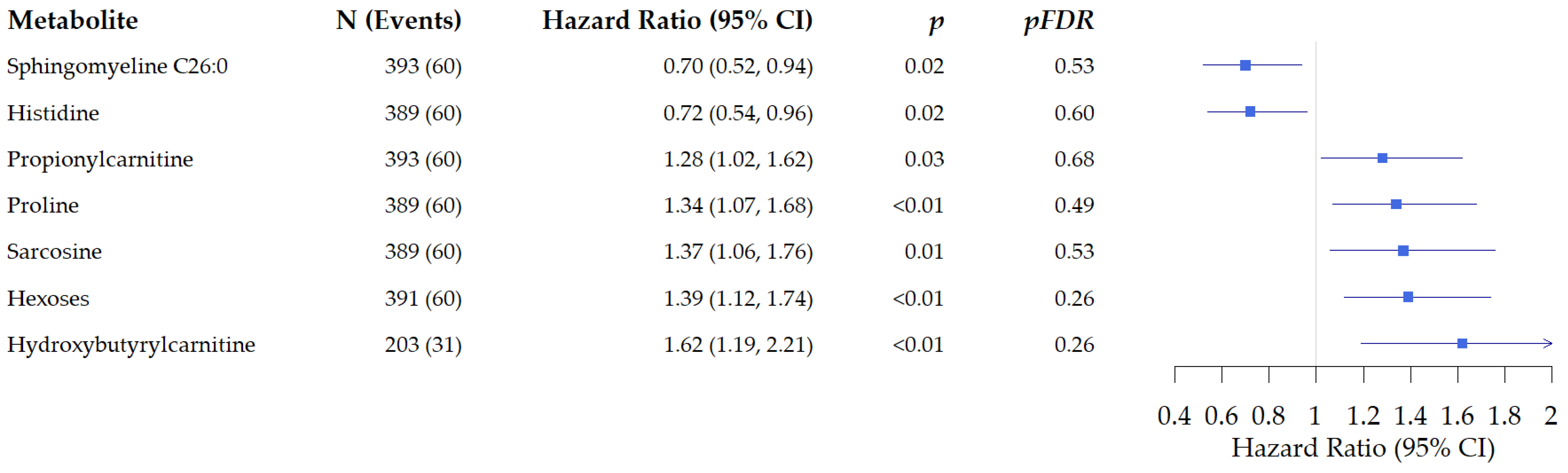
3.2.1. Associations of Metabolites with All-Cause Mortality in Patients with Colon Cancer
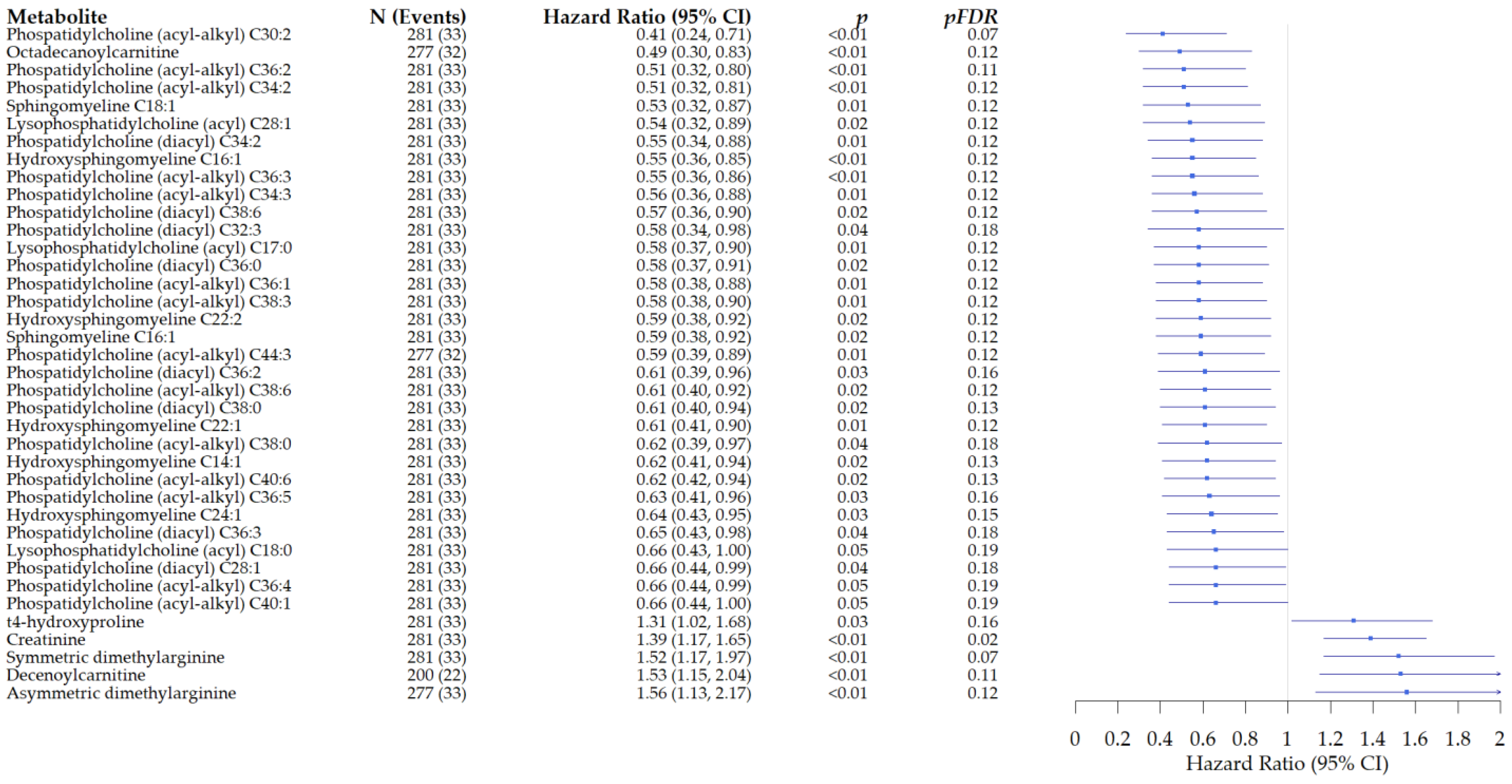
3.2.2. Associations of Metabolites with All-Cause Mortality in Patients with Rectal Cancer
3.2.3. Heterogeneity in the Associations of Metabolites with All-Cause Mortality in Patients with Rectal Cancer Compared with Colon Cancer
3.3. Pathway Analysis
3.3.1. Pathway Analysis in Patients with Colon Cancer
3.3.2. Pathway Analysis in Rectal Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Paschke, S.; Jafarov, S.; Staib, L.; Kreuser, E.D.; Maulbecker-Armstrong, C.; Roitman, M.; Holm, T.; Harris, C.C.; Link, K.H.; Kornmann, M. Are Colon and Rectal Cancer Two Different Tumor Entities? A Proposal to Abandon the Term Colorectal Cancer. Int. J. Mol. Sci. 2018, 19, 2577. [Google Scholar] [CrossRef]
- Eisele, Y.; Mallea, P.M.; Gigic, B.; Stephens, W.Z.; Warby, C.A.; Buhrke, K.; Lin, T.; Boehm, J.; Schrotz-King, P.; Hardikar, S.; et al. Fusobacterium nucleatum and Clinicopathologic Features of Colorectal Cancer: Results From the ColoCare Study. Clin. Color. Cancer 2021, 20, e165–e172. [Google Scholar] [CrossRef]
- Ose, J.; Gigic, B.; Hardikar, S.; Lin, T.; Himbert, C.; Warby, C.A.; Peoples, A.R.; Lindley, C.L.; Boehm, J.; Schrotz-King, P.; et al. Presurgery Adhesion Molecules and Angiogenesis Biomarkers Are Differently Associated with Outcomes in Colon and Rectal Cancer: Results from the ColoCare Study. Cancer Epidemiol. Biomark. Prev. 2022, 31, 1650–1660. [Google Scholar] [CrossRef]
- Zhang, Z.; Yan, X.; Lu, Y.; Guo, X.; Jiao, M.; Wang, W.; Sun, B.; Zhou, Y.; Hu, Q.; Chu, D. The prognostic impact of BMI on colorectal cancer is stratified by tumor location. Front. Oncol. 2022, 12, 987518. [Google Scholar] [CrossRef]
- Wesselink, E.; Valk, A.W.; Kok, D.E.; Lanen, A.V.; de Wilt, J.H.; van Kouwenhoven, E.A.; Schrauwen, R.W.; van Halteren, H.K.; Winkels, R.M.; Balvers, M.G.; et al. Postdiagnostic intake of a more proinflammatory diet is associated with a higher risk of recurrence and all-cause mortality in colorectal cancer survivors. Am. J. Clin. Nutr. 2023, 117, 243–251. [Google Scholar] [CrossRef]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Gowda, G.A.; Raftery, D. Metabolic profiling: Are we en route to better diagnostic tests for cancer? Future Oncol. 2012, 8, 1207–1210. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.M.; Darzi, A.W.; Takats, Z.; Lindon, J.C. Metabolic phenotyping in clinical and surgical environments. Nature 2012, 491, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.W.; Lane, A.N. NMR-based stable isotope resolved metabolomics in systems biochemistry. J. Biomol. NMR 2011, 49, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Bain, J.R.; Stevens, R.D.; Wenner, B.R.; Ilkayeva, O.; Muoio, D.M.; Newgard, C.B. Metabolomics applied to diabetes research: Moving from information to knowledge. Diabetes 2009, 58, 2429–2443. [Google Scholar] [CrossRef]
- Yanes, O.; Tautenhahn, R.; Patti, G.J.; Siuzdak, G. Expanding coverage of the metabolome for global metabolite profiling. Anal. Chem. 2011, 83, 2152–2161. [Google Scholar] [CrossRef]
- Gumpenberger, T.; Brezina, S.; Keski-Rahkonen, P.; Baierl, A.; Robinot, N.; Leeb, G.; Habermann, N.; Kok, D.E.G.; Scalbert, A.; Ueland, P.M.; et al. Untargeted Metabolomics Reveals Major Differences in the Plasma Metabolome between Colorectal Cancer and Colorectal Adenomas. Metabolites 2021, 11, 119. [Google Scholar] [CrossRef]
- Ose, J.; Gigic, B.; Brezina, S.; Lin, T.; Baierl, A.; Geijsen, A.; van Roekel, E.; Robinot, N.; Gicquiau, A.; Achaintre, D.; et al. Targeted Plasma Metabolic Profiles and Risk of Recurrence in Stage II and III Colorectal Cancer Patients: Results from an International Cohort Consortium. Metabolites 2021, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, S.A.; Ahiahonu, P.W.; Jayasinghe, D.; Heath, D.; Liu, J.; Lu, Y.; Jin, W.; Kavianpour, A.; Yamazaki, Y.; Khan, A.M.; et al. Reduced levels of hydroxylated, polyunsaturated ultra long-chain fatty acids in the serum of colorectal cancer patients: Implications for early screening and detection. BMC Med. 2010, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Cai, G.; Su, M.; Chen, T.; Zheng, X.; Xu, Y.; Ni, Y.; Zhao, A.; Xu, L.X.; Cai, S.; et al. Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J. Proteome Res. 2009, 8, 4844–4850. [Google Scholar] [CrossRef]
- Bertini, I.; Cacciatore, S.; Jensen, B.V.; Schou, J.V.; Johansen, J.S.; Kruhoffer, M.; Luchinat, C.; Nielsen, D.L.; Turano, P. Metabolomic NMR fingerprinting to identify and predict survival of patients with metastatic colorectal cancer. Cancer Res. 2012, 72, 356–364. [Google Scholar] [CrossRef]
- Nishiumi, S.; Kobayashi, T.; Ikeda, A.; Yoshie, T.; Kibi, M.; Izumi, Y.; Okuno, T.; Hayashi, N.; Kawano, S.; Takenawa, T.; et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS ONE 2012, 7, e40459. [Google Scholar] [CrossRef]
- Geijsen, A.; Brezina, S.; Keski-Rahkonen, P.; Baierl, A.; Bachleitner-Hofmann, T.; Bergmann, M.M.; Boehm, J.; Brenner, H.; Chang-Claude, J.; van Duijnhoven, F.J.B.; et al. Plasma metabolites associated with colorectal cancer: A discovery-replication strategy. Int. J. Cancer 2019, 145, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Winkels, R.M.; Heine-Broring, R.C.; van Zutphen, M.; van Harten-Gerritsen, S.; Kok, D.E.; van Duijnhoven, F.J.; Kampman, E. The COLON study: Colorectal cancer: Longitudinal, Observational study on Nutritional and lifestyle factors that may influence colorectal tumour recurrence, survival and quality of life. BMC Cancer 2014, 14, 374. [Google Scholar] [CrossRef] [PubMed]
- van Roekel, E.H.; Bours, M.J.; de Brouwer, C.P.; Ten Napel, H.; Sanduleanu, S.; Beets, G.L.; Kant, I.J.; Weijenberg, M.P. The applicability of the international classification of functioning, disability, and health to study lifestyle and quality of life of colorectal cancer survivors. Cancer Epidemiol. Biomark. Prev. 2014, 23, 1394–1405. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, C.M.; Gigic, B.; Bohm, J.; Ose, J.; Viskochil, R.; Schneider, M.; Colditz, G.A.; Figueiredo, J.C.; Grady, W.M.; Li, C.I.; et al. The ColoCare Study: A Paradigm of Transdisciplinary Science in Colorectal Cancer Outcomes. Cancer Epidemiol. Biomark. Prev. 2019, 28, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Gsur, A.; Baierl, A.; Brezina, S. Colorectal Cancer Study of Austria (CORSA): A Population-Based Multicenter Study. Biology 2021, 10, 722. [Google Scholar] [CrossRef] [PubMed]
- Siskos, A.P.; Jain, P.; Romisch-Margl, W.; Bennett, M.; Achaintre, D.; Asad, Y.; Marney, L.; Richardson, L.; Koulman, A.; Griffin, J.L.; et al. Interlaboratory Reproducibility of a Targeted Metabolomics Platform for Analysis of Human Serum and Plasma. Anal. Chem. 2017, 89, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Carayol, M.; Licaj, I.; Achaintre, D.; Sacerdote, C.; Vineis, P.; Key, T.J.; Onland Moret, N.C.; Scalbert, A.; Rinaldi, S.; Ferrari, P. Reliability of Serum Metabolites over a Two-Year Period: A Targeted Metabolomic Approach in Fasting and Non-Fasting Samples from EPIC. PLoS ONE 2015, 10, e0135437. [Google Scholar] [CrossRef] [PubMed]
- Geijsen, A.; van Roekel, E.H.; van Duijnhoven, F.J.B.; Achaintre, D.; Bachleitner-Hofmann, T.; Baierl, A.; Bergmann, M.M.; Boehm, J.; Bours, M.J.L.; Brenner, H.; et al. Plasma metabolites associated with colorectal cancer stage: Findings from an international consortium. Int. J. Cancer 2020, 146, 3256–3266. [Google Scholar] [CrossRef]
- Di Guida, R.; Engel, J.; Allwood, J.W.; Weber, R.J.; Jones, M.R.; Sommer, U.; Viant, M.R.; Dunn, W.B. Non-targeted UHPLC-MS metabolomic data processing methods: A comparative investigation of normalisation, missing value imputation, transformation and scaling. Metab. Off. J. Metab. Soc. 2016, 12, 93. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.E.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, Q.; Ruan, G.T.; Tang, M.; Zhang, X.; Song, M.M.; Zhang, X.W.; Zhang, K.P.; Ge, Y.Z.; Shi, H.P. Association Between Serum Creatinine Concentrations and Overall Survival in Patients with Colorectal Cancer: A Multi-Center Cohort Study. Front. Oncol. 2021, 11, 710423. [Google Scholar] [CrossRef]
- Lv, J.; Jia, H.; Mo, M.; Yuan, J.; Wu, Z.; Zhang, S.; Zhe, F.; Gu, B.; Fan, B.; Li, C.; et al. Changes of serum metabolites levels during neoadjuvant chemoradiation and prediction of the pathological response in locally advanced rectal cancer. Metabolomics 2022, 18, 99. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, D.A.; Kreider, R.B.; Stout, J.R.; Forero, D.A.; Kerksick, C.M.; Roberts, M.D.; Rawson, E.S. Metabolic Basis of Creatine in Health and Disease: A Bioinformatics-Assisted Review. Nutrients 2021, 13, 1238. [Google Scholar] [CrossRef] [PubMed]
- Willegger, M.; Posch, F.; Schieder, S.; Funovics, P.T.; Scharrer, A.; Brodowicz, T.; Ay, C.; Windhager, R.; Panotopoulos, J. Serum creatinine and albumin predict sarcoma-specific survival in patients with myofibroblastic and fibroblastic sarcomas. J. Orthop. Res. 2017, 35, 2815–2824. [Google Scholar] [CrossRef] [PubMed]
- Zyczkowski, M.; Prokopowicz, G.; Taborowski, P.; Nowakowski, K.; Rajwa, P.; Stelmach, P.; Paradysz, A. Basic Parameters of Blood Count, Serum Sodium, and Creatinine as Prognostic Factors for Renal Cell Carcinoma at Five-Year Follow-Up. Med. Sci. Monit. 2018, 24, 3895–3902. [Google Scholar] [CrossRef] [PubMed]
- Lafleur, J.; Hefler-Frischmuth, K.; Grimm, C.; Schwameis, R.; Gensthaler, L.; Reiser, E.; Hefler, L.A. Prognostic Value of Serum Creatinine Levels in Patients with Epithelial Ovarian Cancer. Anticancer Res. 2018, 38, 5127–5130. [Google Scholar] [CrossRef] [PubMed]
- Baxmann, A.C.; Ahmed, M.S.; Marques, N.C.; Menon, V.B.; Pereira, A.B.; Kirsztajn, G.M.; Heilberg, I.P. Influence of muscle mass and physical activity on serum and urinary creatinine and serum cystatin C. Clin. J. Am. Soc. Nephrol. 2008, 3, 348–354. [Google Scholar] [CrossRef]
- Feeney, G.; Sehgal, R.; Sheehan, M.; Hogan, A.; Regan, M.; Joyce, M.; Kerin, M. Neoadjuvant radiotherapy for rectal cancer management. World J. Gastroenterol. 2019, 25, 4850–4869. [Google Scholar] [CrossRef]
- Feather, C.E.; Lees, J.G.; Makker, P.G.S.; Goldstein, D.; Kwok, J.B.; Moalem-Taylor, G.; Polly, P. Oxaliplatin induces muscle loss and muscle-specific molecular changes in Mice. Muscle Nerve 2018, 57, 650–658. [Google Scholar] [CrossRef]
- Qiu, C.; Zhang, Y.; Chen, L. Impaired Metabolic Pathways Related to Colorectal Cancer Progression and Therapeutic Implications. Iran J. Public Health 2020, 49, 56–67. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, P.; Xiao, Z. Resistant starch prevents tumorigenesis of dimethylhydrazine-induced colon tumors via regulation of an ER stress-mediated mitochondrial apoptosis pathway. Int. J. Mol. Med. 2018, 41, 1887–1898. [Google Scholar] [CrossRef]
- Prokopieva, V.D.; Yarygina, E.G.; Bokhan, N.A.; Ivanova, S.A. Use of Carnosine for Oxidative Stress Reduction in Different Pathologies. Oxidative Med. Cell. Longev. 2016, 2016, 2939087. [Google Scholar] [CrossRef] [PubMed]
- Prakash, M.D.; Fraser, S.; Boer, J.C.; Plebanski, M.; de Courten, B.; Apostolopoulos, V. Anti-Cancer Effects of Carnosine-A Dipeptide Molecule. Molecules 2021, 26, 1644. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, S.L.; Li, J.H.; Dong, C.D.; Chen, C.W.; Wu, C.C. Carnosine suppresses human colorectal cancer cell proliferation by inducing necroptosis and autophagy and reducing angiogenesis. Oncol. Lett. 2022, 23, 44. [Google Scholar] [CrossRef] [PubMed]
- Renner, C.; Asperger, A.; Seyffarth, A.; Meixensberger, J.; Gebhardt, R.; Gaunitz, F. Carnosine inhibits ATP production in cells from malignant glioma. Neurol Res. 2010, 32, 101–105. [Google Scholar] [CrossRef]
- Vaughan, R.A.; Gannon, N.P.; Garcia-Smith, R.; Licon-Munoz, Y.; Barberena, M.A.; Bisoffi, M.; Trujillo, K.A. beta-alanine suppresses malignant breast epithelial cell aggressiveness through alterations in metabolism and cellular acidity in vitro. Mol. Cancer 2014, 13, 14. [Google Scholar] [CrossRef]
- Treede, I.; Braun, A.; Sparla, R.; Kuhnel, M.; Giese, T.; Turner, J.R.; Anes, E.; Kulaksiz, H.; Fullekrug, J.; Stremmel, W.; et al. Anti-inflammatory effects of phosphatidylcholine. J. Biol. Chem. 2007, 282, 27155–27164. [Google Scholar] [CrossRef]
- van Roekel, E.H.; Trijsburg, L.; Assi, N.; Carayol, M.; Achaintre, D.; Murphy, N.; Rinaldi, S.; Schmidt, J.A.; Stepien, M.; Kaaks, R. Circulating metabolites associated with alcohol intake in the european prospective investigation into cancer and nutrition cohort. Nutrients 2018, 10, 654. [Google Scholar] [CrossRef] [PubMed]
- Jaremek, M.; Yu, Z.; Mangino, M.; Mittelstrass, K.; Prehn, C.; Singmann, P.; Xu, T.; Dahmen, N.; Weinberger, K.; Suhre, K. Alcohol-induced metabolomic differences in humans. Transl. Psychiatry 2013, 3, e276. [Google Scholar] [CrossRef]
| Patients with Colon Cancer | And with Rectal Cancer | p-Value * | |
|---|---|---|---|
| Colon cancer | Rectal cancer | ||
| Number of participants, n (%) | 393 (58) | 281 (42) | |
| Age at diagnosis, years (median, range) | 68 (62–75) | 63 (56–70) | <0.01 |
| Sex, n (%) | |||
| Male | 243 (62) | 194 (69) | 0.05 |
| Female | 150 (38) | 87 (31) | |
| Vital status, n (%) | |||
| Alive | 333 (85) | 248 (88) | 0.19 |
| Deceased | 60 (15) | 33 (12) | |
| Follow-up time, years (median, range) | |||
| Alive | 4.82 (3.16–6.03) | 4.01 (2.58–5.61) | <0.001 |
| Deceased | 2.79 (1.29–4.45) | 2.84 (0.92–3.51) | |
| Stage of disease, n (%) | |||
| I | 118 (30) | 55 (20) | <0.01 |
| II | 152 (39) | 62 (22) | |
| III | 123 (31) | 164 (58) | |
| Neo-adjuvant treatment, n (%) | |||
| Yes | 4 (1) | 176 (63) | <0.01 |
| No | 389 (99) | 105 (37) | |
| Surgery, n (%) ** | |||
| Yes | 392 (99.7) | 271 (96) | <0.01 |
| No | 1 (0.3) | 10 (4) | |
| Adjuvant treatment, n (%) | |||
| Yes | 116 (30) | 90 (33) | 0.45 |
| No | 268 (70) | 183 (67) | |
| Body mass index | |||
| Continuous, kg/m2 (median, range) | 26.80 (24.20–30.00) | 26.30 (24.00–29.10) | 0.048 |
| Underweight, <18.5, n (%) | 3 (1) | 4 (1) | 0.23 |
| Normal weight, 18.5–24.9, n (%) | 122 (31) | 98 (35) | |
| Overweight, 25–29.9, n (%) | 168 (43) | 125 (45) | |
| Obese, ≥30, n (%) | 100 (25) | 54 (19) | |
| Height, m (median, range) | 1.72 (1.65–1.78) | 1.73 (1.66–1.79) | 0.06 |
| Weight, kg (median, range) | 80.00 (69.5–90.00) | 79.60 (70.00–88.00) | 0.07 |
| Smoking, n (%) | |||
| Current | 43 (11) | 58 (21) | <0.01 |
| Former | 194 (51) | 144 (53) | |
| Never | 144 (38) | 71 (26) | |
| Alcohol intake, n (%) | |||
| Yes | 325 (85%) | 247 (89%) | 0.14 |
| No | 58 (15%) | 31 (11%) |
| HR 95% CI | HR 95% CI | p_Interaction | |
|---|---|---|---|
| Metabolite Name | Colon Cancer | Rectal Cancer | |
| PC_ae_C36_3 | 1.01 (0.77, 1.33) | 0.55 (0.36, 0.86) | 0.007 |
| PC_aa_C36_3 | 1.11 (0.82, 1.49) | 0.65 (0.43, 0.98) | 0.008 |
| PC_ae_C38_3 | 1.04 (0.79, 1.38) | 0.58 (0.38, 0.90) | 0.01 |
| PC_ae_C34_2 | 0.94 (0.72, 1.24) | 0.51 (0.32, 0.81) | 0.01 |
| PC_ae_C36_2 | 0.96 (0.73, 1.28) | 0.51 (0.32, 0.80) | 0.01 |
| PC_aa_C38_3 | 1.21 (0.91, 1.62) | 0.73 (0.47, 1.13) | 0.01 |
| PC_aa_C36_2 | 1.05 (0.77, 1.43) | 0.61 (0.39, 0.96) | 0.02 |
| PC_ae_C44_3 | 0.80 (0.58, 1.10) | 0.56 (0.36, 0.88) | 0.02 |
| PC_ae_C36_1 | 1.01 (0.76, 1.34) | 0.58 (0.38, 0.88) | 0.02 |
| PC_ae_C30_2 | 0.91 (0.68, 1.22) | 0.41 (0.24, 0.71) | 0.03 |
| PC_aa_C34_2 | 0.96 (0.71, 1.31) | 0.55 (0.34, 0.88) | 0.03 |
| lysoPC_a_C18_0 | 0.96 (0.70, 1.30) | 0.66 (0.43, 1.00) | 0.03 |
| lysoPC_a_C17_0 | 0.90 (0.66, 1.23) | 0.58 (0.38, 0.90) | 0.045 |
| Pathway Analysis | Total | Expected | Hits | FDR | Impact |
|---|---|---|---|---|---|
| Arginine and proline metabolism | 77 | 0.19 | 2 | <0.001 | 0.02 |
| Sphingolipid metabolism | 25 | 0.06 | 1 | <0.001 | 0.01 |
| Beta-alanine metabolism | 28 | 0.07 | 1 | <0.001 | <0.01 |
| Glycolysis or Gluconeogenesis | 31 | 0.08 | 1 | <0.001 | <0.01 |
| Pentose phosphate pathway | 32 | 0.08 | 1 | <0.001 | <0.01 |
| Nitrogen metabolism | 39 | 0.10 | 1 | <0.001 | <0.01 |
| Histidine metabolism | 44 | 0.11 | 1 | <0.001 | 0.14 |
| Glycine, serine, and threonine metabolism | 48 | 0.12 | 1 | <0.001 | 0.05 |
| Starch and sucrose metabolism | 50 | 0.12 | 1 | <0.001 | <0.01 |
| Aminoacyl-tRNA biosynthesis | 75 | 0.19 | 1 | <0.001 | <0.01 |
| Pathways related to metabolites associated with increased risk of death | |||||
| Arginine and proline metabolism | 77 | 0.13 | 2 | <0.001 | 0.02 |
| Glycolysis or Gluconeogenesis | 31 | 0.05 | 1 | <0.001 | <0.01 |
| Pentose phosphate pathway | 32 | 0.05 | 1 | <0.001 | <0.01 |
| Glycine, serine, and threonine metabolism | 48 | 0.08 | 1 | <0.001 | 0.05 |
| Starch and sucrose metabolism | 50 | 0.08 | 1 | <0.001 | <0.01 |
| Pathways related to metabolites associated with decreased risk of death | |||||
| Sphingolipid metabolism | 25 | 0.02 | 1 | <0.001 | 0.01 |
| Beta-alanine metabolism | 28 | 0.02 | 1 | <0.001 | <0.01 |
| Nitrogen metabolism | 39 | 0.03 | 1 | <0.001 | <0.01 |
| Histidine metabolism | 44 | 0.04 | 1 | <0.001 | 0.14 |
| Aminoacyl-tRNA biosynthesis | 75 | 0.06 | 1 | <0.001 | <0.01 |
| Pathway Analysis Overall | Total | Expected | Hits | p-Value | Impact |
|---|---|---|---|---|---|
| Glycerophospholipid metabolism | 39 | 0.10 | 2 | <0.001 | 0.10 |
| Arginine und proline metabolism | 77 | 0.19 | 2 | <0.001 | 0.08 |
| Linoleic acid metabolism | 15 | 0.04 | 1 | <0.001 | <0.01 |
| Sphingolipid metabolism | 25 | 0.06 | 1 | <0.001 | 0.01 |
| Alpha-linolenic acid metabolism | 29 | 0.07 | 1 | <0.001 | <0.01 |
| Arachidonic acid metabolism | 62 | 0.15 | 1 | <0.001 | <0.01 |
| Pathways related to metabolites associated with increased risk of death | |||||
| Arginine und proline metabolism | 77 | 0.10 | 2 | <0.001 | 0.08 |
| Pathways related to metabolites associated with decreased risk of death | |||||
| Glycerphospholipid metabolism | 39 | 0.05 | 2 | <0.001 | 0.10 |
| Linoleic acid metabolism | 15 | 0.02 | 1 | <0.001 | <0.01 |
| Sphingolipid metabolism | 25 | 0.03 | 1 | <0.001 | 0.01 |
| Alpha-linolenic acid metabolism | 29 | 0.04 | 1 | <0.001 | <0.01 |
| Arachidonic acid metabolism | 62 | 0.08 | 1 | <0.001 | <0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ose, J.; Gigic, B.; Brezina, S.; Lin, T.; Peoples, A.R.; Schobert, P.P.; Baierl, A.; van Roekel, E.; Robinot, N.; Gicquiau, A.; et al. Higher Plasma Creatinine Is Associated with an Increased Risk of Death in Patients with Non-Metastatic Rectal but Not Colon Cancer: Results from an International Cohort Consortium. Cancers 2023, 15, 3391. https://doi.org/10.3390/cancers15133391
Ose J, Gigic B, Brezina S, Lin T, Peoples AR, Schobert PP, Baierl A, van Roekel E, Robinot N, Gicquiau A, et al. Higher Plasma Creatinine Is Associated with an Increased Risk of Death in Patients with Non-Metastatic Rectal but Not Colon Cancer: Results from an International Cohort Consortium. Cancers. 2023; 15(13):3391. https://doi.org/10.3390/cancers15133391
Chicago/Turabian StyleOse, Jennifer, Biljana Gigic, Stefanie Brezina, Tengda Lin, Anita R. Peoples, Pauline P. Schobert, Andreas Baierl, Eline van Roekel, Nivonirina Robinot, Audrey Gicquiau, and et al. 2023. "Higher Plasma Creatinine Is Associated with an Increased Risk of Death in Patients with Non-Metastatic Rectal but Not Colon Cancer: Results from an International Cohort Consortium" Cancers 15, no. 13: 3391. https://doi.org/10.3390/cancers15133391
APA StyleOse, J., Gigic, B., Brezina, S., Lin, T., Peoples, A. R., Schobert, P. P., Baierl, A., van Roekel, E., Robinot, N., Gicquiau, A., Achaintre, D., Scalbert, A., van Duijnhoven, F. J. B., Holowatyj, A. N., Gumpenberger, T., Schrotz-King, P., Ulrich, A. B., Ulvik, A., Ueland, P.-M., ... Ulrich, C. M. (2023). Higher Plasma Creatinine Is Associated with an Increased Risk of Death in Patients with Non-Metastatic Rectal but Not Colon Cancer: Results from an International Cohort Consortium. Cancers, 15(13), 3391. https://doi.org/10.3390/cancers15133391






