Simple Summary
Pulmonary sarcomatoid carcinoma (PSC) is an aggressive subtype of non-small-cell lung cancer (NSCLC). It does not respond favorably to standard chemotherapy, and the response to PD-1/PD-L1 inhibitors remains modest. The introduction of new therapeutic approaches for this subtype is crucial. Our study demonstrates that PD-L1 expression was significantly higher in the epithelial component than in the sarcomatoid component. Expression of PD-L1 in both components was only seen in 32.1% of patients. However, the majority of PSC patients had at least one immune checkpoint expression in both components. Thus, combination immune checkpoint inhibition based on expression profiles may prove as a personalized and effective treatment strategy. This study also reveals a high rate of MET exon 14 skipping mutation (METex14) in PSC. METex14 selectively induced PD-L1 expression through MAPK or PI3K/Akt pathways. A combination of targeted therapies with immunotherapy in this population also warrants further investigation as a novel treatment approach.
Abstract
Immunotherapy has transformed lung cancer management, but PSC remains an aggressive subtype with a poor prognosis. This study investigates the differential expression of PD-L1 and alternative immune checkpoints (ICs; B7x, B7-H3, and HHLA2), and genetic alterations in PSCs. Tumor specimens of 41 PSC patients were evaluated. PD-L1, B7x, B7-H3, and HHLA2 were positive in 75.0%, 67.6%, 73.0%, and 91.9% of tumors, respectively. PD-L1 expression was significantly higher in the epithelial compared to the sarcomatoid component (median TPS: 50% vs. 0%, p = 0.010). Expression of PD-L1 in both components was only seen in 32.1% of patients. However, at least one IC was expressed in 92.9% of epithelial and 100% of sarcomatoid components. Furthermore, METex14 was detected in 19.5% of patients and was associated with a higher sarcomatoid percentage. Our preclinical studies revealed that METex14 induced PD-L1 expression via MAPK or PI3K/Akt pathways, and MET inhibitors decreased PD-L1 expression. Our findings demonstrate distinct expressions of ICs in PSC subcomponents. Thus, combination IC inhibition as a therapeutic strategy in PSC warrants further exploration. A high percentage of METex14 in PSC and its role in regulating PD-L1 expression reveal different therapeutic targets in this aggressive NSCLC subtype.
1. Introduction
PSC is a rare and aggressive subtype of NSCLC. It accounts for about 0.5% of NSCLC cases diagnosed in the United States and is associated with poor survival rates [1,2]. PSC is a heterogeneous entity encompassing a broad histological spectrum of disease, subcategorized into pleomorphic carcinoma, spindle cell carcinoma, giant cell carcinoma, carcinosarcoma, and pulmonary blastoma [3].
The NSCLC treatment paradigm and clinical outcomes have shifted drastically with the incorporation of targeted therapy and immunotherapy in the last several decades. Despite these advancements, PSC remains a subtype with a poor prognosis necessitating further investigation of underlying oncogenic pathways, which may ultimately lead to new biomarker-driven therapeutic approaches. In particular, the heterogeneous nature of PSC with varying levels of epithelial and sarcomatoid components requires better characterizing the differing immune checkpoint expression and actionable genetic alterations in these distinct compartments, which may offer a more personalized therapeutic approach for these patients.
PSC has been demonstrated to respond poorly to first-line platinum-based chemotherapy [4]. It is highly aggressive and has worse overall survival compared to the other subtypes of NSCLC [5,6]. Hence, the evaluation of novel therapies in PSC is crucial. A better understanding of immune evasion pathways in PSC is fundamental to maximizing the benefits of immunotherapy. PD-L1 expression is reported to be higher in PSC than in conventional NSCLC, leading to the hypothesis of increased immunotherapy efficacy in this subgroup. Multiple retrospective studies demonstrated an overall response rate of 40–55% upon treatment with PD-1/PD-L1 inhibitors in the PSC population [7,8]. In addition, the use of immunotherapy has produced durable responses in rare PSC patients [9]. Nonetheless, despite a high level of PD-L1 expression in PSC, these response rates to PD-1/PD-L1 inhibitors remain modest. Therefore, extending durable immunotherapy benefits to a broader population in PSC is critical. One promising strategy for achieving this is combination immune checkpoint inhibition as a therapeutic approach. In addition to the PD-L1 and CTLA-4 axis, several additional potentially actionable immune checkpoint molecules have recently been discovered, including the group III molecules of the B7-CD28 immune checkpoint family, which are B7x (B7-H4/B7S1), B7-H3 (CD276), and HHLA2 (B7H7/B7-H5/B7y) [10]. These alternate immune checkpoints predominantly act as co-inhibitors of T-cell function but also may have other functions depending on the engagement of different receptors or immune microenvironment [11,12,13]. Understanding the immune evasion pathways of these molecules, especially in NSCLC, is crucial since they are reported to be commonly expressed in 37–69% of this subgroup of solid tumors [14,15,16,17]. Effective inhibition of these alternative immune checkpoints may prove a promising therapeutic strategy, especially in PD-L1 negative NSCLC, which has been shown to express these alternative molecules widely or in association with primary resistance to PD-1/PD-L1 blockade [14]. As the understanding of these alternate immune checkpoints improves and potent inhibitors are developed, preclinical studies for some are now advancing into clinical trials [10,18].
Sarcomatoid carcinomas are hypothesized to evolve from a shared precursor through epithelial–mesenchymal transition (EMT) [19]. Phylogenetic and genomic profiling of two components of PSC demonstrated that the sarcomatous component arises from an epithelial precursor early in the course of cancer evolution and accumulates different genetic alterations [20]. Similarly, another study investigating gene signatures in biphasic PSCs also revealed that loss of the epithelial-associated transcription factor promotes the development of the sarcomatoid phenotype by expressing EMT-driving transcription factors [21]. As these sarcomatoid carcinomas seem to arise and differentiate from their precursor cells, understanding and therapeutically targeting both components are essential to improve clinical outcomes. In fact, the HGF (hepatocyte growth factor)/MET pathway has been hypothesized to affect the motility and differentiation of epithelial cells and play a role in epithelial–mesenchymal interaction [22,23]. Therefore, genetic alterations of the MET gene and their contribution to the pathogenesis of PSC require further elucidation. In fact, studies of genetic alterations in PSC revealed a high percentage of tumor protein p53 (TP53) (74%), KRAS proto-oncogene, GTP-ase (KRAS) (34%), MET proto-oncogene receptor protein kinase (MET) (13.6%), and epidermal growth factor receptor (EGFR) (8.8%) variants [24]. Our group also reported enrichment of METex14 in PSC patients, occurring in about 20% compared to 0.5–3% in other NSCLC subtypes [25,26,27]. Interestingly, similar to the PSC population, METex14 mutant NSCLC has also demonstrated high levels of PD-L1 expression [28,29]. Whole-transcriptome sequencing of a large cohort of METex14 also revealed a highly immunosuppressive tumor microenvironment with a wide expression of immune checkpoints and inflammatory signature due to IFN-γ signaling [29]. Considering these findings, a more detailed understanding of the regulatory mechanisms of immune checkpoints in METex14 variants is crucial for biomarker-driven therapeutic approaches.
This study investigates the heterogeneity of PD-L1 and alternative immune checkpoints B7x, B7-H3, and HHLA2 in differing PSC components harboring genetic alterations of MET, KRAS, EGFR, and ALK. In addition, given early evidence of enrichment of METex14 mutation in PSC and high levels of PD-L1 expression, this study also aims to explore cellular pathways that may explain this association.
2. Materials and Methods
2.1. Patients
Forty-one patients with pathologically confirmed PSC were identified from a previously reported clinical cohort of patients with NSCLC. The clinicopathological data were collected through retrospective chart review, and the tissue microarray method was implemented, as described previously [14]. All protocols were reviewed and approved by the Institutional Review Board.
2.2. Immunohistochemistry
The immunohistochemistry (IHC) staining method used in this study was derived from prior studies [14,15,30]. In brief, tumor tissues were fixed in 10% neutral-buffered formalin at room temperature for 24 h. The formalin fixed lung tumor tissue was baked at 60 °C for 1 h to overnight followed by deparaffinization and rehydration of tumor tissue sections using xylene and ethanol. The slides were immersed in Dako dual endogenous enzyme block for 10 min at room temperature to block endogenous peroxidase activity (Dako corporation, S2003). Dako target retrieval citrate solution pH 6.1 (Agilent Dako code number S1699) was used for the antigen retrieval step. The slides were heated in the microwave at 900 W for 2 min, placed in a steamer for 30 min, and left to cool for 20 min at room temperature. Dako Protein Block Serum-Free (Agilent Dako code number X0909) was used to block nonspecific binding. The primary antibodies used in this study and incubation details are available in Table 1.

Table 1.
Antibodies used for immunohistochemistry.
Dako Envision system-HRP labeled polymer anti-mouse system (Dako Corporation code K4000) was used and incubated for 30 min at room temperature, followed by DAB chromogen staining (Vector laboratories SK-4100) and hematoxylin nuclear counterstaining. PD-L1, B7x, HHLA2, and B7-H3 expression was evaluated by two independent investigators and quantified as 0, 1, 2, and 3 for absent, low, moderate, and high expression. The H-score was calculated by multiplying the percentage of staining (proportion score) measured by Image J and an ordinal value corresponding to the maximum intensity score in the specimen. PD-L1 staining was reported as a tumor proportion score (TPS).
Tumor-infiltrating lymphocyte (TIL) scores were read by visual estimation of the proportion of lymphocytic infiltration in each histospot of the same hematoxylin- and eosin-stained slides, as previously described [15].
2.3. Cell Culture
H292 and H125 cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). H292 and H125 cells were maintained in RPMI-1640 medium and supplemented with 10% FBS fetal bovine serum (FBS) with penicillin and streptomycin (Life Technologies, Carlsbad, CA, USA) at 37 °C under 5% CO2.
2.4. CRISPR
As previously described, two guide RNAs (gRNAs) separately targeting MET Exon 13 and Exon 15 were designed using the CRISPR design tool (http://crispr.mit.edu/, accessed on 1 February 2017) to generate METex14 [34]. gRNA1 (5′-CTTGTTAAAGACGGCTATCA-3′) and gRNA2 (5′-ACCCACTGAGGTATATGTAT-3′) were cloned into the BbsI site of plasmid PX458 (SpCas9-2A-EGFP) (Addgene: #48138). The transfection step was completed using Lipofectamine 3000 (Life Technologies, Carlsbad, CA, USA), and single-cell clones were selected using flow cytometry.
2.5. RNA Sequencing
Following incubation in a serum-free medium for 18 h, the cells were treated with HGF (100 ng/mL) for 0, 6, and 24 h. The cells were collected, and total RNA was extracted using an RNeasy mini kit (QIAGEN, Valencia, CA, USA). RNA-seq libraries were prepared with the TruSeq sample preparation kit (Illumina, San Diego, CA, USA). The sequencing was performed using the BGI2000 platform with paired-end reads of 200 bp. We sequenced 18 samples in total and the reads in all the samples satisfied the quality control by FastQC (v0.11.9) [35]. The reads were mapped to the human reference genome (GRCh38) using STAR aligner (v2.6.1b) [36], and alignments were guided by a human gene annotation (Gencode primary assembly annotation v30). More than 96% of reads in individual samples were successfully and uniquely mapped, and the mismatch rate was less than 0.35%. We employed HTSeq (v0.6.1) [37] to count reads from the output (bam files) of the STAR aligner and identified >17,700 expressed genes per sample. We performed the standard protocols of STAR and HTSeq in this study, with the default parameters. The raw counts of samples were normalized on the basis of their library sizes, and the differential gene expression analyses were performed on the basis of the negative binomial distribution using the ‘DESeq2’ package (v1.26.0) [38].
2.6. Quantitative PCR
Quantitative reverse transcription PCR (qRT–PCR) assays were performed to measure the mRNA expression of multiple immune checkpoints. Total RNA was extracted from treated cells using an RNeasy Mini Kit (QIAGEN, CA). cDNA was synthesized from 1 μg of purified total RNA with the SuperScriptTM IV First-Strand cDNA synthesis system using oligo(dT) primers (Life Technologies, Carlsbad, CA, USA), and qPCR was performed using CFX96 Real-Time PCR Detection System with the following primer sequences (Table 2).

Table 2.
Primer sequences for quantitative PCR.
All gene expression levels were normalized to the reference gene expression, and relative changes in expression were calculated by the 2−ΔΔCT formula.
2.7. Flow Cytometry
For PD-L1, B7-H3, and B7x expression, cells pretreated at the indicated conditions were stained with APC anti-hPD-L1 (clone 29E.2A3), PE/Cy7 anti-hB7-H3 (clone MIH42), PE anti-hB7x (clone MIH43), or isotype controls for 30 min at 4 °C. Fluorophore-conjugated antibodies were purchased from BioLegend. The staining of HHLA2 was performed by first incubating cells with primary mouse anti-hHHLA2 mAb (clone B5B5) [13] or control mouse IgG1 (clone MOPC-21) for 40 min at 4 °C, followed by staining with APC polyclonal goat F(ab′)2 anti-mouse IgG Fc (eBioscience) for 30 min at 4 °C. Cells were washed after staining and analyzed on an LSR II (BD Biosciences, San Jose, CA, USA). Data were analyzed with FlowJo (FlowJo, LLC, Ashland, OR, USA).
2.8. Western Blot
Total proteins were extracted from treated cells with RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 1 mM EDTA) supplemented with the protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, Rockford, IL, USA). The cell lysate was separated by SDS polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to the nitrocellulose membrane (Millipore, St. Louis, MO, USA). The membranes were soaked with 5% skimmed milk (in PBS) for 1 h and then incubated with the primary antibodies (Table 3) at 4 °C overnight.

Table 3.
Primary antibodies for Western blot.
The membranes were incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody (Cell signaling technology catalog no 7074, Danvers, MA, USA) at 1:2000 dilution for 1 h, and the image was developed with an ECL detecting kit (Amersham Biosciences, Piscataway, NJ, USA, catalog no RPN2108).
2.9. Statistical Analysis
The χ2 test or Fisher exact test was used to compare the distribution of each categorical variable. The Wilcoxon rank-sum test was used for the nonparametric comparison of distribution. Logistic regression was used to adjust variables for multivariate analyses. Survival analysis was performed by log-rank test and further adjusted by Cox regression. All tests were completed as two-sided using IBM SPSS Statistics (version 22).
Statistical analysis for in vitro cell-based experiments was performed using GraphPad Prism (version 8.0). The two-sided t-test was used to estimate the statistical significance of differences between the two groups. Two-way ANOVA with Bonferroni correction was used to determine statistical significance for real-time quantitative PCR analysis. Data are presented as the mean ± standard deviation.
Statistical significance is indicated as follows: * p < 0.05, ** p < 0.01, and *** p < 0.001. All p-values ≤0.05 were considered statistically significant.
3. Results
3.1. Clinicopathological Characteristics, Immune Checkpoint Expression, and Genetic Alterations in PSC Patients
The cohort’s median age was 73 years (range: 38–87). Gender distribution was similar with 48.8% of the patients (20/41) being male. Of the patients with known smoking status (n = 38), 7.3% were never smokers. Among the cohort, 29.3%, 41.5%, 24.4%, and 4.9% of patients had stage I, II, III, and IV disease, respectively. Among patients with available subtype information (n = 37), 97.3% had pleomorphic carcinoma, and 2.7% had spindle cell carcinoma. The epithelial histology was the most dominant component across the PSC samples with a median of 70% per sample (interquartile range: 26–85%), followed by a sarcomatoid component of 30% (interquartile range: 15–50%) and spindle cell component of 25% (interquartile range: 15–50%).
METex14 and KRAS mutations were found in 19.5% and 14.6%, respectively, while EGFR/ALK alterations were absent. PD-L1, B7x, B7-H3, and HHLA2 were positive in 75.0% (27/36), 67.6% (25/37), 73.0% (27/37), and 91.9% (34/37) of patients, respectively (representative images in Figure 1). A TIL proportion equal to or greater than 30% was found in 28.6% (10/35) patients. There were no significant associations between clinicopathological factors and the expression of any of the four immune checkpoints (Table 4).
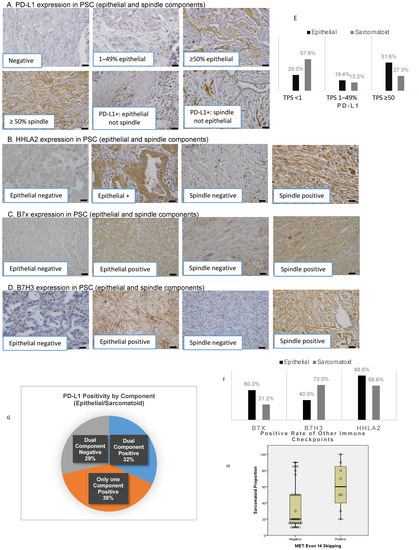
Figure 1.
Immune checkpoint distribution and the correlation of sarcomatoid percentage with genetic alterations. Immunohistochemical staining of PD-L1 (A), HHLA2 (B), B7x (C), and B7-H3 (D) in PSC scale bar 20 µm. Distribution of PD-L1 (E) and alternate immune checkpoint molecules (F) by histologic components. The distribution of PD-L1 positivity in histologic subcomponents (G). The correlation of sarcomatoid percentage with METex14 (H).

Table 4.
Patient characteristics by expression of immune checkpoints.
3.2. Distinct Expression of Immune Checkpoints in Epithelial and Sarcomatoid Components
PD-L1 expression was significantly higher in the epithelial compared to the sarcomatoid component (median TPS: 50% vs. 0%, p = 0.010, H-score 75 vs. 0, p = 0.014, Table 5A). The percentage of TPS <1%, 1–49%, and ≥50% differed between epithelial and sarcomatoid components (29.0% vs. 57.6%, 19.4% vs. 15.2%, and 51.6% vs. 27.3%, Figure 1E). Analogous to PD-L1, B7x expression was more enriched in the epithelial than in the sarcomatoid component (TPS 25 vs. 0, p = 0.008). In contrast, there was a trend toward higher expression of B7-H3 in the sarcomatoid component (TPS 0% vs. 50%, p = 0.053, H-score epithelial 0% vs. 75%, p = 0.176, Table 5A). There was no significant difference found in HHLA2 expression between the two components.

Table 5.
Expression of immune checkpoints in epithelial and sarcomatoid components. A. Differential expression of immune checkpoints. B. PD-L1 co-expression with other immune checkpoints on epithelial and sarcomatoid components. C. Alternative immune checkpoint expression in dual PD-L1-negative tumors.
Although PD-L1 was positive in most tumors, dual expression of PD-L1 in epithelial and sarcomatoid components was only present in 32.1% (9/28) of patients (Figure 1G). PD-L1 was expressed in only one component in 39% of patients, and PD-L1 was negative in both components in 29% of patients.
PD-L1 expression was not associated with the other three immune checkpoints (Table 4). At least one immune checkpoint was expressed in 92.9% of epithelial and 100% of sarcomatoid components (Table 5B). In tumors with negative PD-L1 in both components, HHLA2 was expressed in 100% of epithelial and 66.7% of sarcomatoid components (Table 5C). In tumors with negative PD-L1 in both components and negative HHLA2 in the sarcomatoid component, B7-H3 was 100% positive in the sarcomatoid component.
3.3. Association of Epithelial and Sarcomatoid Distribution in PSC Samples with Oncogenic Genomic Alterations and Immune Checkpoints
Tumors harboring METex14 had significantly higher sarcomatoid percentage (median 60%, interquartile range: 35–88% vs. MET wildtype 20%, interquartile range: 15–50%, p = 0.024, Figure 1H). After adjusting for age, smoking, stage, and gender, a trend toward a higher sarcomatoid percentage in this subpopulation was preserved (p = 0.055). There was no significant association between higher sarcomatoid percentage with KRAS mutation or positivity of immune checkpoints.
3.4. Genomic Alterations and Their Association with Immune Checkpoints and Tumor-Infiltrating Lymphocytes
METex14 and KRAS mutations were mutually exclusive in our cohort. METex14 was associated with a higher B7-H3 H-score in the sarcomatoid component compared to the wildtype (p = 0.017, Supplementary Table S1). METex14 was also associated with a higher HHLA2 H-score in both epithelial (p < 0.001) and sarcomatoid (p = 0.031) components (Supplementary Table S1). METex14 tumors also had numerically higher PD-L1 TPS in the sarcomatoid component compared to MET wildtype (p = 0.079). KRAS mutation was significantly associated with a lower HHLA2 H-score in the epithelial component (p = 0.041).
PSC samples with METex14 were more likely to have higher tumor-infiltrating lymphocytes (p = 0.027, Supplementary Table S2). However, after adjusting for age, gender, smoking, and stage, the association was no longer significant (p = 0.093). There were no significant associations between KRAS mutation and tumor-infiltrating lymphocytes (Supplementary Table S2).
3.5. Survival Analysis
The median survival of the entire cohort was 1020 days (95% CI: 643–1397). After adjusting for age, gender, smoking, stage, genetic mutation, and expression of immune checkpoints, a higher percentage of the sarcomatoid component was associated with shorter survival (continuous variable, HR 1.048, 95% CI: 1.007–1.080). After adjusting for age, gender, smoking, and stage, the lack of B7-H3 expression in the sarcomatoid component was significantly associated with better survival (HR 0.034, 95% CI 0.002–0.741). However, there was no significant association between B7-H3 expression in the epithelial component and patient survival (Supplementary Table S3).
3.6. HGF/METex14 Signaling Specifically Induces PD-L1 Expression
METex14 lung cancer cell line models were established using CRISPR technology, where deletion of MET exon 14 in both alleles was regarded as mutated (METex14), and cell lines with intact MET exon 14 were used as wildtype (Figure 2A). RNA-sequencing analysis was pursued to clarify the global transcriptome signatures underlying HGF/METex14 signaling. As shown in Figure 2B, the expressions of CD276 (B7-H3), CD274 (PD-L1), and programmed cell death 1 ligand 2 (PDCD1LG2, PD-L2) were enhanced post 6 h and 24 h HGF treatment. In contrast, the expressions of V-Set immunoregulatory receptor (VSIR, VISTA), CD86, and V-Set domain containing T-cell activation inhibitor 1 (VTCN1) genes were decreased at 6 h and reversed to a baseline level at 24 h post HGF treatment in both METex14 and MET WT cell lines (Figure 2B). qRT-PCR and flow cytometry analyses also confirmed these observations. As shown in Figure 2C, the expression level of CD274 mRNA was markedly increased at 6 h followed by a decrease at 24 h of HGF treatment in both MET and METex14 cell lines. In addition, CD274 mRNA expression levels were significantly higher in both 6 h and 24 h in METex14 compared to MET cells. However, mRNA expression of CD276, VTCN1, and HHLA2 did not show a significant change in exposure to HGF in MET WT and METex14 cells (Figure 2C). Simultaneously, flow cytometry analyses validated that HGF treatment dramatically enhanced the PD-L1 and B7-H3 expression at all timepoints over 24 h but did not affect B7x and HHLA2 expression in either H292 MET WT or METex14 cell lines. The comparison between METex14 and MET WT cell lines revealed that HGF treatment led to significantly higher PD-L1 but similar B7-H3 expression in METex14 cells (Figure 2D). Similar results were obtained in modified H125 cells except for the lack of increase in B7-H3 by HGF treatment (Figure 2E,F). Furthermore, we also validated that HGF/METex14 signaling promotes PD-L1 expression using a Western blot assay in a time-dependent manner (Figure 2G). These results suggest that HGF/METex14 signaling significantly increases PD-L1 expression in both cell lines, whereas it might not affect B7-H3, B7x, and HHLA2 expression.
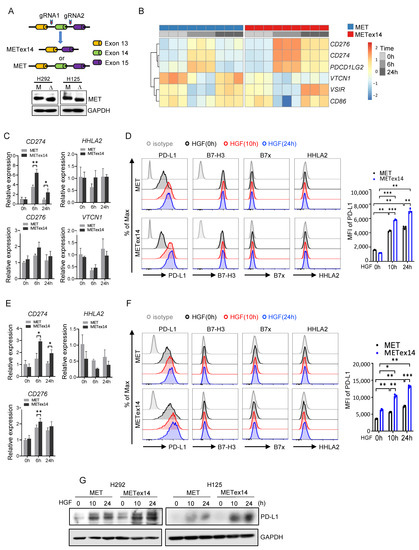
Figure 2.
Effect of HGF/METex14 signaling on expression of multiple immune checkpoints. (A) Schematic diagrams of the establishment of METex14 cell models using CRISPR gene editing technology. Western blot analysis of MET WT and METex14 expression in H292 and H125 cells. M: MET; Δ: METex14. (B) RNA sequencing analysis was pursued on H292 MET WT and METex14 cells treated with HGF at indicated times. The expression profile of a group of immune checkpoints is shown. (C–F) Analysis of expression level of multiple immune checkpoints in MET WT and METex14 cells treated with HGF at indicated timepoints. The fold changes of CD276, CD274, VTCN1, and HHLA2 mRNA expression using qRT-PCR (H292 cells in C, H125 cells in E), and flow cytometry analysis of expression of PD-L1, B7-H3, B7x, and HHLA2 (H292 cells in D, H125 cells in F) are shown. (G) Western blot analysis of PD-L1 expression in MET WT and METex14 cells exposed to HGF at indicated times. MFI: mean fluorescence intensity. The two-sided t-test was used to estimate the statistical significance of differences between the two groups. Two-way ANOVA with Bonferroni correction was used to determine statistical significance for real-time quantitative PCR analysis. * p < 0.05; ** p < 0.01; *** p < 0.001. The uncropped blots are shown in File S1.
3.7. METex14 Drives PD-L1 Expression via PI3K/Akt and MAPK Signaling Cascades, and MET Inhibitors Block PD-L1 Expression
To understand how HGF/METex14 signaling regulates PD-L1 expression, we investigated the PI3K/Akt and MAPK signaling cascades. Similar to our prior work, HGF treatment delayed receptor degradation and significantly increased the activity of PI3K/Akt and MAPK in the METex14 cell line compared to the MET WT cell line (Figure 3A,B) [34]. We next assessed the effect of LY294002 (PI3K inhibitor) and U0126 (MAPK inhibitor) on HGF-dependent PD-L1 expression. As expected, LY294002 and U0126 markedly repressed HGF-driven p-Akt and p-MAPK in both MET and METex14 cell lines (Figure 3C,D). More importantly, both LY294002 and U0126 dramatically inhibited HGF-elicited PD-L1 expression in both MET and METex14 cell lines (Figure 3E,F in H292 cells, and Figure 3G,H in H125 cells). These results indicated that HGF/METex14 signaling significantly increases PD-L1 expression via PI3K/Akt and MAPK pathways.
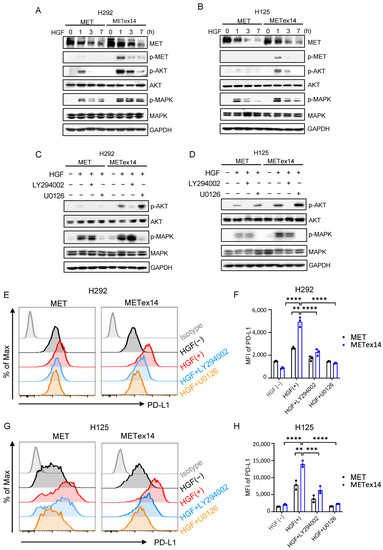
Figure 3.
HGF/METex14 signaling significantly drives the expression of PD-L1 via activation of PI3K/Akt and MAPK pathways. (A,B) Effect of HGF/METex14-mediated downstream signaling. MET WT and METex14 cells (H292 (A) and H125 (B)) treated with HGF (100 ng/mL) at indicated timepoints were harvested, and phosphorylated and total protein levels of MET, Akt, and MAPK were measured using Western blot. (C–H) Blockade of PI3K/Akt and MAPK pathways inhibited HGF/METex14-triggered PD-L1 expression. MET WT and METex14 cells (H292 (C) and H125 (D)) treated with HGF with or without LY294002 (PI3K inhibitor, 20 μM) or U0126 (MAPK inhibitor, 10 μM) for 3 h were harvested. Western blot analysis of phosphorylated and total protein levels of Akt, MAPK, and GAPDH are shown. Flow cytometry analysis of PD-L1 expression in MET and METex14 cells in exposure to HGF at indicated times (H292 (E,F) and H125 (G,H)) are shown. MFI: mean fluorescence intensity. The two-sided t-test was used to estimate the statistical significance of differences between the two groups. Two-way ANOVA with Bonferroni correction was used to determine statistical significance for real-time quantitative PCR analysis. ** p < 0.01; *** p < 0.001; **** p < 0.0001. The uncropped blots are shown in File S1.
These results raise the possibility that inhibition of MET signaling might repress PD-L1 expression. We next assessed this possibility using two MET inhibitors: capmatinib and tepotinib. Capmatinib, similar to our prior work [34], and tepotinib markedly repressed MET receptor phosphorylation and downstream Akt and MAPK phosphorylation (Figure 4A,B). More importantly, capmatinib and tepotinib treatment vigorously suppressed HGF-induced PD-L1 expression in both MET and METex14 cell lines (Figure 4C,D in H292 cells, Figure 4E,F in H125 cells). These results demonstrate that MET inhibitor reduces HGF/METex14 signaling-driven PD-L1 expression in both H292 and H125 NSCLC cell lines.
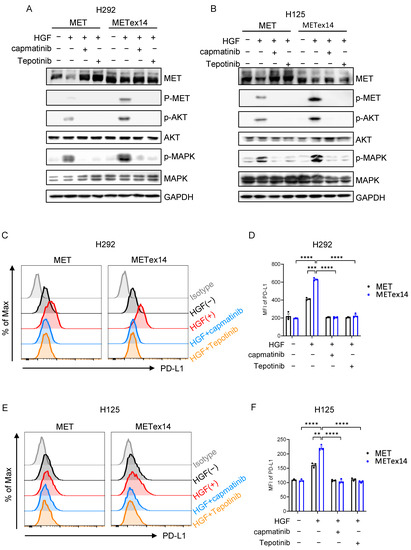
Figure 4.
Effect of MET inhibitors, capmatinib and tepotinib, on HGF/METex14-triggered PD-L1 expression. (A,B) Effect of MET inhibitors, tepotinib or capmatinib, on METex14-mediated receptor degradation and downstream signaling. MET WT and METex14 cells treated with HGF (100 ng/mL) with or without capmatinib (100 nM) or tepotinib (100 nM) for 1 h were lysed, and phosphorylated and total protein levels of MET, AKT, and MAPK were measured using Western blot (H292 (A) and H125 (B)). (C–F) The effect of capmatinib or tepotinib on HGF/METex14-triggered PD-L1 expression. Flow cytometry analysis of PD-L1 expression in MET and METex14 cells in exposure to HGF with capmatinib or tepotinib at indicated times (H292 (C,D) and H125 (E,F)) are shown. MFI: mean fluorescence intensity. The two-sided t-test was used to estimate the statistical significance of differences between the two groups. Two-way ANOVA with Bonferroni correction was used to determine statistical significance for real-time quantitative PCR analysis. ** p < 0.01; *** p < 0.001; **** p < 0.0001. The uncropped blots are shown in File S1.
4. Discussion
Given the known heterogeneity of PSC, our study investigated the differential expression of immune checkpoints in epithelial and sarcomatoid components, as well as their associations with genetic alterations. Prior studies have reported high PD-L1 expression in the PSC population, consistent with our study results [39,40]. However, the reports of the distribution of PD-L1 expression in epithelial and sarcomatoid components of PSC are few and demonstrated variable findings [20,41]. Our study is the first to report that PD-L1 expression is more enriched in the epithelial compared to the sarcomatoid component. These differences may stem from different patient characteristics or antibodies.
In our cohort, dual component positivity of PD-L1 was only seen in 32% of tumors. While the intricate mechanism of differential PD-L1 expression in these two components currently remains unclear, it may result in important therapeutic consequences. For example, despite higher PD-L1 TPS in PSC, the heterogeneity of PD-L1 expression in the various components may limit the efficacy of single-agent immunotherapy. Our group previously showed that the majority of PD-L1 negative NSCLC expresses alternative immune checkpoints, such as B7x and HHLA2 [14]. In addition, recently, HHLA2’s interaction with KIR3DL3 (killer cell Ig-like receptor, three Ig domains, and long cytoplasmic tail 3) was demonstrated to inhibit the function of CD8+ T and NK cells, and a first-in-class monoclonal antibody, NPX267, targeting KIR3DL3 enhanced antitumor immunity in HHLA2+ tumors [13,42]. In this PSC cohort, over 90% of PSC tumors expressed at least one immune checkpoint (B7x, B7-H3, HHLA2, or PD-L1) in both components. Of note, HHLA2 was almost universally present across the whole cohort, with positive rates above 90%. A pooled analysis of 90 PSC patients treated with single-agent PD-1/PD-L1 inhibitors revealed an overall response rate (partial or complete response) of 54.5% even though 74.2% of patients had PD-L1 TPS ≥ 50% [7]. Thus, despite high PD-L1 expression, increasing the response rates in this aggressive tumor type remains a challenge. According to our study findings, treating PSC patients with combination immune checkpoint inhibitors regimens tailored based on detailed histopathologic reporting of component-specific immune checkpoint expressions may overcome immunotherapy resistance and increase treatment efficacy.
In addition to delineating the heterogeneity of PSC, our study demonstrates that these subcomponents and immune checkpoint expression levels may affect clinical outcomes. A higher sarcomatoid percentage is associated with worse survival, independent of age, gender, stage, genetic mutation, and expression of immune checkpoints. In addition, the lack of B7-H3 in the sarcomatoid component was independently associated with better survival.
These findings argue for more detailed pathological reporting of PSC, including the percentages of subcomponents due to potential correlations with survival and varying immune checkpoint molecule expression that may guide therapeutic management. Furthermore, reporting PD-L1 TPS across the whole specimen could overestimate the potential benefit of PD-1/PD-L1 monotherapy since the sarcomatoid component may express a disproportionately low PD-L1 level. As prospective data regarding the biomarker value of B7x, B7-H3, and HHLA2 molecules and the efficacy of their inhibitors accumulate, pathology reports of PSC may need to include the immune checkpoint expression levels by histologic components.
Our PSC cohort was enriched in METex14 variants (19.5%), followed by KRAS mutations (14.6%). In contrast to other studies reporting ALK and EGFR alterations in PSC, we did not find any ALK/EGFR alterations, possibly due to the higher smoking rate and lesser frequency of Asian patients in our cohort [24,27]. In addition, patients with METex14 had a numerically higher sarcomatoid percentage, which could be secondary to the role of the HGF/METex14 pathway in epithelial–mesenchymal transition and evolution of sarcomatoid components in PSC (Figure 1H). Interestingly, the METex14 variant also showed higher PD-L1 expression, yet the underlying cellular pathways remain elusive. Using a pair of MET WT and METex14 lung cancer cell lines models, we demonstrated that the HGF/MET signaling pathway plays a critical role in enhancing PD-L1 expression with higher levels of PD-L1 in METex14 cell lines compared to MET WT. Of note, by using CRISPR technology to develop METex14 cell line models, our study enables investigation of the specific effect of METex14 alteration independent from that of MET amplification. Intriguingly, our results highlight the role of METex14 signaling in selectively increasing the expression of the immune checkpoint, i.e., PD-L1 expression, compared to the other immune checkpoints, B7x, B7-H3, and HHLA2. The low baseline expression of these alternative immune checkpoints in parental cells and the regulation of expression in an HGF/METex14-independent pathway may explain these findings.
In this study, HGF/METex14 signaling significantly increased the activation of downstream PI3K/Akt and MAPK pathways. Moreover, inhibition of PI3K and MAPK pathway markedly repressed HGF-dependent PD-L1 expression, suggesting that either PI3K/Akt or MAPK cascades are tightly involved in the upregulation of PD-L1 expression. These results may partially elucidate the mechanism underlying high PD-L1 expression in METex14 variants and, therefore, could offer a rationale for combination therapy strategies consisting of PD-L1 and MAPK or PI3K/Akt pathway inhibition as a treatment strategy in this subpopulation. Multiple clinical trials investigating the effect and safety of MAPK pathway inhibitors with PD-L1/PD-1 inhibitors in NSCLC are currently ongoing (NCT03581487, NCT03600701). Subgroup analyses for METex14 patients in these trials should be pursued as these patients may demonstrate better clinical outcomes with combination therapy.
MET-inhibiting TKIs, such as capmatinib and tepotinib, have shown promising results in clinical trials for NSCLC patients with METex14 or MET amplification, leading to accelerated FDA approvals [43,44]. In our study, both capmatinib and tepotinib rigorously repressed METex14-mediated PD-L1 expression. These results revealed that MET inhibitors at least partially inhibit PD-1/PD-L1 signaling. However, despite high PD-L1 expression in METex14 NSCLC, the efficacy of PD-1/PD-L1 inhibitors seems lower than expected, with reported overall response rates ranging between 16% and 43% [28,45,46]. On the basis of the demonstration of the dynamic nature of PD-L1 expression with MET inhibitors demonstrated in our study, the timing and sequence of targeted and immunotherapy warrant further investigation.
Our study is limited by the small sample size; however, this is expected given the low prevalence of PSC as a rare subtype of NSCLC. Yet another limitation is the retrospective nature of our study. Although we separately reported the IHC of two components, tumors were not microdissected for further genetic studies.
5. Conclusions
Heterogeneity of immune checkpoint expression exists between epithelial and sarcomatoid components in PSCs, with only one-third of PSCs expressing PD-L1 in both components. Alternate immune checkpoints are also widely expressed in PSCs, and further studies exploring the use of combinations of new immune checkpoints in this population are necessary. Furthermore, given the association of METex14 and higher sarcomatoid percentage, further studies investigating the underlying mechanism of sarcomatoid differentiation in this subpopulation are warranted. Our study also sheds light on the mechanisms underlying the upregulation of PD-L1 by HGF/METex14 signaling via MAPK and PI3K/Akt pathways. In addition, by demonstrating the dynamic nature of PD-L1 levels via MET inhibition in the METex14 variant, future investigators may be better poised to explore new targeted and immunotherapy combinations and optimize their timing.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15133372/s1: Table S1. Immune checkpoint expression by genetic alterations and histologic subcomponents; Table S2. Association of tumor-infiltrating lymphocytes with genetic alterations; Table S3. Full Cox regression model for survival analysis; File S1: Uncropped Western blot images.
Author Contributions
Conceptualization, H.C., A.B., B.H. and X.Z.; methodology, F.W., A.E.C.D., A.B., B.H. and H.C.; validation F.W., A.E.C.D., A.B., B.H. and H.C.; formal analysis, L.D., F.W., A.B., A.E.C.D., B.H. and H.C.; investigation, F.W., A.E.C.D., L.D., J.Y., J.S., C.S. and S.L.; resources, H.C., X.Z. and A.B.; writing—original draft preparation, F.W. and A.E.C.D.; writing—review and editing, F.W., A.E.C.D., L.D., J.Y., J.S., C.S., S.L., H.C., A.B., B.H. and X.Z.; visualization, F.W. and A.E.C.D.; supervision, H.C.; funding acquisition, H.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by American Lung Association, Lung Cancer Discovery Award, grant number LCD-687010.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of Montefiore Medical Center/Albert Einstein College of Medicine (IRB number: 2013-2570, the latest version approval date: February 2023).
Informed Consent Statement
Patient consent was waived due to the retrospective, observational nature of the study.
Data Availability Statement
The supporting data are not publicly available due to research participant privacy restrictions.
Conflicts of Interest
X. Zang is an inventor of two granted patents (HHLA2 as a novel inhibitor of human immune system and uses thereof; antibodies to human B7x for treatment of metastatic cancer) and of two pending patents (monoclonal antibodies against IgV domain of B7-H3 and uses thereof; chimeric antigen receptors targeting B7-H3 (CD276) and associated methods). H. Cheng receives grants from Astra Zeneca, Genentech, and honoraria from Janssen, G1 therapeutics. B. Halmos has grants or contracts from Boehringer Ingelheim, Astra Zeneca, Merck, BMS, Advaxis, Amgen, AbbVie, Daiichi, Pfizer, GSK, Beigene, Janssen, Numab, and Arrivent and is on the monitoring/advisory boards of Astra Zeneca, Boehringer Ingelheim, Apollomics, Janssen, Takeda, Merck, BMS, Genentech, Pfizer, Eli-Lilly, TPT, and Arcus.
References
- Rahouma, M.; Kamel, M.; Narula, N.; Nasar, A.; Harrison, S.; Lee, B.; Stiles, B.; Altorki, N.K.; Port, J.L. Pulmonary Sarcomatoid Carcinoma: An Analysis of a Rare Cancer from the Surveillance, Epidemiology, and End Results Database. Eur. J. Cardiothorac. Surg. 2018, 53, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Maneenil, K.; Xue, Z.; Liu, M.; Boland, J.; Wu, F.; Stoddard, S.M.; Molina, J.; Yang, P. Sarcomatoid Carcinoma of the Lung: The Mayo Clinic Experience in 127 Patients. Clin. Lung Cancer 2018, 19, e323–e333. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.M.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef]
- Vieira, T.; Girard, N.; Ung, M.; Monnet, I.; Cazes, A.; Bonnette, P.; Duruisseaux, M.; Mazieres, J.; Antoine, M.; Cadranel, J.; et al. Efficacy of First-Line Chemotherapy in Patients with Advanced Lung Sarcomatoid Carcinoma. J. Thorac. Oncol. 2013, 8, 1574–1577. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, D.; Liu, H.; Chen, J. Pulmonary Sarcomatoid Carcinoma: Progress, Treatment and Expectations. Ther. Adv. Med. Oncol. 2020, 12, 1758835920950207. [Google Scholar] [CrossRef] [PubMed]
- Shum, E.; Stuart, M.; Borczuk, A.; Wang, F.; Cheng, H.; Halmos, B. Recent Advances in the Management of Pulmonary Sarcomatoid Carcinoma. Expert Rev. Respir. Med. 2016, 10, 407–416. [Google Scholar] [CrossRef]
- Babacan, N.A.; Pina, I.B.; Signorelli, D.; Prelaj, A.; Garassino, M.C.; Tanvetyanon, T. Relationship Between Programmed Death Receptor-Ligand 1 Expression and Response to Checkpoint Inhibitor Immunotherapy in Pulmonary Sarcomatoid Carcinoma: A Pooled Analysis. Clin. Lung Cancer 2020, 21, e456–e463. [Google Scholar] [CrossRef]
- Domblides, C.; Leroy, K.; Monnet, I.; Mazières, J.; Barlesi, F.; Gounant, V.; Baldacci, S.; Mennecier, B.; Toffart, A.-C.; Audigier-Valette, C.; et al. Efficacy of Immune Checkpoint Inhibitors in Lung Sarcomatoid Carcinoma. J. Thorac. Oncol. 2020, 15, 860–866. [Google Scholar] [CrossRef]
- Sukrithan, V.; Sandler, J.; Gucalp, R.; Gralla, R.; Halmos, B. Immune Checkpoint Blockade Is Associated with Durable Responses in Pulmonary Sarcomatoid Carcinoma. Clin. Lung Cancer 2019, 20, e242–e246. [Google Scholar] [CrossRef]
- Janakiram, M.; Shah, U.A.; Liu, W.; Zhao, A.; Schoenberg, M.P.; Zang, X. The Third Group of the B7-CD28 Immune Checkpoint Family: HHLA2, TMIGD2, B7x, and B7-H3. Immunol. Rev. 2017, 276, 26–39. [Google Scholar] [CrossRef]
- Suh, W.-K.; Gajewska, B.U.; Okada, H.; Gronski, M.A.; Bertram, E.M.; Dawicki, W.; Duncan, G.S.; Bukczynski, J.; Plyte, S.; Elia, A.; et al. The B7 Family Member B7-H3 Preferentially down-Regulates T Helper Type 1–Mediated Immune Responses. Nat. Immunol. 2003, 4, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Picarda, E.; Galbo, P.M.; Zong, H.; Rajan, M.R.; Wallenius, V.; Zheng, D.; Börgeson, E.; Singh, R.; Pessin, J.; Zang, X. The Immune Checkpoint B7-H3 (CD276) Regulates Adipocyte Progenitor Metabolism and Obesity Development. Sci. Adv. 2022, 8, eabm7012. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Ren, X.; Galbo, P.M.; Moerdler, S.; Wang, H.; Sica, R.A.; Etemad-Gilbertson, B.; Shi, L.; Zhu, L.; Tang, X.; et al. KIR3DL3-HHLA2 Is a Human Immunosuppressive Pathway and a Therapeutic Target. Sci. Immunol. 2021, 6, eabf9792. [Google Scholar] [CrossRef]
- Cheng, H.; Borczuk, A.; Janakiram, M.; Ren, X.; Lin, J.; Assal, A.; Halmos, B.; Perez-Soler, R.; Zang, X. Wide Expression and Significance of Alternative Immune Checkpoint Molecules, B7x and HHLA2, in PD-L1 Negative Human Lung Cancers. Clin. Cancer Res. 2018, 24, 1954–1964. [Google Scholar] [CrossRef]
- Cheng, H.; Janakiram, M.; Borczuk, A.; Lin, J.; Qiu, W.; Liu, H.; Chinai, J.M.; Halmos, B.; Perez-Soler, R.; Zang, X. HHLA2, a New Immune Checkpoint Member of the B7 Family, Is Widely Expressed in Human Lung Cancer and Associated with EGFR Mutational Status. Clin. Cancer Res. 2017, 23, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Li, W.; Chen, K.; Xie, Y.; Liu, Q.; Yao, M.; Duan, W.; Zhou, X.; Liang, R.; Tao, M. B7-H1 and B7-H3 Are Independent Predictors of Poor Prognosis in Patients with Non-Small Cell Lung Cancer. Oncotarget 2014, 6, 3452–3461. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Y.; Zhao, J.; Gu, M.; Giscombe, R.; Lefvert, A.K.; Wang, X. B7-H3 and B7-H4 Expression in Non-Small-Cell Lung Cancer. Lung Cancer 2006, 53, 143–151. [Google Scholar] [CrossRef]
- John, P.; Wei, Y.; Liu, W.; Du, M.; Guan, F.; Zang, X. The B7x Immune Checkpoint Pathway: From Discovery to Clinical Trial. Trends Pharmacol. Sci. 2019, 40, 883–896. [Google Scholar] [CrossRef]
- Pang, A.; Carbini, M.; Moreira, A.L.; Maki, R.G. Carcinosarcomas and Related Cancers: Tumors Caught in the Act of Epithelial-Mesenchymal Transition. JCO 2018, 36, 210–216. [Google Scholar] [CrossRef]
- Nakagomi, T.; Goto, T.; Hirotsu, Y.; Shikata, D.; Yokoyama, Y.; Higuchi, R.; Amemiya, K.; Okimoto, K.; Oyama, T.; Mochizuki, H.; et al. New Therapeutic Targets for Pulmonary Sarcomatoid Carcinomas Based on Their Genomic and Phylogenetic Profiles. Oncotarget 2018, 9, 10635–10649. [Google Scholar] [CrossRef]
- Manzotti, G.; Torricelli, F.; Benedetta, D.; Lococo, F.; Sancisi, V.; Rossi, G.; Piana, S.; Ciarrocchi, A. An Epithelial-to-Mesenchymal Transcriptional Switch Triggers Evolution of Pulmonary Sarcomatoid Carcinoma (PSC) and Identifies Dasatinib as New Therapeutic Option. Clin. Cancer Res. 2019, 25, 2348–2360. [Google Scholar] [CrossRef] [PubMed]
- Stoker, M.; Gherardi, E.; Perryman, M.; Gray, J. Scatter Factor Is a Fibroblast-Derived Modulator of Epithelial Cell Mobility. Nature 1987, 327, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P. Epithelial–Mesenchymal Transitions in Tumour Progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef]
- Schrock, A.B.; Li, S.D.; Frampton, G.M.; Suh, J.; Braun, E.; Mehra, R.; Buck, S.C.; Bufill, J.A.; Peled, N.; Karim, N.A.; et al. Pulmonary Sarcomatoid Carcinomas Commonly Harbor Either Potentially Targetable Genomic Alterations or High Tumor Mutational Burden as Observed by Comprehensive Genomic Profiling. J. Thorac. Oncol. 2017, 12, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Frampton, G.M.; Ali, S.M.; Rosenzweig, M.; Chmielecki, J.; Lu, X.; Bauer, T.M.; Akimov, M.; Bufill, J.A.; Lee, C.; Jentz, D.; et al. Activation of MET via Diverse Exon 14 Splicing Alterations Occurs in Multiple Tumor Types and Confers Clinical Sensitivity to MET Inhibitors. Cancer Discov. 2015, 5, 850–859. [Google Scholar] [CrossRef]
- Awad, M.M.; Oxnard, G.R.; Jackman, D.M.; Savukoski, D.O.; Hall, D.; Shivdasani, P.; Heng, J.C.; Dahlberg, S.E.; Jänne, P.A.; Verma, S.; et al. MET Exon 14 Mutations in Non-Small-Cell Lung Cancer Are Associated with Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J. Clin. Oncol. 2016, 34, 721–730. [Google Scholar] [CrossRef]
- Liu, X.; Jia, Y.; Stoopler, M.B.; Shen, Y.; Cheng, H.; Chen, J.; Mansukhani, M.; Koul, S.; Halmos, B.; Borczuk, A.C. Next-Generation Sequencing of Pulmonary Sarcomatoid Carcinoma Reveals High Frequency of Actionable MET Gene Mutations. J. Clin. Oncol. 2016, 34, 794–802. [Google Scholar] [CrossRef]
- Sabari, J.K.; Leonardi, G.C.; Shu, C.A.; Umeton, R.; Montecalvo, J.; Ni, A.; Chen, R.; Dienstag, J.; Mrad, C.; Bergagnini, I.; et al. PD-L1 Expression, Tumor Mutational Burden, and Response to Immunotherapy in Patients with MET Exon 14 Altered Lung Cancers. Ann. Oncol. 2018, 29, 2085–2091. [Google Scholar] [CrossRef]
- Kim, S.Y.; Yin, J.; Bohlman, S.; Walker, P.; Dacic, S.; Kim, C.; Khan, H.; Liu, S.V.; Ma, P.C.; Nagasaka, M.; et al. Characterization of MET Exon 14 Skipping Alterations (in NSCLC) and Identification of Potential Therapeutic Targets Using Whole Transcriptome Sequencing. JTO Clin. Res. Rep. 2022, 3, 100381. [Google Scholar] [CrossRef]
- Zhang, Z.; Kobayashi, S.; Borczuk, A.C.; Leidner, R.S.; LaFramboise, T.; Levine, A.D.; Halmos, B. Dual Specificity Phosphatase 6 (DUSP6) Is an ETS-Regulated Negative Feedback Mediator of Oncogenic ERK Signaling in Lung Cancer Cells. Carcinogenesis 2010, 31, 577–586. [Google Scholar] [CrossRef]
- Jeon, H.; Vigdorovich, V.; Garrett-Thomson, S.C.; Janakiram, M.; Ramagopal, U.A.; Abadi, Y.M.; Lee, J.S.; Scandiuzzi, L.; Ohaegbulam, K.C.; Chinai, J.M.; et al. Structure and Cancer Immunotherapy of the B7 Family Member B7x. Cell Rep. 2014, 9, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Chinai, J.M.; Buhl, S.; Scandiuzzi, L.; Ray, A.; Jeon, H.; Ohaegbulam, K.C.; Ghosh, K.; Zhao, A.; Scharff, M.D.; et al. HHLA2 Is a Member of the B7 Family and Inhibits Human CD4 and CD8 T-Cell Function. Proc. Natl. Acad. Sci. USA 2013, 110, 9879–9884. [Google Scholar] [CrossRef] [PubMed]
- Zang, X.; Thompson, R.H.; Al-Ahmadie, H.A.; Serio, A.M.; Reuter, V.E.; Eastham, J.A.; Scardino, P.T.; Sharma, P.; Allison, J.P. B7-H3 and B7x Are Highly Expressed in Human Prostate Cancer and Associated with Disease Spread and Poor Outcome. Proc. Natl. Acad. Sci. USA 2007, 104, 19458–19463. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, Y.; Qiu, W.; Shum, E.; Feng, M.; Zhao, D.; Zheng, D.; Borczuk, A.; Cheng, H.; Halmos, B. Functional Analysis of MET Exon 14 Skipping Alteration in Cancer Invasion and Metastatic Dissemination. Cancer Res. 2022, 82, 1365–1379. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 8 December 2020).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.; Anders, S.; Huber, W. Differential Analysis of Count Data–the DESeq2 Package. Genome Biol. 2014, 15, 10–1186. [Google Scholar]
- Vieira, T.; Antoine, M.; Hamard, C.; Fallet, V.; Duruisseaux, M.; Rabbe, N.; Rodenas, A.; Cadranel, J.; Wislez, M. Sarcomatoid Lung Carcinomas Show High Levels of Programmed Death Ligand-1 (PD-L1) and Strong Immune-Cell Infiltration by TCD3 Cells and Macrophages. Lung Cancer 2016, 98, 51–58. [Google Scholar] [CrossRef]
- Wu, X.; Huang, Y.; Li, Y.; Wang, Q.; Wang, H.; Jiang, L. 18F-FDG PET/CT Imaging in Pulmonary Sarcomatoid Carcinoma and Correlation with Clinical and Genetic Findings. Ann. Nucl. Med. 2019, 33, 647–656. [Google Scholar] [CrossRef]
- Kim, S.; Kim, M.-Y.; Koh, J.; Go, H.; Lee, D.S.; Jeon, Y.K.; Chung, D.H. Programmed Death-1 Ligand 1 and 2 Are Highly Expressed in Pleomorphic Carcinomas of the Lung: Comparison of Sarcomatous and Carcinomatous Areas. Eur. J. Cancer 2015, 51, 2698–2707. [Google Scholar] [CrossRef]
- Lamb, M.; Wei, Y.; Ren, X.; O’Connor, R.; Dulak, A.; Rausch, M.; Strand, J.; Etemad-Gilbertson, B.; Gilligan, R.; Chappel, S.; et al. 489 NPX267, a First-in-Class Monoclonal Antibody Targeting KIR3DL3, Blocks HHLA2-Mediated Immunosuppression and Potentiates T and NK Cell-Mediated Antitumor Immunity. J. Immunother. Cancer 2022, 10. [Google Scholar] [CrossRef]
- Wolf, J.; Seto, T.; Han, J.-Y.; Reguart, N.; Garon, E.B.; Groen, H.J.M.; Tan, D.S.W.; Hida, T.; de Jonge, M.; Orlov, S.V.; et al. Capmatinib in MET Exon 14-Mutated or MET-Amplified Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 944–957. [Google Scholar] [CrossRef] [PubMed]
- Paik, P.K.; Felip, E.; Veillon, R.; Sakai, H.; Cortot, A.B.; Garassino, M.C.; Mazieres, J.; Viteri, S.; Senellart, H.; Van Meerbeeck, J.; et al. Tepotinib in Non-Small-Cell Lung Cancer with MET Exon 14 Skipping Mutations. N. Engl. J. Med. 2020, 383, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Mazieres, J.; Drilon, A.; Lusque, A.; Mhanna, L.; Cortot, A.B.; Mezquita, L.; Thai, A.A.; Mascaux, C.; Couraud, S.; Veillon, R.; et al. Immune Checkpoint Inhibitors for Patients with Advanced Lung Cancer and Oncogenic Driver Alterations: Results from the IMMUNOTARGET Registry. Ann. Oncol. 2019, 30, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Guisier, F.; Descourt, R.; Babey, H.; Huchot, E.; Falchero, L.; Veillon, R.; Cortot, A.B.; Tissot, C.; Chouaid, C.; Decroisette, C. Brief Report: First-Line Pembrolizumab in Metastatic Non-Small Cell Lung Cancer Habouring MET Exon 14 Skipping Mutation and PD-L1 ≥50% (GFPC 01-20 Study). Clin. Lung Cancer 2022, 23, e545–e549. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).