Multi-Omics Analyses Identify Signatures in Patients with Liver Cirrhosis and Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Fecal Bacterial 16S rRNA Gene Sequencing
2.3. Fecal DNA Virus Genome Sequencing
2.4. Plasma Endotoxemia Evaluation and Zonulin Measurement
2.5. Plasma Aqueous Phase Metabolite Measurement
2.6. Plasma Cytokine/Chemokine Profiles
2.7. Statistical Analysis
3. Results
3.1. Comparison of Study Cohort Characteristics
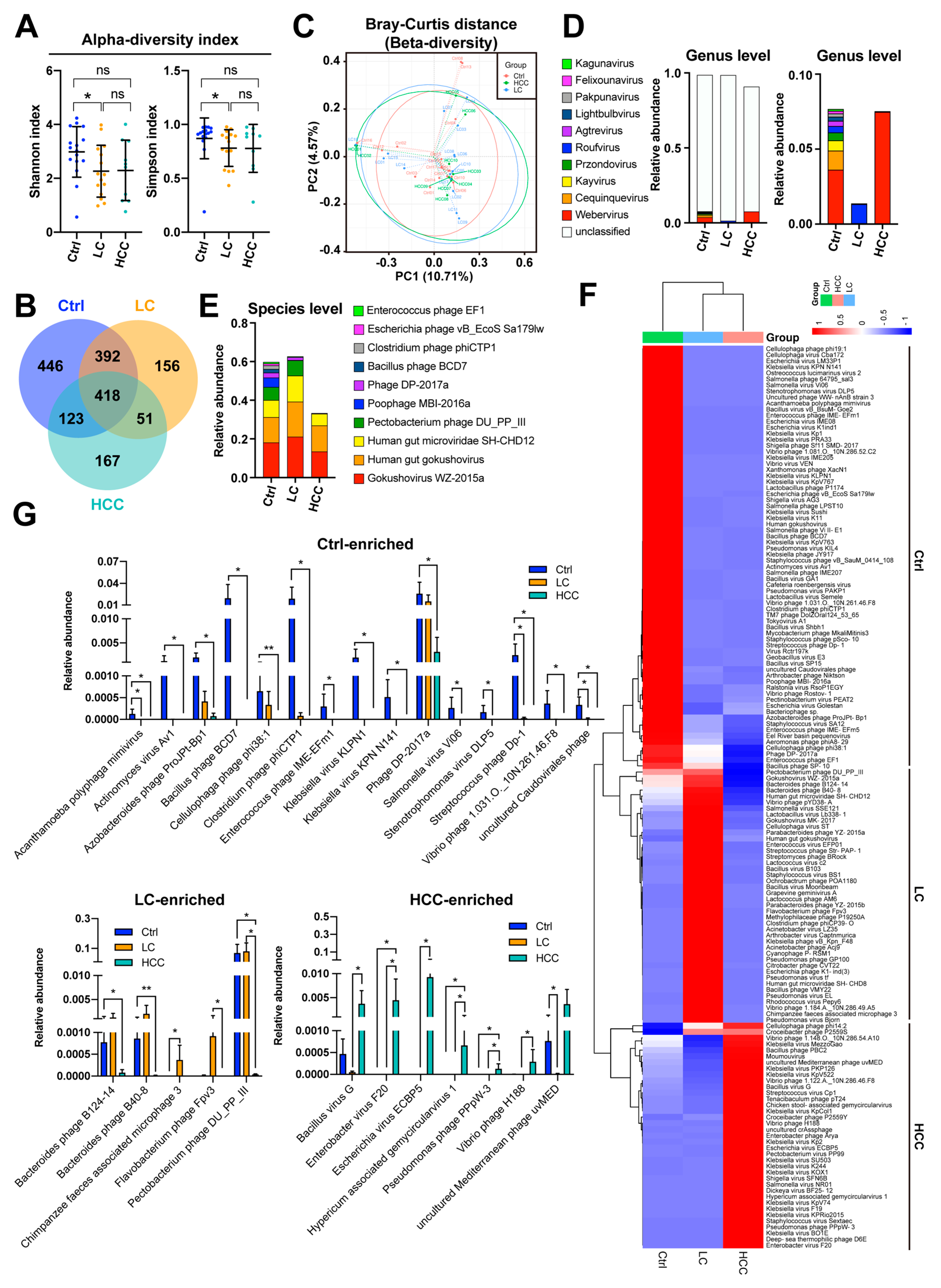
3.2. 16S rRNA Gene Composition of Gut Bacteria in Subjects
3.3. Comparison of Gut Viral Community in Subjects
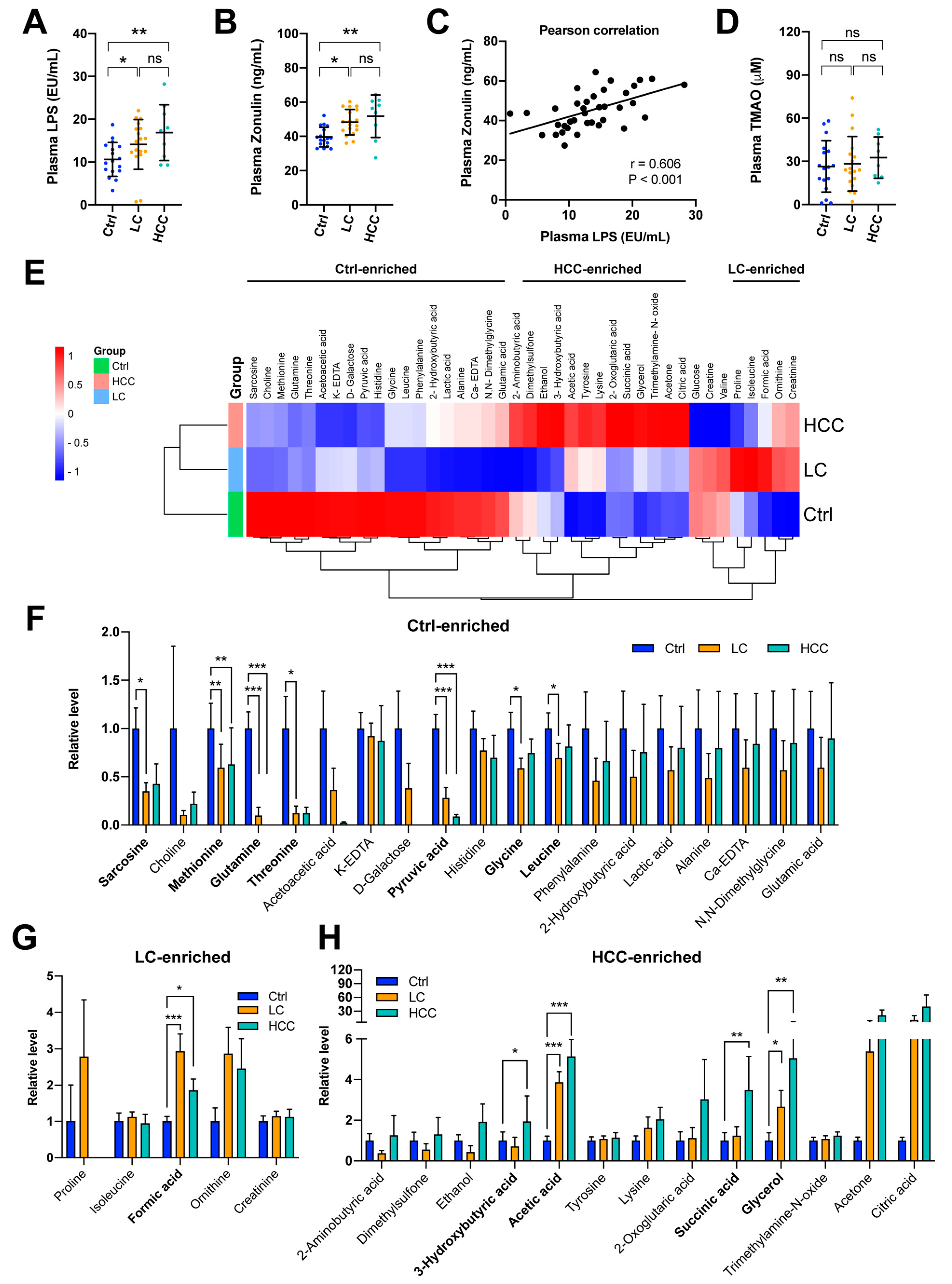
3.4. Increased Levels of LPS and Zonulin, but Not Trimethylamine N-oxide (TMAO) in the LC and HCC Subjects
3.5. Signature of Metabolic Changes in Subjects
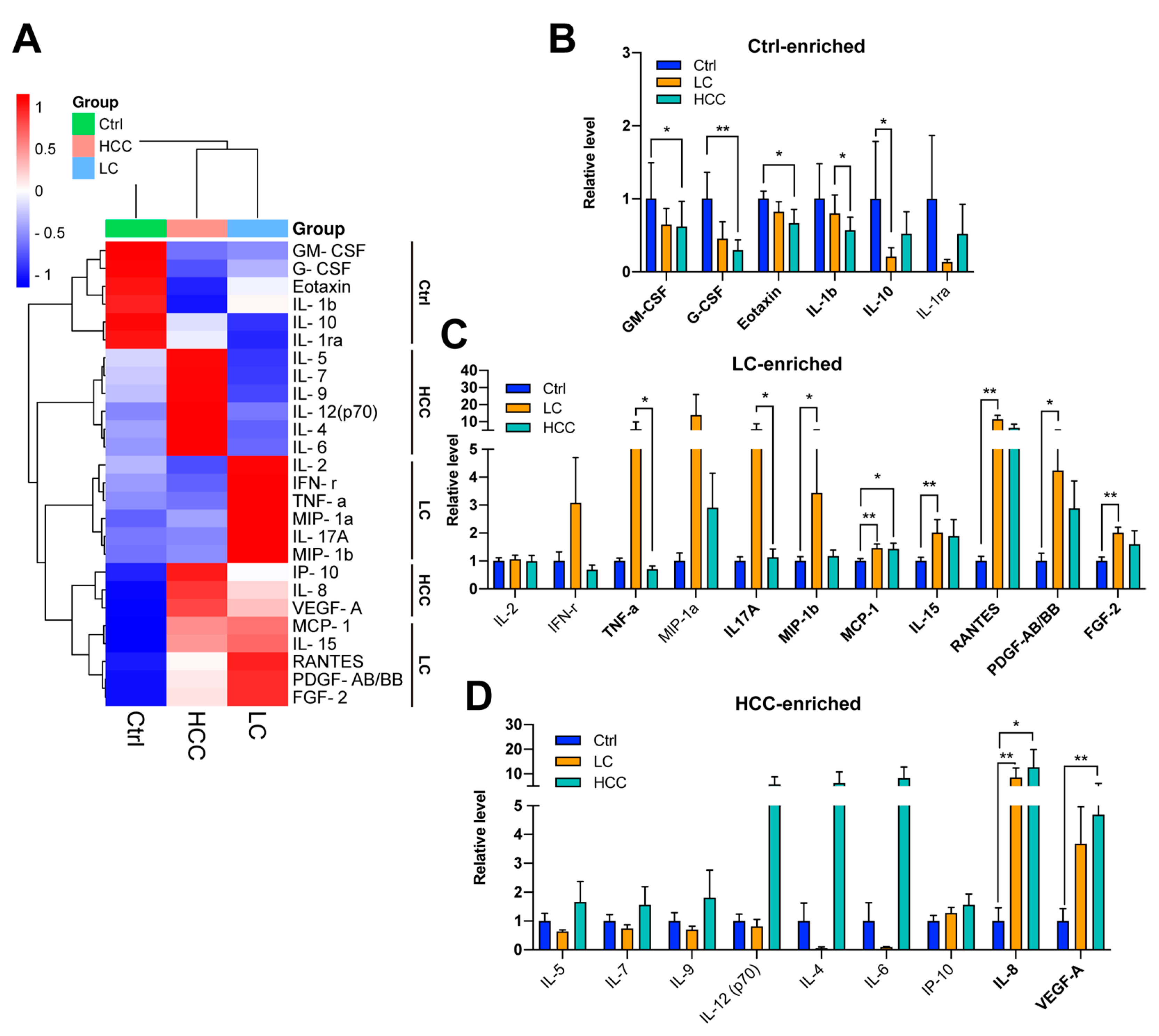
3.6. Profiling of a Panel of Cytokines/Chemokines in Subjects
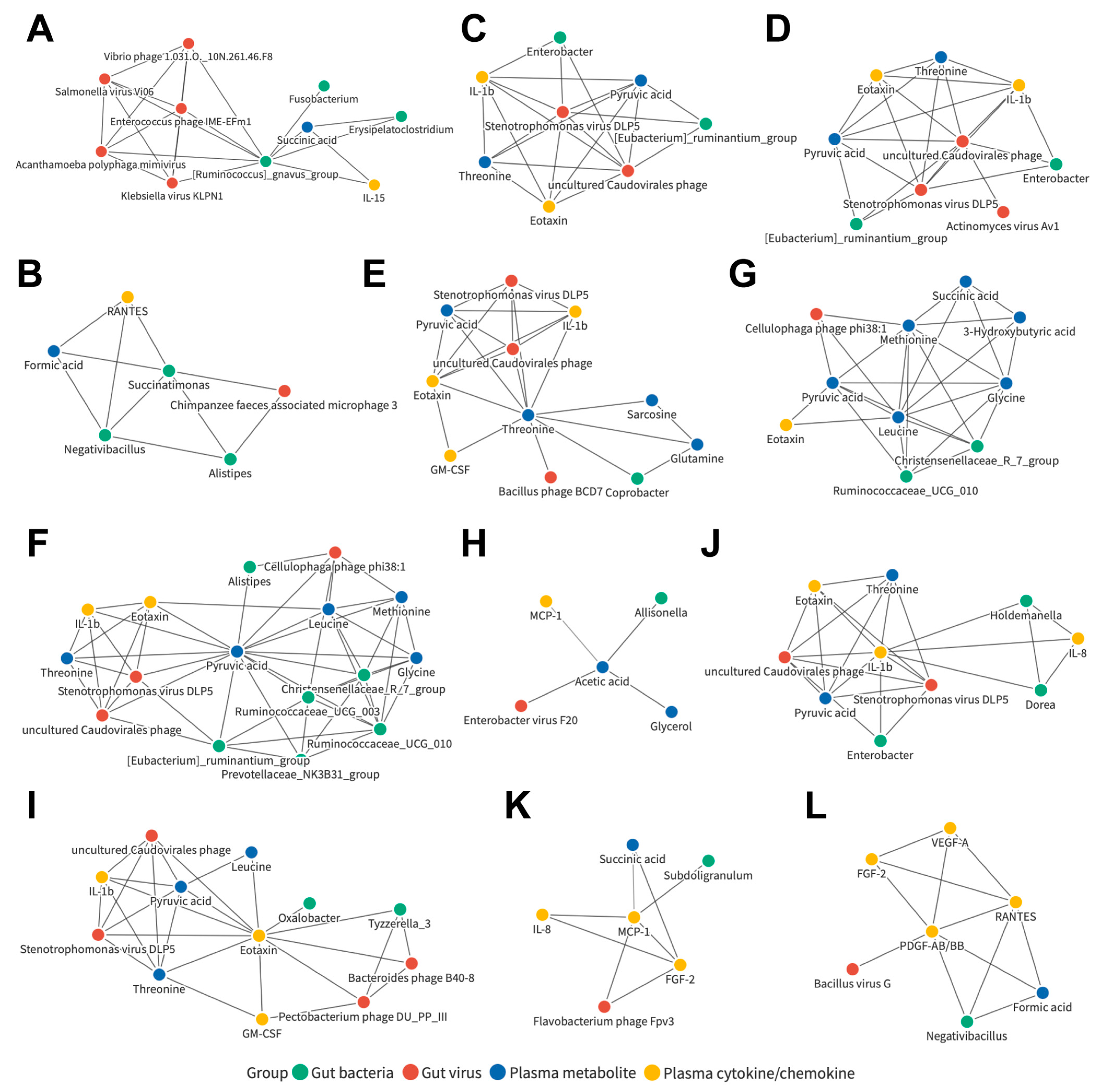
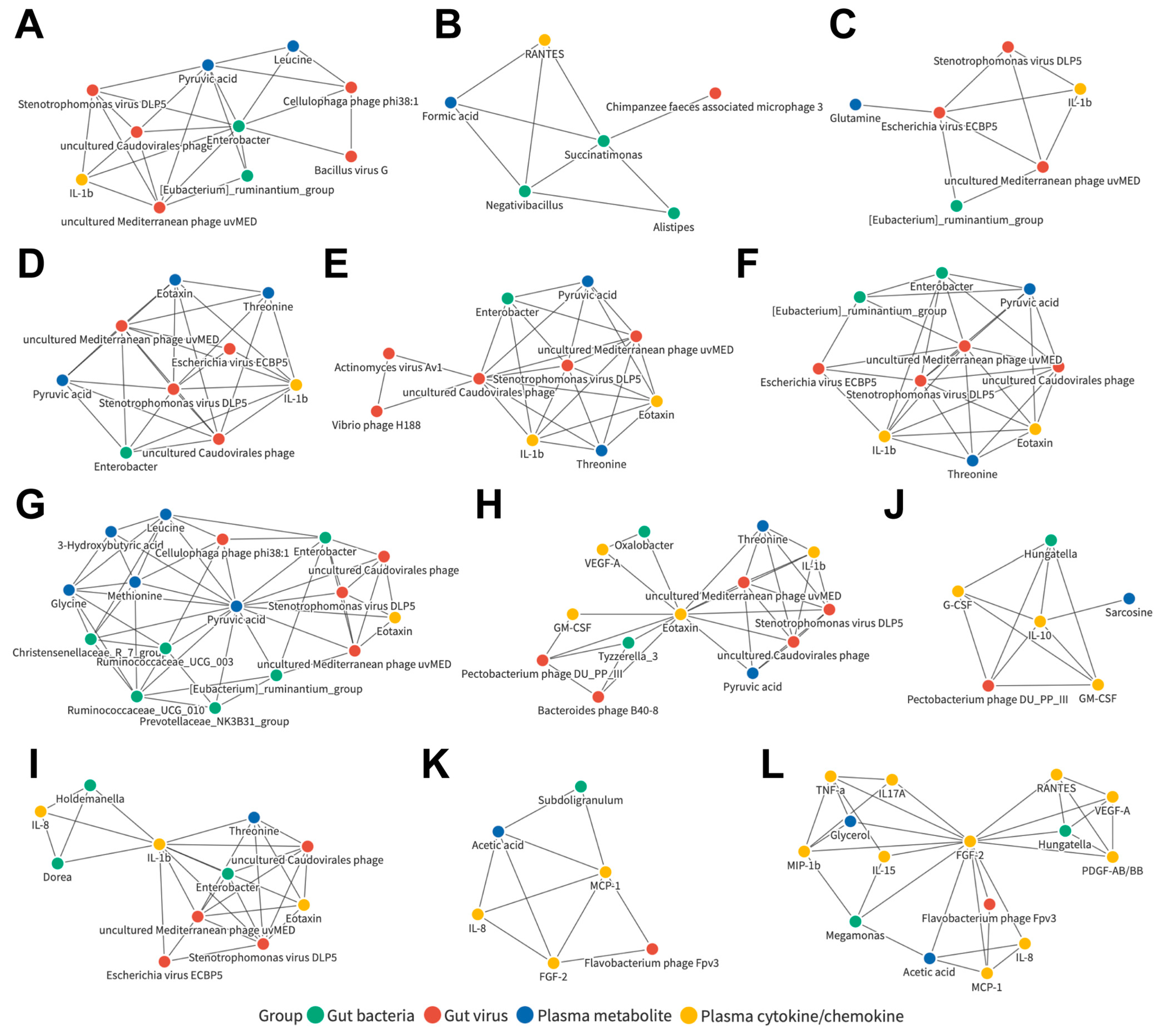
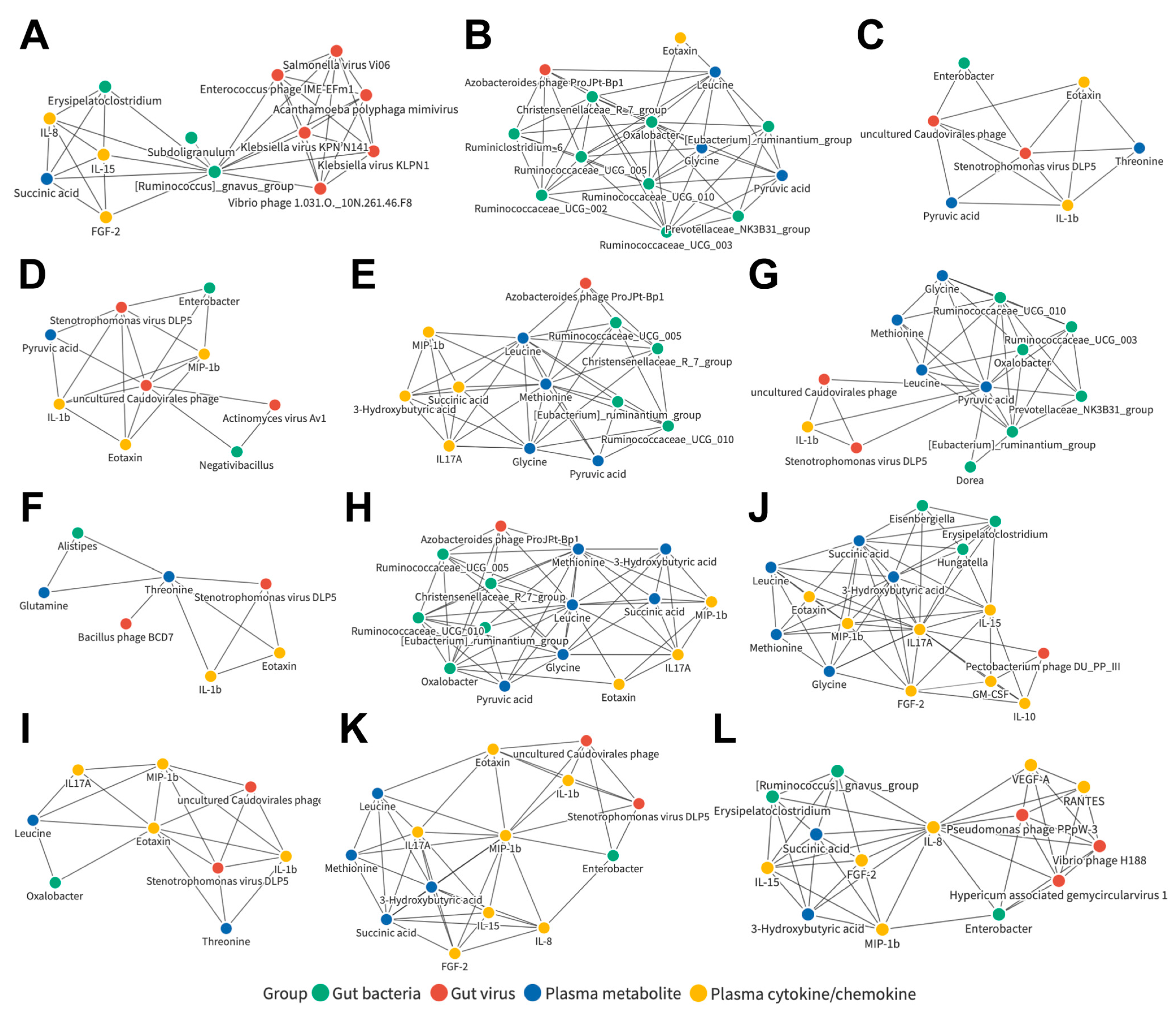
3.7. Joint Correlation Analysis of Gut Microbiota, Plasma Metabolite, and Plasma Cytokine/Chemokine Signatures in All Subjects
3.8. Joint Correlation Analysis of Gut Microbiota, Plasma Metabolite, and Plasma Cytokine/Chemokine Signatures in the Control and LC Cohorts
3.9. Joint Correlation Analysis of Gut Microbiota, Plasma Metabolite, and Plasma Cytokine/Chemokine Signatures in the Control and HCC Subjects
3.10. Joint Correlation Analysis of Gut Microbiota, Plasma Metabolite, and Plasma Cytokine/Chemokine Signatures in the LC and HCC Subjects
3.11. Identification of Liver Disease Severity-Associated, LC or HCC Exclusive, or Common Biomarker Networks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Ganne-Carrie, N.; Nahon, P. Hepatocellular carcinoma in the setting of alcohol-related liver disease. J. Hepatol. 2019, 70, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, N.; Fricker, Z.; Hubbard, R.A.; Ioannou, G.N.; Lewis, J.D.; Taddei, T.H.; Rothstein, K.D.; Serper, M.; Goldberg, D.S.; Kaplan, D.E. Risk Prediction Models for Post-Operative Mortality in Patients With Cirrhosis. Hepatology 2021, 73, 204–218. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Debelius, J.; Brenner, D.A.; Karin, M.; Loomba, R.; Schnabl, B.; Knight, R. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.X.; Yan, H.X.; Liu, Q.; Yang, W.; Wu, H.P.; Dong, W.; Tang, L.; Lin, Y.; He, Y.Q.; Zou, S.S.; et al. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology 2010, 52, 1322–1333. [Google Scholar] [CrossRef] [PubMed]
- Dapito, D.H.; Mencin, A.; Gwak, G.Y.; Pradere, J.P.; Jang, M.K.; Mederacke, I.; Caviglia, J.M.; Khiabanian, H.; Adeyemi, A.; Bataller, R.; et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 2012, 21, 504–516. [Google Scholar] [CrossRef]
- Caussy, C.; Tripathi, A.; Humphrey, G.; Bassirian, S.; Singh, S.; Faulkner, C.; Bettencourt, R.; Rizo, E.; Richards, L.; Xu, Z.Z.; et al. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat. Commun. 2019, 10, 1406. [Google Scholar] [CrossRef]
- Ponziani, F.R.; Bhoori, S.; Castelli, C.; Putignani, L.; Rivoltini, L.; Del Chierico, F.; Sanguinetti, M.; Morelli, D.; Paroni Sterbini, F.; Petito, V.; et al. Hepatocellular Carcinoma Is Associated With Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 2019, 69, 107–120. [Google Scholar] [CrossRef]
- Loomba, R.; Seguritan, V.; Li, W.; Long, T.; Klitgord, N.; Bhatt, A.; Dulai, P.S.; Caussy, C.; Bettencourt, R.; Highlander, S.K.; et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017, 25, 1054–1062.e5. [Google Scholar] [CrossRef]
- Liu, J.; Tang, W.; Budhu, A.; Forgues, M.; Hernandez, M.O.; Candia, J.; Kim, Y.; Bowman, E.D.; Ambs, S.; Zhao, Y.; et al. A Viral Exposure Signature Defines Early Onset of Hepatocellular Carcinoma. Cell 2020, 182, 317–328.e10. [Google Scholar] [CrossRef]
- Lang, S.; Demir, M.; Martin, A.; Jiang, L.; Zhang, X.; Duan, Y.; Gao, B.; Wisplinghoff, H.; Kasper, P.; Roderburg, C.; et al. Intestinal Virome Signature Associated With Severity of Nonalcoholic Fatty Liver Disease. Gastroenterology 2020, 159, 1839–1852. [Google Scholar] [CrossRef]
- Ma, C.; Han, M.; Heinrich, B.; Fu, Q.; Zhang, Q.; Sandhu, M.; Agdashian, D.; Terabe, M.; Berzofsky, J.A.; Fako, V.; et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018, 360, eaan5931. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, F.; Lu, H.; Wang, B.; Chen, Y.; Lei, D.; Wang, Y.; Zhu, B.; Li, L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 2011, 54, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Wu, Z.; Xu, W.; Yang, J.; Chen, Y.; Li, L. Intestinal Microbiota Was Assessed in Cirrhotic Patients with Hepatitis B Virus Infection. Microb. Ecol. 2011, 61, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Sikaroodi, M.; Shamsaddini, A.; Henseler, Z.; Santiago-Rodriguez, T.; Acharya, C.; Fagan, A.; Hylemon, P.B.; Fuchs, M.; Gavis, E.; et al. Interaction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy. Gut 2021, 70, 1162–1173. [Google Scholar] [CrossRef] [PubMed]
- Behary, J.; Amorim, N.; Jiang, X.T.; Raposo, A.; Gong, L.; McGovern, E.; Ibrahim, R.; Chu, F.; Stephens, C.; Jebeili, H.; et al. Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma. Nat. Commun. 2021, 12, 187. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Escapa, I.F.; Chen, T.; Huang, Y.; Gajare, P.; Dewhirst, F.E.; Lemon, K.P. New Insights into Human Nostril Microbiome from the Expanded Human Oral Microbiome Database (eHOMD): A Resource for the Microbiome of the Human Aerodigestive Tract. mSystems 2018, 3, e00187-18. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glockner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, D61–D65. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, S.; Thiemermann, C. Role of Metabolic Endotoxemia in Systemic Inflammation and Potential Interventions. Front. Immunol. 2020, 11, 594150. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A. All disease begins in the (leaky) gut: Role of zonulin-mediated gut permeability in the pathogenesis of some chronic inflammatory diseases. F1000Research 2020, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, N.; Kawada, N. Role of the Gut-Liver Axis in Liver Inflammation, Fibrosis, and Cancer: A Special Focus on the Gut Microbiota Relationship. Hepatol. Commun. 2019, 3, 456–470. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.H.; Chen, C.L.; Hsu, C.Y.; Chien, K.L.; Kao, J.H.; Chen, P.J.; Chen, T.H.; Chen, C.H. Long-term effectiveness of population-wide multifaceted interventions for hepatocellular carcinoma in Taiwan. J. Hepatol. 2021, 75, 132–141. [Google Scholar] [CrossRef] [PubMed]
- De la Cuesta-Zuluaga, J.; Kelley, S.T.; Chen, Y.; Escobar, J.S.; Mueller, N.T.; Ley, R.E.; McDonald, D.; Huang, S.; Swafford, A.D.; Knight, R.; et al. Age- and Sex-Dependent Patterns of Gut Microbial Diversity in Human Adults. mSystems 2019, 4, e00261-19. [Google Scholar] [CrossRef]
- Yen, Y.H.; Chang, K.C.; Tsai, M.C.; Tseng, P.L.; Lin, M.T.; Wu, C.K.; Lin, J.T.; Hu, T.H.; Wang, J.H.; Chen, C.H. Elevated body mass index is a risk factor associated with possible liver cirrhosis across different etiologies of chronic liver disease. J. Formos. Med. Assoc. 2018, 117, 268–275. [Google Scholar] [CrossRef]
- Komiyama, S.; Yamada, T.; Takemura, N.; Kokudo, N.; Hase, K.; Kawamura, Y.I. Profiling of tumour-associated microbiota in human hepatocellular carcinoma. Sci. Rep. 2021, 11, 10589. [Google Scholar] [CrossRef]
- Lapidot, Y.; Amir, A.; Nosenko, R.; Uzan-Yulzari, A.; Veitsman, E.; Cohen-Ezra, O.; Davidov, Y.; Weiss, P.; Bradichevski, T.; Segev, S.; et al. Alterations in the Gut Microbiome in the Progression of Cirrhosis to Hepatocellular Carcinoma. mSystems 2020, 5, e00153-20. [Google Scholar] [CrossRef]
- Sutton, T.D.S.; Hill, C. Gut Bacteriophage: Current Understanding and Challenges. Front. Endocrinol. 2019, 10, 784. [Google Scholar] [CrossRef]
- Liu, B.; Zhou, Z.; Jin, Y.; Lu, J.; Feng, D.; Peng, R.; Sun, H.; Mu, X.; Li, C.; Chen, Y. Hepatic stellate cell activation and senescence induced by intrahepatic microbiota disturbances drive progression of liver cirrhosis toward hepatocellular carcinoma. J. Immunother. Cancer 2022, 10, e003069. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Y.; Tan, X.Y.; Li, Q.J.; Liao, G.C.; Fang, A.P.; Zhang, D.M.; Chen, P.Y.; Wang, X.Y.; Luo, Y.; Long, J.A.; et al. Trimethylamine N-oxide, a gut microbiota-dependent metabolite of choline, is positively associated with the risk of primary liver cancer: A case-control study. Nutr. Metab. 2018, 15, 81. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hong, Z.; Tan, G.; Dong, X.; Yang, G.; Zhao, L.; Chen, X.; Zhu, Z.; Lou, Z.; Qian, B.; et al. NMR and LC/MS-based global metabolomics to identify serum biomarkers differentiating hepatocellular carcinoma from liver cirrhosis. Int. J. Cancer 2014, 135, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, A.; Higashi, T.; Sakata, T.; Nagashima, H. Serum amino acid levels in patients with hepatocellular carcinoma. Cancer 1984, 54, 1875–1882. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Lu, Q.; Liu, X.; Cong, H.; Zhao, L.; Wang, H.; Lin, D. Application of 1H NMR-based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci. 2009, 100, 782–785. [Google Scholar] [CrossRef]
- Al Bander, Z.; Nitert, M.D.; Mousa, A.; Naderpoor, N. The Gut Microbiota and Inflammation: An Overview. Int. J. Environ. Res. Public Health 2020, 17, 7618. [Google Scholar] [CrossRef]
- Korbecki, J.; Kojder, K.; Siminska, D.; Bohatyrewicz, R.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of the Ligands of Receptors CCR1, CCR2, CCR3, and CCR4. Int. J. Mol. Sci. 2020, 21, 8412. [Google Scholar] [CrossRef]
- Zajkowska, M.; Mroczko, B. Eotaxins and Their Receptor in Colorectal Cancer-A Literature Review. Cancers 2020, 12, 1383. [Google Scholar] [CrossRef]
- Kurys-Denis, E.; Prystupa, A.; Luchowska-Kocot, D.; Krupski, W.; Bis-Wencel, H.; Panasiuk, L. PDGF-BB homodimer serum level—A good indicator of the severity of alcoholic liver cirrhosis. Ann. Agric. Environ. Med. 2020, 27, 80–85. [Google Scholar] [CrossRef]
- Queck, A.; Bode, H.; Uschner, F.E.; Brol, M.J.; Graf, C.; Schulz, M.; Jansen, C.; Praktiknjo, M.; Schierwagen, R.; Klein, S.; et al. Systemic MCP-1 Levels Derive Mainly From Injured Liver and Are Associated With Complications in Cirrhosis. Front. Immunol. 2020, 11, 354. [Google Scholar] [CrossRef]
- Seitz, T.; Hellerbrand, C. Role of fibroblast growth factor signalling in hepatic fibrosis. Liver Int. 2021, 41, 1201–1215. [Google Scholar] [CrossRef] [PubMed]
- Dagouassat, M.; Suffee, N.; Hlawaty, H.; Haddad, O.; Charni, F.; Laguillier, C.; Vassy, R.; Martin, L.; Schischmanoff, P.O.; Gattegno, L.; et al. Monocyte chemoattractant protein-1 (MCP-1)/CCL2 secreted by hepatic myofibroblasts promotes migration and invasion of human hepatoma cells. Int. J. Cancer 2010, 126, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Gao, B. Hepatoprotective and anti-inflammatory cytokines in alcoholic liver disease. J. Gastroenterol. Hepatol. 2012, 27 (Suppl. 2), 89–93. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Qian, X.; Jiang, R.; Liu, Q.; Wang, Y.; Chen, C.; Wang, X.; Ryffel, B.; Sun, B. IL-17A plays a critical role in the pathogenesis of liver fibrosis through hepatic stellate cell activation. J. Immunol. 2013, 191, 1835–1844. [Google Scholar] [CrossRef]
- Zimmermann, H.W.; Seidler, S.; Gassler, N.; Nattermann, J.; Luedde, T.; Trautwein, C.; Tacke, F. Interleukin-8 is activated in patients with chronic liver diseases and associated with hepatic macrophage accumulation in human liver fibrosis. PLoS ONE 2011, 6, e21381. [Google Scholar] [CrossRef]
- Morris, R.M.; Mortimer, T.O.; O’Neill, K.L. Cytokines: Can Cancer Get the Message? Cancers 2022, 14, 2178. [Google Scholar] [CrossRef]


| Parameter | Ctrl (n = 17) † | LC (n = 18) † | HCC (n = 10) † | PCtrl-LC ‡ | PCtrl-HCC ‡ | PLC-HCC ‡ |
|---|---|---|---|---|---|---|
| Age (Year) | 56.7 ± 9.8 | 65.2 ± 11.4 | 70.2 ± 5.0 | 0.024 | <0.001 | 0.202 |
| Sex (Male:Female) | 7:10 | 12:6 | 6:4 | 0.241 | 0.585 | 0.724 |
| BMI | 23.1 ± 1.51 | 26.2 ± 3.6 | 25.0 ± 4.1 | 0.002 | 0.094 | 0.480 |
| Etiology(B:C:other) | - | 11:4:3 | 6:4:0 | - | - | 0.304 |
| AST (U/L) | 30.9 ± 3.5 | 35.6 ± 10.8 | 43.3 ± 17.1 | 0.097 | 0.007 | 0.155 |
| ALT (U/L) | 28.5 ± 4.7 | 30.0 ± 17.9 | 48.1 ± 30.0 | 0.740 | 0.013 | 0.055 |
| Bilirubin (mg/dL) | - | 1.1 ± 0.5 | 1.2 ± 0.7 | - | - | 0.664 |
| Albumin (g/dL) | - | 4.4 ± 0.4 | 3.6 ± 0.5 | - | - | <0.001 |
| T-Cholesterol (mg/dL) | - | 166.6 ± 26.1 | 150.0 ± 40.3 | - | - | 0.196 |
| Triglyceride (mg/dL) | - | 108.6 ± 54.8 | 99.4 ± 59.1 | - | - | 0.682 |
| HbA1c (%) | - | 6.1 ± 1.2 | 6.1 ± 0.9 | - | - | 0.999 |
| AFP (ng/mL) | - | 10.9 (1.0–25.1) | 101.8 (1.0–250.5) | - | - | 0.075 |
| Platelet (1000/μL) | - | 120.2 ± 45.8 | 100.7 ± 43.6 | - | - | 0.283 |
| NAs treatment (Yes:No) | 10:8 | 6:4 | 0.820 | |||
| DAA treatment (Yes:No) | 4:14 | 4:6 | 0.575 | |||
| Child-Pugh score | 5.1 ± 0.2 | 6.0 ± 1.6 | 0.158 |
| Metabolite | Ctrl (n = 17) † | LC (n = 18) † | HCC (n = 10) † | PCtrl-LC ‡ | PCtrl-HCC ‡ | PLC-HCC ‡ |
|---|---|---|---|---|---|---|
| Ethanol, μM | 82.4 ± 95.4 | 35.1 ± 112.8 | 158.0 ± 205.1 | 0.168 | 0.830 | 0.189 |
| Trimethylamine-N-oxide, μM | 26.5 ± 17.9 | 28.3 ± 19.0 | 32.6 ± 14.3 | 0.975 | 0.393 | 0.402 |
| 2-Aminobutyric acid, μM | 46.9 ± 64.7 | 17.7 ± 27.6 | 58.8 ± 130.3 | 0.179 | 0.401 | 0.824 |
| Alanine, mM | 19.7 ± 32.5 | 9.6 ± 21.4 | 15.7 ± 32.9 | 0.587 | 0.574 | 0.892 |
| Creatine, μM | 43.5 ± 26.5 | 45.4 ± 106.8 | 21.5 ± 21.6 | 0.072 | 0.086 | 0.898 |
| Creatinine, μM | 58.0 ± 35.2 | 66.0 ± 36.7 | 64.9 ± 36.2 | 0.560 | 0.582 | 0.927 |
| Glutamic acid, mM | 26.0 ± 41.3 | 15.5 ± 34.7 | 23.4 ± 42.4 | 0.103 | 0.027 | 0.348 |
| Glutamine, μM | 336.4 ± 238.9 | 32.9 ± 125.6 | 0.0 ± 0.0 | <0.001 | <0.001 | 0.617 |
| Glycine, μM | 372.6 ± 259.2 | 219.2 ± 164.9 | 278.1 ± 154.3 | 0.022 | 0.594 | 0.197 |
| Histidine, μM | 63.9 ± 47.8 | 49.4 ± 33.3 | 44.5 ± 42.1 | 0.354 | 0.329 | 0.804 |
| Isoleucine, μM | 40.3 ± 38.6 | 876.1 ± 3525.0 | 38.0 ± 29.1 | 0.171 | 0.755 | 0.438 |
| Leucine, μM | 203.5 ± 135.8 | 141.2 ± 131.4 | 165.3 ± 129.9 | 0.027 | 0.346 | 0.416 |
| Lysine, μM | 98.3 ± 93.2 | 160.0 ± 222.1 | 200.4 ± 163.8 | 0.722 | 0.129 | 0.212 |
| Methionine, μM | 150.4 ± 162.3 | 89.3 ± 155.0 | 94.5 ± 161.4 | 0.008 | 0.003 | 0.397 |
| N,N-Dimethylglycine, mM | 29.4 ± 47.0 | 16.7 ± 38.4 | 25.0 ± 46.3 | 0.200 | 0.136 | 0.627 |
| Ornithine, μM | 28.1 ± 42.5 | 80.3 ± 86.8 | 68.9 ± 65.4 | 0.092 | 0.180 | 0.991 |
| Phenylalanine, mM | 1.5 ± 2.3 | 0.7 ± 1.4 | 1.0 ± 1.7 | 0.683 | 0.584 | 0.380 |
| Proline, μM | 13.0 ± 53.6 | 36.2 ± 86.0 | 0.0 ± 0.0 | 0.291 | 0.633 | 0.186 |
| Sarcosine, μM | 3.8 ± 3.4 | 1.3 ± 1.5 | 1.6 ± 2.3 | 0.032 | 0.101 | 0.957 |
| Threonine, μM | 40.0 ± 54.8 | 4.9 ± 12.7 | 4.9 ± 7.2 | 0.038 | 0.261 | 0.608 |
| Tyrosine, μM | 37.7 ± 28.2 | 40.7 ± 23.8 | 43.1 ± 26.4 | 0.967 | 0.691 | 0.664 |
| Valine, μM | 181.6 ± 121.4 | 190.7 ± 86.7 | 146.8 ± 96.6 | 0.804 | 0.318 | 0.417 |
| 2-Hydroxybutyric acid, mM | 29.2 ± 46.6 | 14.6 ± 34.1 | 22.0 ± 41.0 | 0.258 | 0.946 | 0.406 |
| Acetic acid, μM | 39.1 ± 34.9 | 150.8 ± 87.2 | 200.5 ± 94.2 | <0.001 | <0.001 | 0.343 |
| Citric acid, mM | 0.1 ± 0.1 | 1.3 ± 4.8 | 4.7 ± 8.7 | 0.891 | 0.785 | 0.701 |
| Formic acid, μM | 50.7 ± 28.2 | 148.6 ± 104.6 | 94.0 ± 45.1 | <0.001 | 0.035 | 0.505 |
| Lactic acid, mM | 32.0 ± 44.8 | 18.2 ± 32.6 | 25.6 ± 38.9 | 0.449 | 0.948 | 0.591 |
| Succinic acid, μM | 45.5 ± 73.7 | 56.2 ± 86.2 | 158.1 ± 214.1 | 0.059 | 0.008 | 0.248 |
| Choline, μM | 168.9 ± 594.2 | 17.6 ± 33.8 | 37.1 ± 58.3 | 0.333 | 0.766 | 0.284 |
| 2-Oxoglutaric acid, μM | 41.6 ± 74.5 | 46.6 ± 92.3 | 125.9 ± 231.3 | 0.991 | 0.791 | 0.796 |
| 3-Hydroxybutyric acid, mM | 12.8 ± 22.6 | 9.1 ± 24.7 | 24.9 ± 45.8 | 0.143 | 0.023 | 0.257 |
| Acetoacetic acid, μM | 169.8 ± 271.3 | 61.4 ± 165.5 | 4.5 ± 3.6 | 0.714 | 0.478 | 0.672 |
| Acetone, μM | 26.8 ± 19.2 | 144.1 ± 451.8 | 544.3 ± 935.1 | 0.148 | 0.130 | 0.707 |
| Pyruvic acid, μM | 339.6 ± 206.6 | 95.5 ± 156.2 | 29.5 ± 21.0 | <0.001 | <0.001 | 0.333 |
| D-Galactose, mM | 28.9 ± 46.0 | 11.0 ± 31.9 | 0.0 ± 0.1 | 0.180 | 0.164 | 0.736 |
| Glucose, mM | 3.3 ± 2.8 | 3.3 ± 2.8 | 2.4 ± 2.2 | 0.962 | 0.555 | 0.578 |
| Glycerol, μM | 15.3 ± 24.1 | 40.6 ± 52.5 | 77.3 ± 97.4 | 0.027 | 0.005 | 0.272 |
| Dimethylsulfone, μM | 58.7 ± 98.0 | 32.4 ± 72.9 | 76.1 ± 138.9 | 0.948 | 0.829 | 0.867 |
| Ca-EDTA, mM | 30.6 ± 45.4 | 18.2 ± 37.7 | 25.7 ± 45.3 | 0.012 | 0.013 | 0.611 |
| K-EDTA, mM | 3.8 ± 2.6 | 3.5 ± 2.2 | 3.4 ± 4.0 | 0.237 | 0.222 | 0.771 |
| Cytokine/Chemokine | Ctrl (n = 17) † | LC (n = 18) † | HCC (n = 10) † | PCtrl-LC ‡ | PCtrl-HCC ‡ | PLC-HCC ‡ |
|---|---|---|---|---|---|---|
| IL-1b (pg/mL) | 2.4 ± 4.8 | 1.9 ± 2.6 | 1.4 ± 1.4 | 0.466 | 0.196 | 0.048 |
| IL-1ra (ng/mL) | 128.4 ± 458.2 | 17.1 ± 21.0 | 66.6 ± 165.5 | 0.857 | 0.348 | 0.426 |
| IL-2 (pg/mL) | 1.4 ± 0.7 | 1.4 ± 0.9 | 1.3 ± 1.0 | 0.587 | 0.121 | 0.270 |
| IL-4 (pg/mL) | 9.4 ± 24.1 | 0.5 ± 1.9 | 57.6 ± 135.5 | 0.971 | 0.249 | 0.257 |
| IL-5 (pg/mL) | 1.7 ± 1.9 | 1.1 ± 0.4 | 2.8 ± 3.9 | 0.306 | 0.700 | 0.625 |
| IL-6 (pg/mL) | 2.2 ± 5.9 | 0.2 ± 0.3 | 18.3 ± 32.3 | 0.474 | 0.592 | 0.248 |
| IL-7 (pg/mL) | 3.9 ± 3.7 | 2.9 ± 2.1 | 6.1 ± 7.9 | 0.494 | 0.436 | 0.842 |
| IL-8 (pg/mL) | 3.6 ± 6.9 | 30.6 ± 60.0 | 45.7 ± 82.8 | <0.001 | 0.018 | 0.598 |
| IL-9 (pg/mL) | 0.5 ± 0.6 | 0.3 ± 0.3 | 0.9 ± 1.5 | 0.193 | 0.160 | 0.762 |
| IL-10 (pg/mL) | 3.2 ± 10.5 | 0.7 ± 1.7 | 1.7 ± 3.1 | 0.016 | 0.080 | 0.766 |
| IL-12 (p70) (pg/mL) | 2.0 ± 1.9 | 1.6 ± 2.1 | 11.0 ± 19.8 | 0.059 | 0.835 | 0.159 |
| IL-15 (pg/mL) | 1.5 ± 0.8 | 3.0 ± 2.9 | 2.8 ± 2.8 | 0.006 | 0.154 | 0.368 |
| IL-17a (pg/mL) | 1.6 ± 1.0 | 9.2 ± 22.5 | 1.9 ± 1.6 | 0.283 | 0.364 | 0.048 |
| FGF-2 (pg/mL) | 32.4 ± 19.2 | 65.0 ± 28.3 | 51.6 ± 50.7 | 0.002 | 0.4011 | 0.073 |
| G-CSF (pg/mL) | 4.7 ± 7.0 | 2.1 ± 4.6 | 1.4 ± 2.1 | 0.109 | 0.006 | 0.156 |
| GM-CSF (pg/mL) | 6.6 ± 13.4 | 4.3 ± 6.2 | 4.1 ± 7.2 | 0.277 | 0.039 | 0.250 |
| IFN-γ(pg/mL) | 3.7 ± 5.0 | 11.5 ± 25.7 | 2.6 ± 2.0 | 0.731 | 0.339 | 0.207 |
| MCP-1 (pg/mL) | 236.2 ± 85.3 | 343.7 ± 152.8 | 337.3 ± 160.0 | 0.008 | 0.047 | 0.690 |
| MIP-1a (pg/mL) | 2.7 ± 3.2 | 37.9 ± 140.1 | 7.9 ± 10.6 | 0.662 | 0.159 | 0.295 |
| MIP-1b (pg/mL) | 40.4 ± 25.3 | 138.8 ± 320.5 | 47.3 ± 28.4 | 0.046 | 0.669 | 0.258 |
| Eotaxin (pg/mL) | 76.7 ± 33.6 | 63.0 ± 45.3 | 51.1 ± 45.6 | 0.116 | 0.046 | 0.500 |
| IP-10 (ng/mL) | 0.6 ± 0.4 | 0.7 ± 0.5 | 0.9 ± 0.7 | 0.219 | 0.316 | 0.968 |
| TNF-α (pg/mL) | 9.6 ± 4.0 | 53.4 ± 174.2 | 6.8 ± 3.6 | 0.565 | 0.190 | 0.049 |
| VEGF-a (pg/mL) | 22.9 ± 40.5 | 84.1 ± 125.3 | 107.2 ± 105.8 | 0.064 | 0.007 | 0.626 |
| PDGF-AB/BB (ng/mL) | 17.9 ± 20.7 | 75.9 ± 73.1 | 51.6 ± 56.2 | 0.015 | 0.101 | 0.666 |
| RANTES (ng/mL) | 52.5 ± 35.9 | 595.2 ± 542.2 | 333.5 ± 364.8 | 0.007 | 0.435 | 0.123 |
| Network Centered on | All Subjects | Ctrl-LC | Ctrl-HCC | Exclusive or Common Signature | Liver Disease Severity-Associated (LC-HCC) |
|---|---|---|---|---|---|
| Ruminococcus gnavus group | Yes | - | Yes | HCC specific | - |
| Succinatimonas | Yes | Yes | - | LC specific | - |
| Enterobacter | - | Yes | - | LC specific | - |
| Oxalobacter | - | - | Yes | HCC specific | - |
| Ruminococcaceae UCG 002 | - | - | - | - | Yes |
| Ruminococcaceae UCG 005 | - | - | - | - | Yes |
| Negativibacillus | - | - | - | - | Yes |
| Tyzzerella 3 | - | - | - | - | Yes |
| Stenotrophomonas virus DLP5 | Yes | Yes | Yes | Common | - |
| Uncultured Caudovirales phage | Yes | Yes | Yes | Common | Yes |
| Escherichia virus ECBP5 | - | Yes | - | LC specific | - |
| Uncultured Mediterranean phage uvMED | - | Yes | - | LC specific | - |
| Actinomyces virus Av1 | - | - | - | - | Yes |
| Azobacteroides phage ProJPt-Bp1 | - | - | - | - | Yes |
| Bacteroides phage B124-14 | - | - | - | - | Yes |
| Bacteroides phage B40-8 | - | - | - | - | Yes |
| Clostridium phage phiCTP1 | - | - | - | - | Yes |
| Flavobacterium phage Fpv3 | - | - | - | - | Yes |
| Pectobacterium phage DU_PP_III | - | - | - | - | Yes |
| Streptococcus phage Dp-1 | - | - | - | - | Yes |
| Threonine | Yes | - | Yes | HCC specific | Yes |
| pyruvic acid | Yes | Yes | Yes | Common | - |
| Leucine | Yes | - | Yes | HCC specific | - |
| Acetic acid | Yes | - | - | Common * | - |
| Methionine | - | - | Yes | HCC specific | - |
| Formic acid | - | - | - | - | Yes |
| 3-hydroxybutyric acid | - | - | - | - | Yes |
| Succinic acid | - | - | - | - | Yes |
| Eotaxin | Yes | Yes | Yes | Common | - |
| IL-1b | Yes | Yes | - | LC specific | - |
| MCP-1 | Yes | Yes | - | LC specific | Yes |
| PDGF-AB/BB | Yes | - | - | Common * | - |
| IL-10 | - | Yes | - | LC specific | Yes |
| FGF-2 | - | Yes | - | LC specific | - |
| IL-17A | - | - | Yes | HCC specific | - |
| MIP-1b | - | - | Yes | HCC specific | - |
| IL-8 | - | - | Yes | HCC specific | - |
| GM-CSF | - | - | - | - | Yes |
| RANTES | - | - | - | - | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, M.-W.; Chu, Y.-D.; Hsu, C.-W.; Chen, Y.-C.; Liang, K.-H.; Yeh, C.-T. Multi-Omics Analyses Identify Signatures in Patients with Liver Cirrhosis and Hepatocellular Carcinoma. Cancers 2023, 15, 210. https://doi.org/10.3390/cancers15010210
Lai M-W, Chu Y-D, Hsu C-W, Chen Y-C, Liang K-H, Yeh C-T. Multi-Omics Analyses Identify Signatures in Patients with Liver Cirrhosis and Hepatocellular Carcinoma. Cancers. 2023; 15(1):210. https://doi.org/10.3390/cancers15010210
Chicago/Turabian StyleLai, Ming-Wei, Yu-De Chu, Chao-Wei Hsu, Yi-Cheng Chen, Kung-Hao Liang, and Chau-Ting Yeh. 2023. "Multi-Omics Analyses Identify Signatures in Patients with Liver Cirrhosis and Hepatocellular Carcinoma" Cancers 15, no. 1: 210. https://doi.org/10.3390/cancers15010210
APA StyleLai, M.-W., Chu, Y.-D., Hsu, C.-W., Chen, Y.-C., Liang, K.-H., & Yeh, C.-T. (2023). Multi-Omics Analyses Identify Signatures in Patients with Liver Cirrhosis and Hepatocellular Carcinoma. Cancers, 15(1), 210. https://doi.org/10.3390/cancers15010210






