Cancer Cells Upregulate Tau to Gain Resistance to DNA Damaging Agents
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Plasmids
2.2. Cell Culture and Transfection
2.3. Cellular Extracts and Western Blotting
2.4. Cell Fractionation into Cytosolic and Microtubule Fractions
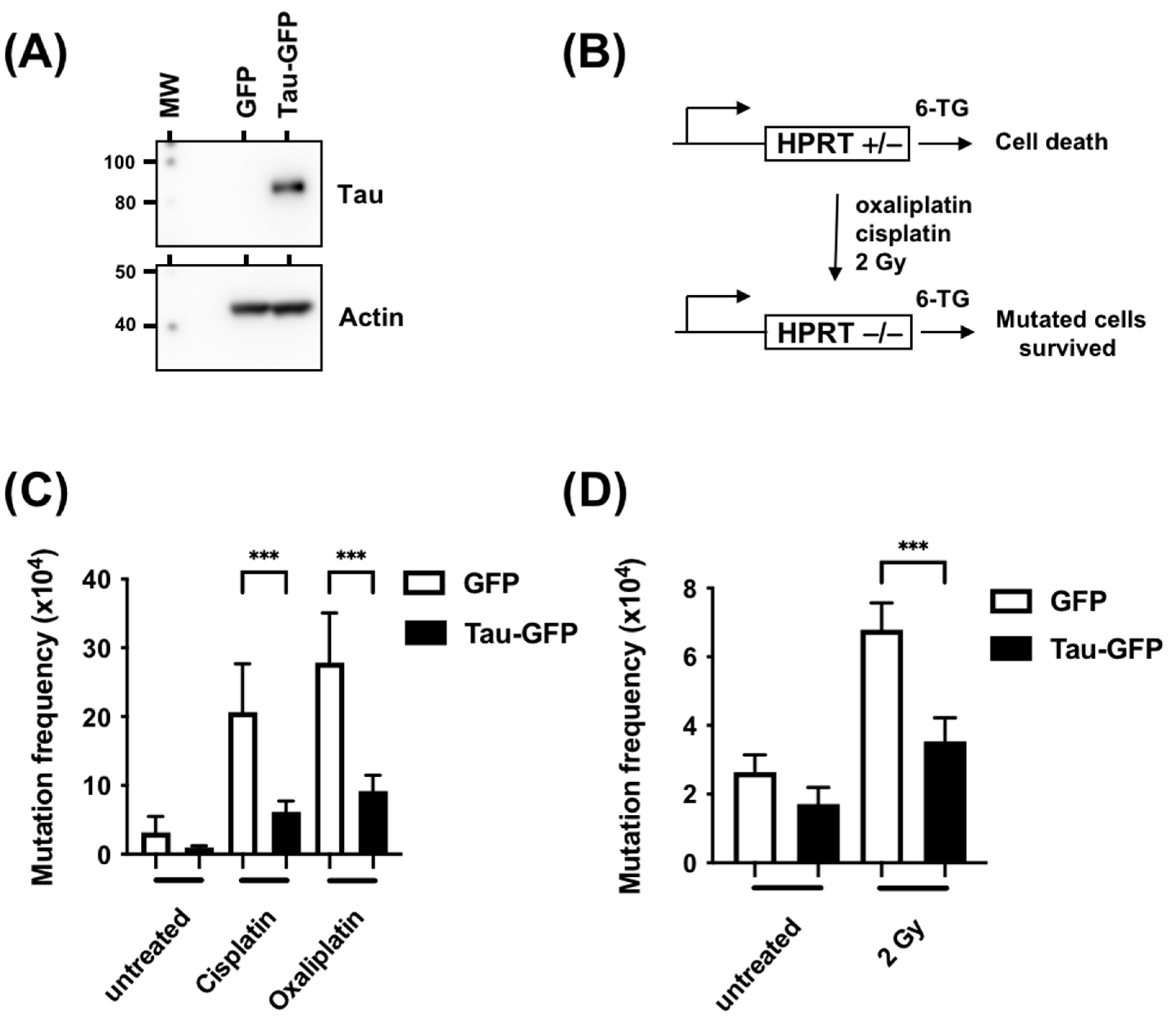
2.5. Hprt Mutant Frequency Determination
2.6. Immunofluorescence
2.7. Proximity Ligation Assay
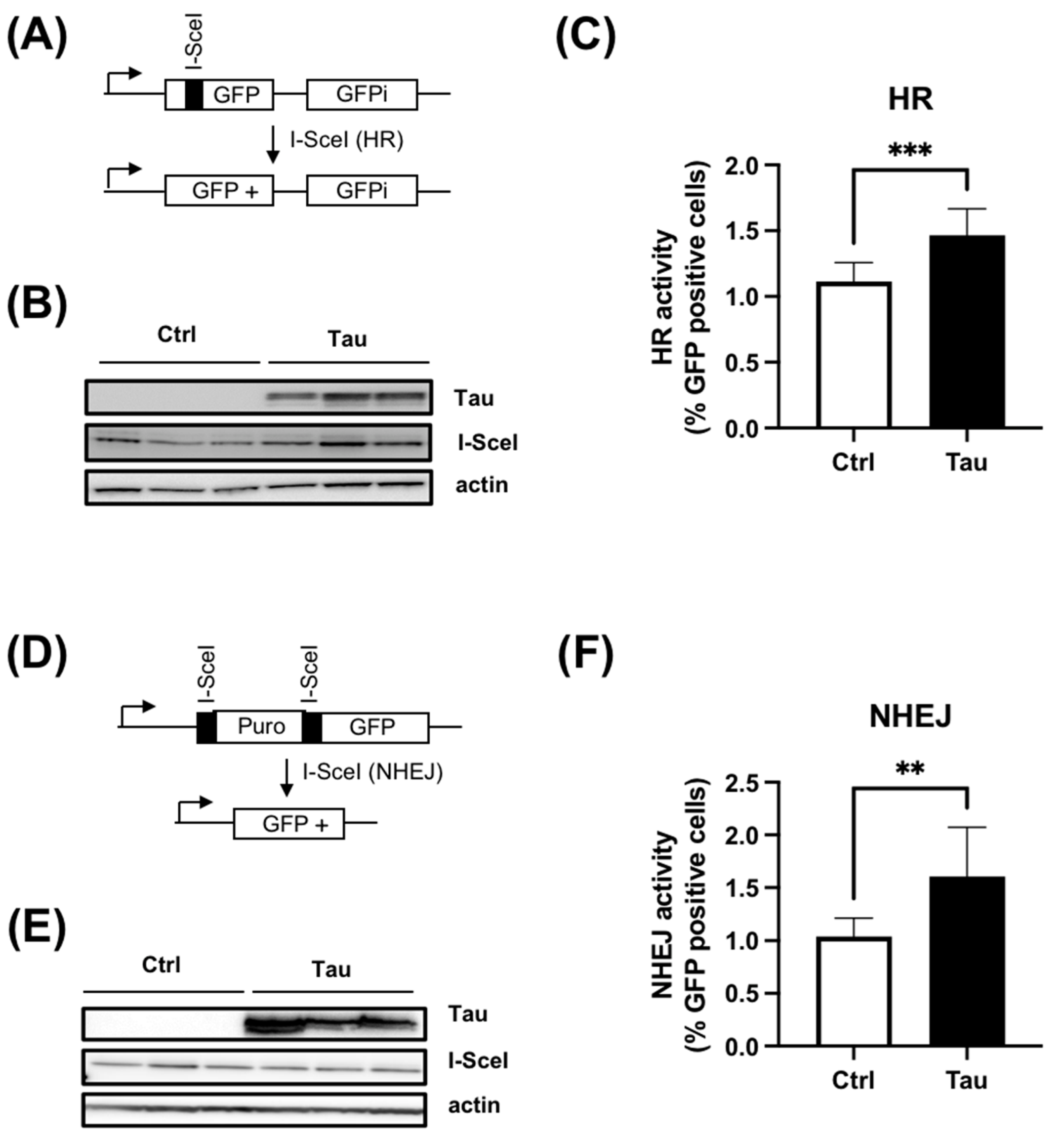
2.8. DNA Repair Reporter Assays
2.9. Xenograft Studies
2.10. Statistical Analysis
3. Results
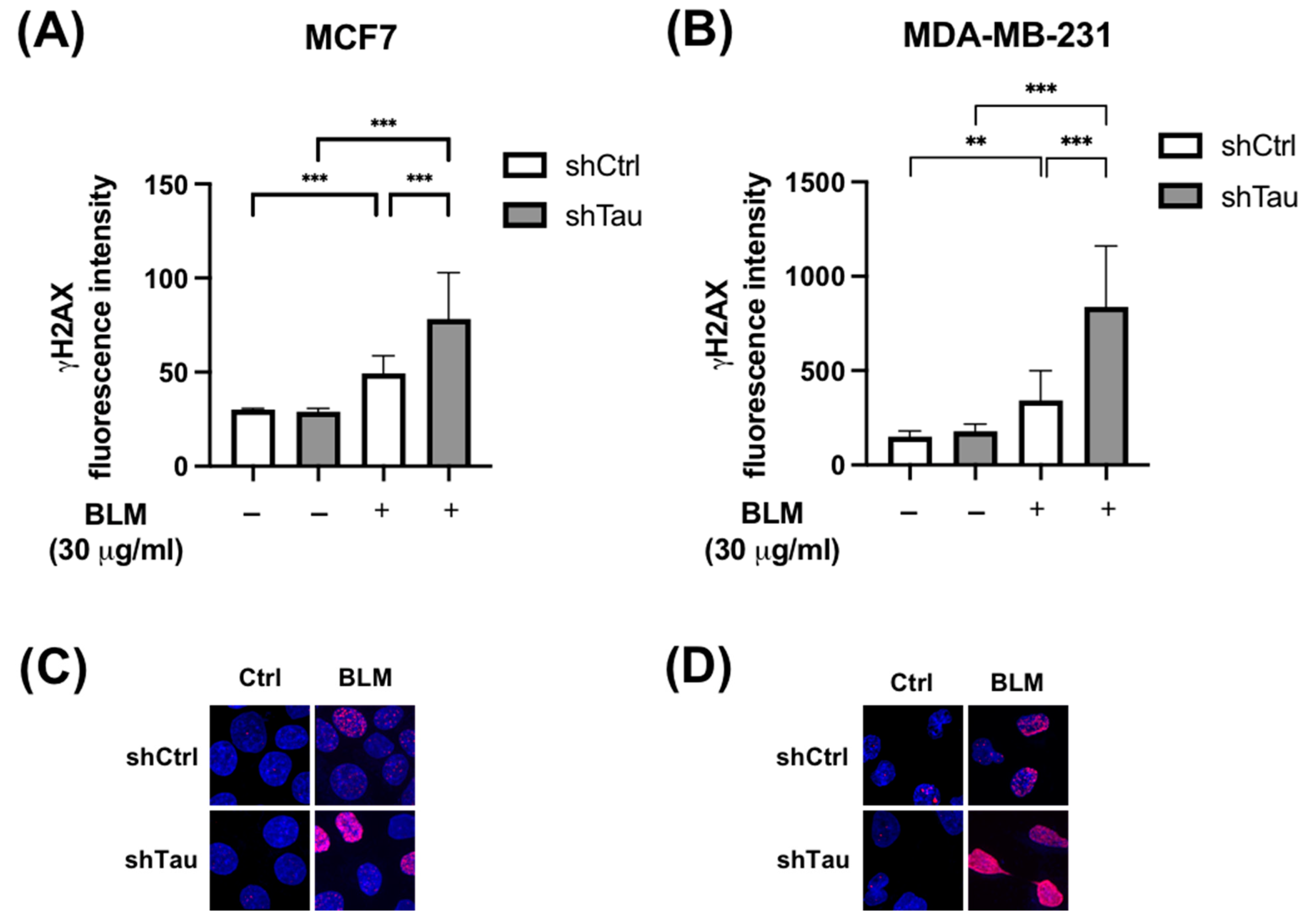
3.1. Tau Aids Clearance of Double-Strand Breaks
3.2. Tau Decreases the Mutation Rate Induced by DNA Damaging Agents
3.3. Tau Increases HR and cNHEJ Activities
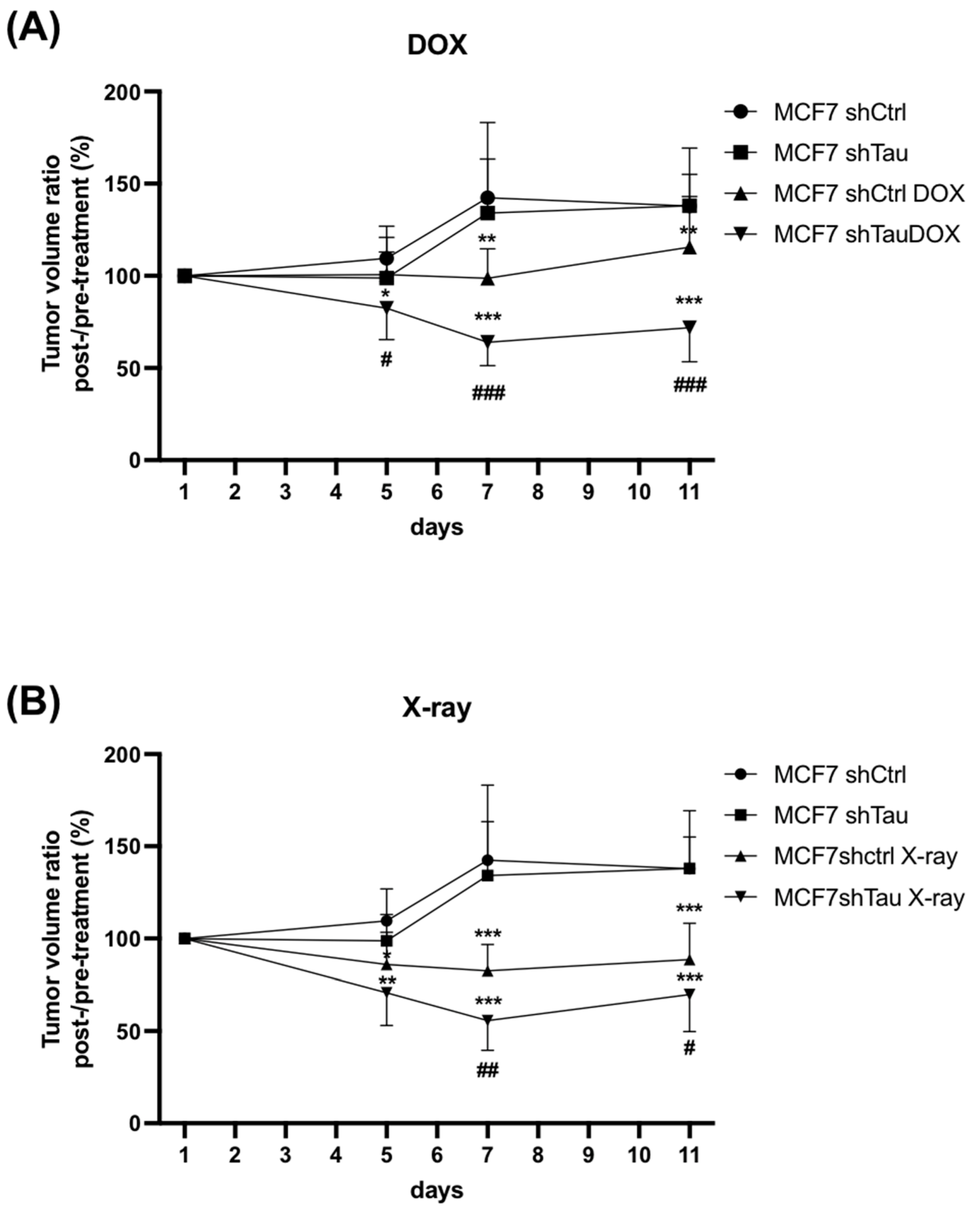
3.4. shTau Tumors Are More Sensitive to DNA Damage
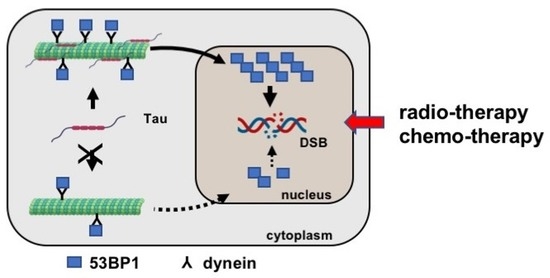
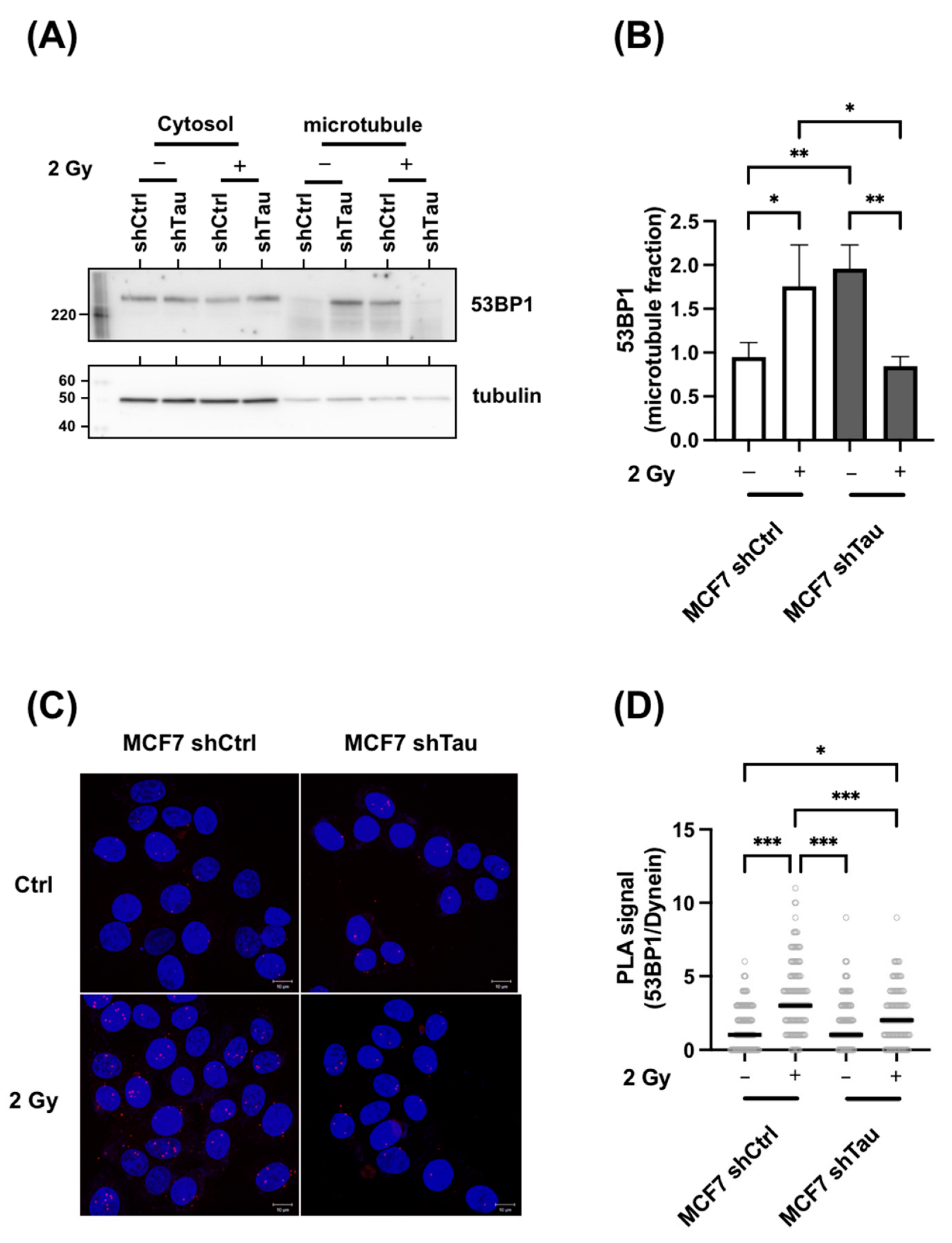
3.5. Tau Regulates 53BP1 Nuclear Localization
3.6. Tau Silencing Alters 53BP1 Trafficking on Microtubules
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Trenner, A.; Sartori, A.A. Harnessing DNA double-strand break repair for cancer treatment. Front. Oncol. 2019, 9, 1388. [Google Scholar] [CrossRef] [PubMed]
- Khanna, K.K.; Jackson, S.P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 2001, 27, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Her, J.; Bunting, S.F. How cells ensure correct repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10502–10511. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef]
- Clouaire, T.; Legube, G. A snapshot on the cis chromatin response to DNA double-strand breaks. Trends Genet. 2019, 35, 330–345. [Google Scholar] [CrossRef]
- Chapman, J.R.; Sossick, A.J.; Boulton, S.J.; Jackson, S.P. Brca1-associated exclusion of 53bp1 from DNA damage sites underlies temporal control of DNA repair. J. Cell Sci. 2012, 125, 3529–3534. [Google Scholar] [CrossRef]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair 2008, 7, 1765–1771. [Google Scholar] [CrossRef]
- Beucher, A.; Birraux, J.; Tchouandong, L.; Barton, O.; Shibata, A.; Conrad, S.; Goodarzi, A.A.; Krempler, A.; Jeggo, P.A.; Lobrich, M. Atm and artemis promote homologous recombination of radiation-induced DNA double-strand breaks in g2. EMBO J. 2009, 28, 3413–3427. [Google Scholar] [CrossRef]
- Poruchynsky, M.S.; Komlodi-Pasztor, E.; Trostel, S.; Wilkerson, J.; Regairaz, M.; Pommier, Y.; Zhang, X.; Kumar Maity, T.; Robey, R.; Burotto, M.; et al. Microtubule-targeting agents augment the toxicity of DNA-damaging agents by disrupting intracellular trafficking of DNA repair proteins. Proc. Natl. Acad. Sci. USA 2015, 112, 1571–1576. [Google Scholar] [CrossRef]
- Lottersberger, F.; Karssemeijer, R.A.; Dimitrova, N.; de Lange, T. 53bp1 and the linc complex promote microtubule-dependent dsb mobility and DNA repair. Cell 2015, 163, 880–893. [Google Scholar] [CrossRef]
- Weingarten, M.D.; Lockwood, A.H.; Hwo, S.Y.; Kirschner, M.W. A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. USA 1975, 72, 1858–1862. [Google Scholar] [CrossRef] [PubMed]
- Sotiropoulos, I.; Galas, M.C.; Silva, J.M.; Skoulakis, E.; Wegmann, S.; Maina, M.B.; Blum, D.; Sayas, C.L.; Mandelkow, E.M.; Mandelkow, E.; et al. Atypical, non-standard functions of the microtubule associated tau protein. Acta Neuropathol. Commun. 2017, 5, 91. [Google Scholar] [CrossRef] [PubMed]
- Frost, B.; Hemberg, M.; Lewis, J.; Feany, M.B. Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 2014, 17, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Rico, T.; Gilles, M.; Chauderlier, A.; Comptdaer, T.; Magnez, R.; Chwastyniak, M.; Drobecq, H.; Pinet, F.; Thuru, X.; Buee, L.; et al. Tau stabilizes chromatin compaction. Front. Cell Dev. Biol. 2021, 9, 740550. [Google Scholar] [CrossRef]
- Colnaghi, L.; Rondelli, D.; Muzi-Falconi, M.; Sertic, S. Tau and DNA damage in neurodegeneration. Brain Sci. 2020, 10, 946. [Google Scholar] [CrossRef] [PubMed]
- Violet, M.; Delattre, L.; Tardivel, M.; Sultan, A.; Chauderlier, A.; Caillierez, R.; Talahari, S.; Nesslany, F.; Lefebvre, B.; Bonnefoy, E.; et al. A major role for tau in neuronal DNA and rna protection in vivo under physiological and hyperthermic conditions. Front. Cell Neurosci. 2014, 8, 84. [Google Scholar] [CrossRef]
- Violet, M.; Chauderlier, A.; Delattre, L.; Tardivel, M.; Chouala, M.S.; Sultan, A.; Marciniak, E.; Humez, S.; Binder, L.; Kayed, R.; et al. Prefibrillar tau oligomers alter the nucleic acid protective function of tau in hippocampal neurons in vivo. Neurobiol. Dis. 2015, 82, 540–551. [Google Scholar] [CrossRef]
- Wei, Y.; Qu, M.H.; Wang, X.S.; Chen, L.; Wang, D.L.; Liu, Y.; Hua, Q.; He, R.Q. Binding to the minor groove of the double-strand, tau protein prevents DNA from damage by peroxidation. PLoS ONE 2008, 3, e2600. [Google Scholar] [CrossRef]
- Qi, H.; Cantrelle, F.X.; Benhelli-Mokrani, H.; Smet-Nocca, C.; Buee, L.; Lippens, G.; Bonnefoy, E.; Galas, M.C.; Landrieu, I. Nuclear magnetic resonance spectroscopy characterization of interaction of tau with DNA and its regulation by phosphorylation. Biochemistry 2015, 54, 1525–1533. [Google Scholar] [CrossRef]
- Hedna, R.; Kovacic, H.; Pagano, A.; Peyrot, V.; Robin, M.; Devred, F.; Breuzard, G. Tau protein as therapeutic target for cancer? Focus on glioblastoma. Cancers 2022, 14, 5386. [Google Scholar] [CrossRef]
- Rossi, G.; Redaelli, V.; Perego, P.; Ferrari, R.; Giaccone, G.; Tagliavini, F. Tau mutations as a novel risk factor for cancer-response. Cancer Res. 2018, 78, 6525. [Google Scholar] [CrossRef] [PubMed]
- Papin, S.; Paganetti, P. Emerging evidences for an implication of the neurodegeneration-associated protein tau in cancer. Brain Sci. 2020, 10, 862. [Google Scholar] [CrossRef] [PubMed]
- Delobel, P.; Flament, S.; Hamdane, M.; Jakes, R.; Rousseau, A.; Delacourte, A.; Vilain, J.P.; Goedert, M.; Buee, L. Functional characterization of ftdp-17 tau gene mutations through their effects on xenopus oocyte maturation. J. Biol. Chem. 2002, 277, 9199–9205. [Google Scholar] [CrossRef]
- Chauderlier, A.; Gilles, M.; Spolcova, A.; Caillierez, R.; Chwastyniak, M.; Kress, M.; Drobecq, H.; Bonnefoy, E.; Pinet, F.; Weil, D.; et al. Tau/ddx6 interaction increases microrna activity. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 762–772. [Google Scholar] [CrossRef]
- Bennardo, N.; Cheng, A.; Huang, N.; Stark, J.M. Alternative-nhej is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008, 4, e1000110. [Google Scholar] [CrossRef] [PubMed]
- Pierce, A.J.; Johnson, R.D.; Thompson, L.H.; Jasin, M. Xrcc3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999, 13, 2633–2638. [Google Scholar] [CrossRef]
- Derisbourg, M.; Leghay, C.; Chiappetta, G.; Fernandez-Gomez, F.J.; Laurent, C.; Demeyer, D.; Carrier, S.; Buee-Scherrer, V.; Blum, D.; Vinh, J.; et al. Role of the tau n-terminal region in microtubule stabilization revealed by new endogenous truncated forms. Sci. Rep. 2015, 5, 9659. [Google Scholar] [CrossRef]
- Silva, M.J.; Costa, P.; Dias, A.; Valente, M.; Louro, H.; Boavida, M.G. Comparative analysis of the mutagenic activity of oxaliplatin and cisplatin in the hprt gene of cho cells. Environ. Mol. Mutagen. 2005, 46, 104–115. [Google Scholar] [CrossRef]
- Atkinson, J.; Bezak, E.; Kempson, I. Imaging DNA double-strand breaks—Are we there yet? Nat. Rev. Mol. Cell Biol. 2022, 23, 579–580. [Google Scholar] [CrossRef]
- Galas, M.C.; Bonnefoy, E.; Buee, L.; Lefebvre, B. Emerging connections between tau and nucleic acids. Adv. Exp. Med. Biol. 2019, 1184, 135–143. [Google Scholar]
- Sultan, A.; Nesslany, F.; Violet, M.; Begard, S.; Loyens, A.; Talahari, S.; Mansuroglu, Z.; Marzin, D.; Sergeant, N.; Humez, S.; et al. Nuclear tau, a key player in neuronal DNA protection. J. Biol. Chem. 2011, 286, 4566–4575. [Google Scholar] [CrossRef] [PubMed]
- Asada-Utsugi, M.; Uemura, K.; Ayaki, T.; T Uemura, M.; Minamiyama, S.; Hikiami, R.; Morimura, T.; Shodai, A.; Ueki, T.; Takahashi, R.; et al. Failure of DNA double-strand break repair by tau mediates alzheimer’s disease pathology in vitro. Commun. Biol. 2022, 5, 358. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Suarez, I.; Redwood, A.B.; Grotsky, D.A.; Neumann, M.A.; Cheng, E.H.; Stewart, C.L.; Dusso, A.; Gonzalo, S. A new pathway that regulates 53bp1 stability implicates cathepsin l and vitamin d in DNA repair. EMBO J. 2011, 30, 3383–3396. [Google Scholar] [CrossRef] [PubMed]
- Lester, E.; Parker, R. The tau of nuclear-cytoplasmic transport. Neuron 2018, 99, 869–871. [Google Scholar] [CrossRef] [PubMed]
- Diez, L.; Wegmann, S. Nuclear transport deficits in tau-related neurodegenerative diseases. Front. Neurol. 2020, 11, 1056. [Google Scholar] [CrossRef]
- Paonessa, F.; Evans, L.D.; Solanki, R.; Larrieu, D.; Wray, S.; Hardy, J.; Jackson, S.P.; Livesey, F.J. Microtubules deform the nuclear membrane and disrupt nucleocytoplasmic transport in tau-mediated frontotemporal dementia. Cell Rep. 2019, 26, 582–593.e5. [Google Scholar] [CrossRef]
- Suomalainen, M.; Nakano, M.Y.; Keller, S.; Boucke, K.; Stidwill, R.P.; Greber, U.F. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 1999, 144, 657–672. [Google Scholar] [CrossRef]
- Chaudhary, A.R.; Berger, F.; Berger, C.L.; Hendricks, A.G. Tau directs intracellular trafficking by regulating the forces exerted by kinesin and dynein teams. Traffic 2018, 19, 111–121. [Google Scholar] [CrossRef]
- Oshidari, R.; Strecker, J.; Chung, D.K.C.; Abraham, K.J.; Chan, J.N.Y.; Damaren, C.J.; Mekhail, K. Nuclear microtubule filaments mediate non-linear directional motion of chromatin and promote DNA repair. Nat. Commun. 2018, 9, 2567. [Google Scholar] [CrossRef]
- Mekhail, K. Defining the damaged DNA mobility paradox as revealed by the study of telomeres, dsbs, microtubules and motors. Front. Genet. 2018, 9, 95. [Google Scholar] [CrossRef]
- Giannakakou, P.; Sackett, D.L.; Ward, Y.; Webster, K.R.; Blagosklonny, M.V.; Fojo, T. P53 is associated with cellular microtubules and is transported to the nucleus by dynein. Nat. Cell Biol. 2000, 2, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Giannakakou, P.; Nakano, M.; Nicolaou, K.C.; O’Brate, A.; Yu, J.; Blagosklonny, M.V.; Greber, U.F.; Fojo, T. Enhanced microtubule-dependent trafficking and p53 nuclear accumulation by suppression of microtubule dynamics. Proc. Natl. Acad. Sci. USA 2002, 99, 10855–10860. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Martinez, J.D. P53 is transported into the nucleus via an hsf1-dependent nuclear localization mechanism. Mol. Carcinog. 2011, 50, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Krishnamurthy, S. Cellular responses to cisplatin-induced DNA damage. J. Nucleic Acids 2010, 2010, 201367. [Google Scholar] [CrossRef]
- Kurihara, M.; Mano, T.; Saito, Y.; Murayama, S.; Toda, T.; Iwata, A. Colocalization of brca1 with tau aggregates in human tauopathies. Brain Sci. 2019, 10, 7. [Google Scholar] [CrossRef]
- Nakamura, M.; Kaneko, S.; Dickson, D.W.; Kusaka, H. Aberrant accumulation of brca1 in alzheimer disease and other tauopathies. J. Neuropathol. Exp. Neurol. 2020, 79, 22–33. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rico, T.; Denechaud, M.; Caillierez, R.; Comptdaer, T.; Adriaenssens, E.; Buée, L.; Lefebvre, B. Cancer Cells Upregulate Tau to Gain Resistance to DNA Damaging Agents. Cancers 2023, 15, 116. https://doi.org/10.3390/cancers15010116
Rico T, Denechaud M, Caillierez R, Comptdaer T, Adriaenssens E, Buée L, Lefebvre B. Cancer Cells Upregulate Tau to Gain Resistance to DNA Damaging Agents. Cancers. 2023; 15(1):116. https://doi.org/10.3390/cancers15010116
Chicago/Turabian StyleRico, Thomas, Marine Denechaud, Raphaelle Caillierez, Thomas Comptdaer, Eric Adriaenssens, Luc Buée, and Bruno Lefebvre. 2023. "Cancer Cells Upregulate Tau to Gain Resistance to DNA Damaging Agents" Cancers 15, no. 1: 116. https://doi.org/10.3390/cancers15010116
APA StyleRico, T., Denechaud, M., Caillierez, R., Comptdaer, T., Adriaenssens, E., Buée, L., & Lefebvre, B. (2023). Cancer Cells Upregulate Tau to Gain Resistance to DNA Damaging Agents. Cancers, 15(1), 116. https://doi.org/10.3390/cancers15010116