Circulating miRNAs in Breast Cancer Diagnosis and Prognosis
Abstract
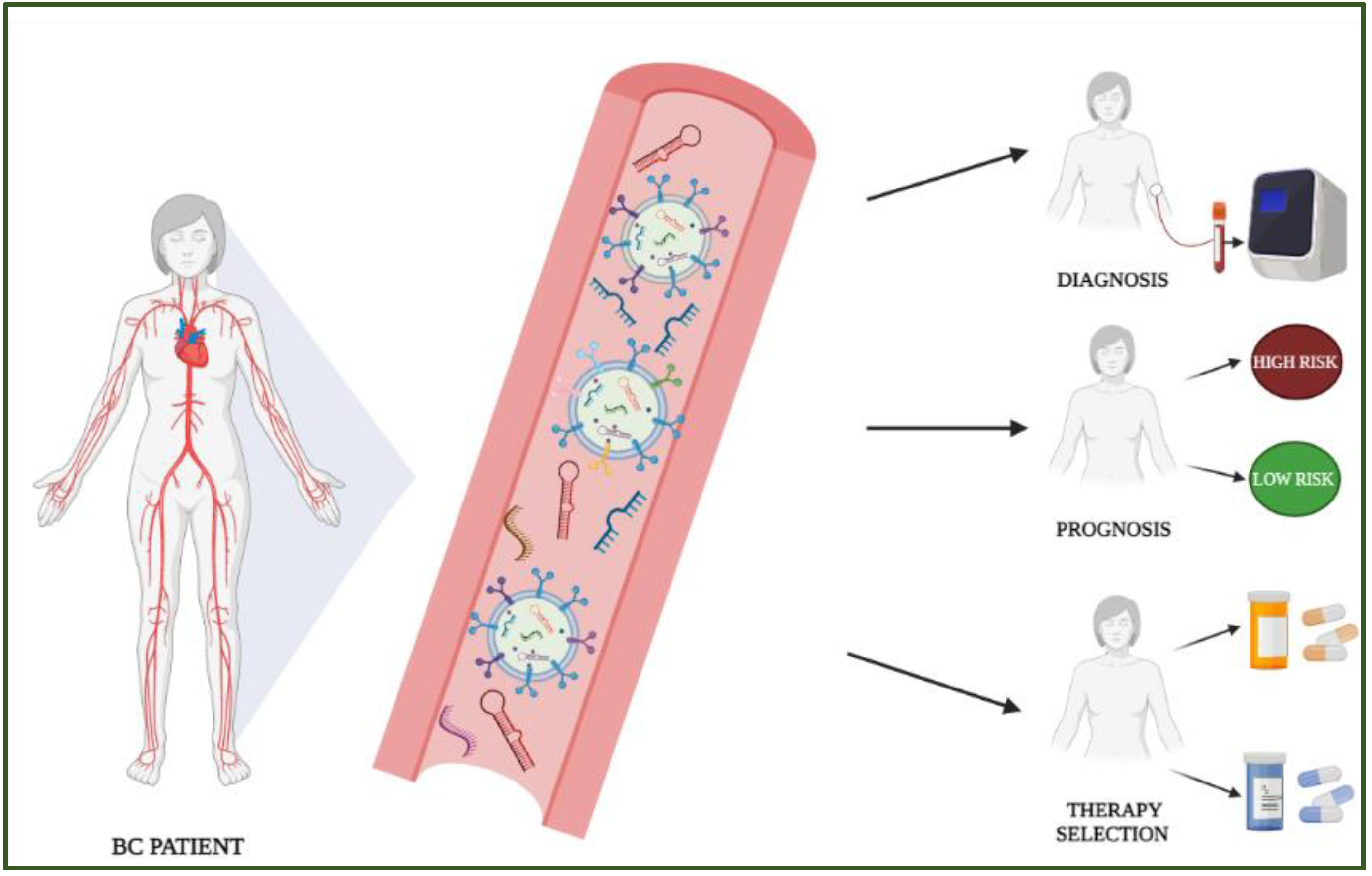
Simple Summary
Abstract
1. Introduction
1.1. Free Circulating miRNAs: Pre-Clinical Studies
1.2. Extracellular Vesicle (EV)-Associated Circulating miRNAs: Pre-Clinical Studies
1.3. Free Circulating miRNAs: Clinical Studies
1.4. EV-Associated Circulating miRNAs: Clinical Studies
2. Discussion
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Momenimovahed, Z.; Salehiniya, H. Epidemiological characteristics of and risk factors for breast cancer in the world. Breast Cancer Targets Ther. 2019, 11, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Perou, C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2011, 5, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.J.; Albain, K.S.; André, F.; Bergh, J.; et al. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef]
- Lianidou, E.; Pantel, K. Liquid biopsies. Genes Chromosom. Cancer 2019, 58, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Mar-Aguilar, F.; Mendoza-Ramírez, J.A.; Malagón-Santiago, I.; Espino-Silva, P.K.; Santuario-Facio, S.K.; Ruiz-Flores, P.; Rodríguez-Padilla, C.; Reséndez-Pérez, D. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis. Markers 2013, 34, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Mandujano-Tinoco, E.A.; García-Venzor, A.; Melendez-Zajgla, J.; Maldonado, V. New emerging roles of microRNAs in breast cancer. Breast Cancer Res. Treat. 2018, 171, 247–259. [Google Scholar] [CrossRef]
- Lakshmi, S.; Hughes, T.A.; Priya, S. Exosomes and exosomal RNAs in breast cancer: A status update. Eur. J. Cancer 2021, 144, 252–268. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015, 25, 137–147. [Google Scholar] [CrossRef]
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, V.; Naqvi, A.R.; Uttamani, J.R.; Nares, S. MiRNA-Target Interaction Reveals Cell-Specific Post-Transcriptional Regulation in Mammalian Cell Lines. Int. J. Mol. Sci. 2016, 17, 72. [Google Scholar] [CrossRef]
- Vasudevan, S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip. Rev. RNA 2012, 3, 311–330. [Google Scholar] [CrossRef]
- Orang, A.V.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int. J. Genom. 2014, 2014, 970607. [Google Scholar] [CrossRef]
- Lai, X.; Wolkenhauer, O.; Vera, J. Understanding microRNA-mediated gene regulatory networks through mathematical modelling. Nucleic Acids Res. 2016, 44, 6019–6035. [Google Scholar] [CrossRef]
- Yang, Z.; Jakymiw, A.; Wood, M.R.; Eystathioy, T.; Rubin, R.L.; Fritzler, M.J.; Chan, E.K.L. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J. Cell Sci. 2004, 117, 5567–5578. [Google Scholar] [CrossRef]
- Sampson, V.B.; Rong, N.H.; Han, J.; Yang, Q.; Aris, V.; Soteropoulos, P.; Petrelli, N.J.; Dunn, S.P.; Krueger, L.J. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007, 67, 9762–9770. [Google Scholar] [CrossRef]
- Bueno, M.J.; Gómez de Cedrón, M.; Laresgoiti, U.; Fernández-Piqueras, J.; Zubiaga, A.M.; Malumbres, M. Multiple E2F-induced microRNAs prevent replicative stress in response to mitogenic signaling. Mol. Cell. Biol. 2010, 30, 2983–2995. [Google Scholar] [CrossRef]
- Hu, W.; Chan, C.S.; Wu, R.; Zhang, C.; Sun, Y.; Song, J.S.; Tang, L.H.; Levine, A.J.; Feng, Z. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol. Cell 2010, 38, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Huang, S.; Wu, S.; Guo, W.; Li, J.; He, X. MicroRNA-1285 inhibits the expression of p53 by directly targeting its 3’ untranslated region. Biochem. Biophys. Res. Commun. 2010, 396, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Chim, S.S.C.; Shing, T.K.F.; Hung, E.C.W.; Leung, T.Y.; Lau, T.K.; Chiu, R.W.K.; Lo, Y.M.D. Detection and characterization of placental microRNAs in maternal plasma. Clin. Chem. 2008, 54, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, X.; Li, L. Biogenesis and function of extracellular miRNAs. ExRNA 2019, 1, 1–9. [Google Scholar] [CrossRef]
- Mussbacher, M.; Pirabe, A.; Brunnthaler, L.; Schrottmaier, W.C.; Assinger, A. Horizontal MicroRNA Transfer by Platelets–Evidence and Implications. Front. Physiol. 2021, 12, 781. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, H.; Jin, F.; Yan, X.; Xu, G.; Hu, H.; Liang, G.; Zhan, S.; Hu, X.; Zhao, Q.; et al. Injured liver-released miRNA-122 elicits acute pulmonary inflammation via activating alveolar macrophage TLR7 signaling pathway. Proc. Natl. Acad. Sci. USA 2019, 116, 6162–6171. [Google Scholar] [CrossRef]
- Su, Z.; Yang, Z.; Xu, Y.; Chen, Y.; Yu, Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget 2015, 6, 8474. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The Majority of MicroRNAs Detectable in Serum and Saliva Is Concentrated in Exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef]
- Munir, J.; Yoon, J.K.; Ryu, S. Therapeutic miRNA-Enriched Extracellular Vesicles: Current Approaches and Future Prospects. Cells 2020, 9, 2271. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Montani, F.; Bianchi, F. Circulating Cancer Biomarkers: The Macro-revolution of the Micro-RNA. EBioMedicine 2016, 5, 4. [Google Scholar] [CrossRef]
- Orlandella, F.M.; Auletta, L.; Greco, A.; Zannetti, A.; Salvatore, G. Preclinical Imaging Evaluation of miRNAs’ Delivery and Effects in Breast Cancer Mouse Models: A Systematic Review. Cancers 2021, 13, 6020. [Google Scholar] [CrossRef]
- Gervin, E.; Shin, B.; Opperman, R.; Cullen, M.; Feser, R.; Maiti, S.; Majumder, M. Chemically Induced Hypoxia Enhances miRNA Functions in Breast Cancer. Cancers 2020, 12, 2008. [Google Scholar] [CrossRef]
- Ouyang, Q.; Cui, Y.; Yang, S.; Wei, W.; Zhang, M.; Zeng, J.; Qu, F. lncRNA MT1JP Suppresses Biological Activities of Breast Cancer Cells in vitro and in vivo by Regulating the miRNA-214/RUNX3 Axis. Onco. Targets. Ther. 2020, 13, 5033. [Google Scholar] [CrossRef]
- Meng, Q.; Wang, L.; Lv, Y.; Wu, J.; Shi, W. Deletion of HNF1A-AS1 Suppresses the Malignant Phenotypes of Breast Cancer Cells in Vitro and in Vivo through Targeting miRNA-20a-5p/TRIM32 Axis. Cancer Biother. Radiopharm. 2021, 36, 23–35. [Google Scholar] [CrossRef]
- Li, L.; Yuan, L.; Luo, J.; Gao, J.; Guo, J.; Xie, X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin. Exp. Med. 2013, 13, 109–117. [Google Scholar] [CrossRef]
- Li, Z.H.; Weng, X.; Xiong, Q.Y.; Tu, J.H.; Xiao, A.; Qiu, W.; Gong, Y.; Hu, E.W.; Huang, S.; Cao, Y.L. MiR-34a expression in human breast cancer is associated with drug resistance. Oncotarget 2017, 8, 106270–106282. [Google Scholar] [CrossRef] [PubMed]
- Si, W.; Li, Y.; Shao, H.; Hu, R.; Wang, W.; Zhang, K.; Yang, Q. MiR-34a Inhibits Breast Cancer Proliferation and Progression by Targeting Wnt1 in Wnt/β-Catenin Signaling Pathway. Am. J. Med. Sci. 2016, 352, 191–199. [Google Scholar] [CrossRef]
- Surapaneni, S.K.; Bhat, Z.R.; Tikoo, K. MicroRNA-941 regulates the proliferation of breast cancer cells by altering histone H3 Ser 10 phosphorylation. Sci. Reports 2020, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Belizário, J.E. Immunodeficient Mouse Models: An Overview. Open Immunol. J. 2009, 2, 79–85. [Google Scholar] [CrossRef]
- Chdiwa, T.; Kawai, K.; Noguchi, A.; Sato, H.; Hayashi, A.; Cho, H.; Shiozawa, M.; Kishida, T.; Morinaga, S.; Yokose, T.; et al. Establishment of patient-derived cancer xenografts in immunodeficient NOG mice. Int. J. Oncol. 2015, 47, 61–70. [Google Scholar] [CrossRef]
- Fujii, E.; Suzuki, M.; Matsubara, K.; Watanabe, M.; Chen, Y.J.; Adachi, K.; Ohnishi, Y.; Tanigawa, M.; Tsuchiya, M.; Tamaoki, N. Establishment and characterization of in vivo human tumor models in the NOD/SCID/gamma(c)(null) mouse. Pathol. Int. 2008, 58, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409. [Google Scholar] [CrossRef]
- Dan, T.; Shastri, A.A.; Palagani, A.; Buraschi, S.; Neill, T.; Savage, J.E.; Kapoor, A.; Deangelis, T.; Addya, S.; Camphausen, K.; et al. miR-21 Plays a Dual Role in Tumor Formation and Cytotoxic Response in Breast Tumors. Cancers 2021, 13, 1–16. [Google Scholar] [CrossRef]
- Shen, S.J.; Song, Y.; Ren, X.Y.; Xu, Y.L.; Zhou, Y.D.; Liang, Z.Y.; Sun, Q. MicroRNA-27b-3p Promotes Tumor Progression and Metastasis by Inhibiting Peroxisome Proliferator-Activated Receptor Gamma in Triple-Negative Breast Cancer. Front. Oncol. 2020, 10, 1371. [Google Scholar] [CrossRef]
- Wu, Z.; Cai, X.; Huang, C.; Xu, J.; Liu, A. MiR-497 suppresses angiogenesis in breast carcinoma by targeting HIF-1α. Oncol. Rep. 2016, 35, 1696–1702. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, S.; Zhu, J.; Yang, Q.; Dong, H.; Huang, J. MicroRNA-544 down-regulates both Bcl6 and Stat3 to inhibit tumor growth of human triple negative breast cancer. Biol. Chem. 2016, 397, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zheng, X.Y. MicroRNA-490 inhibits tumorigenesis and progression in breast cancer. Onco. Targets. Ther. 2016, 9, 4505–4516. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shibuya, N.; Kakeji, Y.; Shimono, Y. MicroRNA-93 targets WASF3 and functions as a metastasis suppressor in breast cancer. Cancer Sci. 2020, 111, 2093. [Google Scholar] [CrossRef]
- Oh, K.; Lee, D.-S. In vivo validation of metastasis-regulating microRNA-766 in human triple-negative breast cancer cells. Lab. Anim. Res. 2017, 33, 256. [Google Scholar] [CrossRef]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.T.; Bora, A.; Lässer, C.; Lötvall, J.; Nolte-’t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Chen, Y.; Li, G.; Liu, M.L. Microvesicles as Emerging Biomarkers and Therapeutic Targets in Cardiometabolic Diseases. Genomics. Proteom. Bioinform. 2018, 16, 50–62. [Google Scholar] [CrossRef]
- Battistelli, M.; Falcieri, E. Apoptotic Bodies: Particular Extracellular Vesicles Involved in Intercellular Communication. Biology 2020, 9, 21. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Groot, M.; Lee, H. Sorting Mechanisms for MicroRNAs into Extracellular Vesicles and Their Associated Diseases. Cells 2020, 9, 1044. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Cheng, L.; Shao, Q.; Chen, Z.; Lv, X.; Li, J.; He, L.; Sun, Y.; Ji, Q.; Lu, P.; et al. Characterization of serum small extracellular vesicles and their small RNA contents across humans, rats, and mice. Sci. Rep. 2020, 10, 1–16. [Google Scholar] [CrossRef]
- Fujita, Y.; Kuwano, K.; Ochiya, T.; Takeshita, F. The Impact of Extracellular Vesicle-Encapsulated Circulating MicroRNAs in Lung Cancer Research. Biomed Res. Int. 2014, 2014, 486413. [Google Scholar] [CrossRef]
- De Miguel Pérez, D.; Rodriguez Martínez, A.; Ortigosa Palomo, A.; Delgado Ureña, M.; Garcia Puche, J.L.; Robles Remacho, A.; Exposito Hernandez, J.; Lorente Acosta, J.A.; Ortega Sánchez, F.G.; Serrano, M.J. Extracellular vesicle-miRNAs as liquid biopsy biomarkers for disease identification and prognosis in metastatic colorectal cancer patients. Sci. Rep. 2020, 10, 3974. [Google Scholar] [CrossRef] [PubMed]
- Le, M.T.N.; Hamar, P.; Guo, C.; Basar, E.; Perdigão-Henriques, R.; Balaj, L.; Lieberman, J. MiR-200–containing extracellular vesicles promote breast cancer cell metastasis. J. Clin. Invest. 2014, 124, 5109. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.T.; Peng, B.; Zhang, D.X.; Ma, V.; Mathey-Andrews, C.A.; Lam, C.K.; Kiomourtzis, T.; Jin, J.; McReynolds, L.; Huang, L.; et al. Tumor-secreted extracellular vesicles promote the activation of cancer-associated fibroblasts via the transfer of microRNA-125b. J. Extracell. Vesicles 2019, 8, 1599680. [Google Scholar] [CrossRef]
- Ren, Z.; Lv, M.; Yu, Q.; Bao, J.; Lou, K.; Li, X. MicroRNA-370-3p shuttled by breast cancer cell-derived extracellular vesicles induces fibroblast activation through the CYLD/Nf-κB axis to promote breast cancer progression. FASEB J. 2021, 35, e21383. [Google Scholar] [CrossRef]
- Zheng, X.; Lu, S.; He, Z.; Huang, H.; Yao, Z.; Miao, Y.; Cai, C.; Zou, F. MCU-dependent negative sorting of miR-4488 to extracellular vesicles enhances angiogenesis and promotes breast cancer metastatic colonization. Oncogene 2020, 39, 6975–6989. [Google Scholar] [CrossRef]
- Lu, C.; Zhao, Y.; Wang, J.; Shi, W.; Dong, F.; Xin, Y.; Zhao, X.; Liu, C. Breast cancer cell-derived extracellular vesicles transfer miR-182-5p and promote breast carcinogenesis via the CMTM7/EGFR/AKT axis. Mol. Med. 2021, 27, 78. [Google Scholar] [CrossRef]
- Farahani, M.; Rubbi, C.; Liu, L.; Slupsky, J.R.; Kalakonda, N. CLL Exosomes Modulate the Transcriptome and Behaviour of Recipient Stromal Cells and Are Selectively Enriched in miR-202-3p. PLoS ONE 2015, 10, e0141429. [Google Scholar] [CrossRef]
- Ostenfeld, M.S.; Jeppesen, D.K.; Laurberg, J.R.; Boysen, A.T.; Bramsen, J.B.; Primdal-Bengtson, B.; Hendrix, A.; Lamy, P.; Dagnaes-Hansen, F.; Rasmussen, M.H.; et al. Cellular disposal of miR23b by RAB27-dependent exosome release is linked to acquisition of metastatic properties. Cancer Res. 2014, 74, 5758–5771. [Google Scholar] [CrossRef] [PubMed]
- Heneghan, H.M.; Miller, N.; Lowery, A.J.; Sweeney, K.J.; Newell, J.; Kerin, M.J. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann. Surg. 2010, 251, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.M.; Said, M.M.; Hilal, A.M.; Medhat, A.M.; Abd Elsalam, I.M. Candidate circulating microRNAs as potential diagnostic and predictive biomarkers for the monitoring of locally advanced breast cancer patients. Tumour Biol. 2020, 42, 1010428320963811. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, P.M.M.; Jucoski, T.S.; Vieira, E.; Carvalho, T.M.; Malheiros, D.; De Souza Fonseca Ribeiro, E.M. Liquid biopsy for breast cancer using extracellular vesicles and cell-free microRNAs as biomarkers. Transl. Res. 2020, 223, 40–60. [Google Scholar] [CrossRef]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef]
- Baffa, R.; Fassan, M.; Volinia, S.; O’Hara, B.; Liu, C.G.; Palazzo, J.P.; Gardiman, M.; Rugge, M.; Gomella, L.G.; Croce, C.M.; et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J. Pathol. 2009, 219, 214–221. [Google Scholar] [CrossRef]
- Frankel, L.B.; Christoffersen, N.R.; Jacobsen, A.; Lindow, M.; Krogh, A.; Lund, A.H. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008, 283, 1026–1033. [Google Scholar] [CrossRef]
- Qian, B.; Katsaros, D.; Lu, L.; Preti, M.; Durando, A.; Arisio, R.; Mu, L.; Yu, H. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res. Treat. 2009, 117, 131–140. [Google Scholar] [CrossRef]
- Jinling, W.; Sijing, S.; Jie, Z.; Guinian, W. Prognostic value of circulating microRNA-21 for breast cancer: A systematic review and meta-analysis. Artif. Cells Nanomed. Biotechnol. 2017, 45, 1216–1221. [Google Scholar] [CrossRef]
- Faraoni, I.; Antonetti, F.R.; Cardone, J.; Bonmassar, E. MiR-155 gene: A typical multifunctional microRNA. Biochim. Biophys. Acta 2009, 1792, 497–505. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, H.W.; Lu, M.H.; He, X.H.; Li, Y.; Gu, H.; Liu, M.F.; Wang, E.D. MicroRNA-155 functions as an oncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010, 70, 3119–3127. [Google Scholar] [CrossRef] [PubMed]
- O’Donovan, P.J.; Livingston, D.M. BRCA1 and BRCA2: Breast/ovarian cancer susceptibility gene products and participants in DNA double-strand break repair. Carcinogenesis 2010, 31, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Eichelser, C.; Flesch-Janys, D.; Chang-Claude, J.; Pantel, K.; Schwarzenbach, H. Deregulated Serum Concentrations of Circulating Cell–Free MicroRNAs miR-17, miR-34a, miR-155, and miR-373 in Human Breast Cancer Development and Progression. Clin. Chem. 2013, 59, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Hagrass, H.A.; Sharaf, S.; Pasha, H.F.; Tantawy, E.A.; Mohamed, R.H.; Kassem, R. Circulating microRNAs-a new horizon in molecular diagnosis of breast cancer. Genes Cancer 2015, 6, 281–287. [Google Scholar] [CrossRef]
- Altuvia, Y.; Landgraf, P.; Lithwick, G.; Elefant, N.; Pfeffer, S.; Aravin, A.; Brownstein, M.J.; Tuschl, T.; Margalit, H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005, 33, 2697–2706. [Google Scholar] [CrossRef]
- Huber, M.A.; Azoitei, N.; Baumann, B.; Grünert, S.; Sommer, A.; Pehamberger, H.; Kraut, N.; Beug, H.; Wirth, T. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J. Clin. Invest. 2004, 114, 569–581. [Google Scholar] [CrossRef]
- Jurmeister, S.; Baumann, M.; Balwierz, A.; Keklikoglou, I.; Ward, A.; Uhlmann, S.; Zhang, J.D.; Wiemann, S.; Sahin, Ö. MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol. Cell. Biol. 2012, 32, 633–651. [Google Scholar] [CrossRef]
- Heyn, H.; Engelmann, M.; Schreek, S.; Ahrens, P.; Lehmann, U.; Kreipe, H.; Schlegelberger, B.; Beger, C. MicroRNA miR-335 is crucial for the BRCA1 regulatory cascade in breast cancer development. Int. J. Cancer 2011, 129, 2797–2806. [Google Scholar] [CrossRef]
- Negrini, M.; Calin, G.A. Breast cancer metastasis: A microRNA story. Breast Cancer Res. 2008, 10, 303. [Google Scholar] [CrossRef]
- Zheng, J.Z.; Huang, Y.N.; Yao, L.; Liu, Y.R.; Liu, S.; Hu, X.; Liu, Z.B.; Shao, Z.M. Elevated miR-301a expression indicates a poor prognosis for breast cancer patients. Sci. Rep. 2018, 8, 2225. [Google Scholar] [CrossRef]
- Cuk, K.; Zucknick, M.; Madhavan, D.; Schott, S.; Golatta, M.; Heil, J.; Marmé, F.; Turchinovich, A.; Sinn, P.; Sohn, C.; et al. Plasma microRNA panel for minimally invasive detection of breast cancer. PLoS ONE 2013, 8, e76729. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Hu, Q.; Schrauder, M.; Yan, L.; Wang, D.; Medico, L.; Guo, Y.; Yao, S.; Zhu, Q.; Liu, B.; et al. Circulating miR-148b and miR-133a as biomarkers for breast cancer detection. Oncotarget 2014, 5, 5284–5294. [Google Scholar] [CrossRef] [PubMed]
- Zearo, S.; Kim, E.; Zhu, Y.; Zhao, J.T.; Sidhu, S.B.; Robinson, B.G.; Soon, P.S.H. MicroRNA-484 is more highly expressed in serum of early breast cancer patients compared to healthy volunteers. BMC Cancer 2014, 14, 200. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Chen, X.; Zhu, D.; Luo, Z.; Yang, M. Low expression of circulating microRNA-34c is associated with poor prognosis in triple-negative breast cancer. Yonsei Med. J. 2017, 58, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Sahlberg, K.K.; Bottai, G.; Naume, B.; Burwinkel, B.; Calin, G.A.; Børresen-Dale, A.L.; Santarpia, L. A serum microRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients. Clin. Cancer Res. 2015, 21, 1207–1214. [Google Scholar] [CrossRef]
- Huo, D.; Clayton, W.M.; Yoshimatsu, T.F.; Chen, J.; Olopade, O.I. Identification of a circulating microRNA signature to distinguish recurrence in breast cancer patients. Oncotarget 2016, 7, 55231–55248. [Google Scholar] [CrossRef]
- Di Cosimo, S.; Appierto, V.; Pizzamiglio, S.; Tiberio, P.; Iorio, M.V.; Hilbers, F.; De Azambuja, E.; De La Peña, L.; Izquierdo, M.; Huober, J.; et al. Plasma miRNA Levels for Predicting Therapeutic Response to Neoadjuvant Treatment in HER2-positive Breast Cancer: Results from the NeoALTTO Trial. Clin. Cancer Res. 2019, 25, 3887–3895. [Google Scholar] [CrossRef]
- Li, Q.; Liu, M.; Ma, F.; Luo, Y.; Cai, R.; Wang, L.; Xu, N.; Xu, B. Circulating miR-19a and miR-205 in serum may predict the sensitivity of luminal A subtype of breast cancer patients to neoadjuvant chemotherapy with epirubicin plus paclitaxel. PLoS ONE 2014, 9, e104870. [Google Scholar] [CrossRef]
- Wang, H.; Tan, G.; Dong, L.; Cheng, L.; Li, K.; Wang, Z.; Luo, H. Circulating MiR-125b as a marker predicting chemoresistance in breast cancer. PLoS ONE 2012, 7, e34210. [Google Scholar] [CrossRef]
- Li, H.; Liu, J.; Chen, J.; Wang, H.; Yang, L.; Chen, F.; Fan, S.; Wang, J.; Shao, B.; Yin, D.; et al. A serum microRNA signature predicts trastuzumab benefit in HER2-positive metastatic breast cancer patients. Nat. Commun. 2018, 9, 1614. [Google Scholar] [CrossRef]
- Matamala, N.; Vargas, M.T.; González-Cámpora, R.; Miñambres, R.; Arias, J.; Menéndez, P.; Andrés-León, E.; Mez-López, G.G.; Yanowsky, K.; Calvete-Candenas, J.; et al. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin. Chem. 2015, 61, 1098–1106. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.Y.; Gong, H.T.; Li, B.F.; Lv, C.L.; Wang, H.T.; Zhou, H.H.; Li, X.X.; Xie, S.Y.; Jiang, B.F. Higher expression of circulating miR-182 as a novel biomarker for breast cancer. Oncol. Lett. 2013, 6, 1681–1686. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zou, X.; Xia, T.; Wang, T.; Liu, P.; Zhou, X.; Wang, S.; Zhu, W. A five-miRNA panel in plasma was identified for breast cancer diagnosis. Cancer Med. 2019, 8, 7006–7017. [Google Scholar] [CrossRef]
- Satomi-Tsushita, N.; Shimomura, A.; Matsuzaki, J.; Yamamoto, Y.; Kawauchi, J.; Takizawa, S.; Aoki, Y.; Sakamoto, H.; Kato, K.; Shimizu, C.; et al. Serum microRNA-based prediction of responsiveness to eribulin in metastatic breast cancer. PLoS ONE 2019, 14, e0222024. [Google Scholar] [CrossRef]
- McAnena, P.; Tanriverdi, K.; Curran, C.; Gilligan, K.; Freedman, J.E.; Brown, J.A.L.; Kerin, M.J. Circulating microRNAs miR-331 and miR-195 differentiate local luminal a from metastatic breast cancer. BMC Cancer 2019, 19, 436. [Google Scholar] [CrossRef]
- Papadaki, C.; Stoupis, G.; Tsalikis, L.; Monastirioti, A.; Papadaki, M.; Maliotis, N.; Stratigos, M.; Mastrostamatis, G.; Mavroudis, D.; Agelaki, S.; et al. Circulating miRNAs as a marker of metastatic disease and prognostic factor in metastatic breast cancer. Oncotarget 2019, 10, 966–981. [Google Scholar] [CrossRef]
- Jang, J.Y.; Kim, Y.S.; Kang, K.N.; Kim, K.H.; Park, Y.J.; Kim, C.W. Multiple microRNAs as biomarkers for early breast cancer diagnosis. Mol. Clin. Oncol. 2021, 14, 31. [Google Scholar] [CrossRef]
- Li, M.; Zhou, Y.; Xia, T.; Zhou, X.; Huang, Z.; Zhang, H.; Zhu, W.; Ding, Q.; Wang, S. Circulating microRNAs from the miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res. Treat. 2018, 170, 257–270. [Google Scholar] [CrossRef]
- Lopes, B.C.; Braga, C.Z.; Ventura, F.V.; de Oliveira, J.G.; Kato-Junior, E.M.; Bordin-Junior, N.A.; Zuccari, D.A.P.C. MiR-210 and miR-152 as Biomarkers by Liquid Biopsy in Invasive Ductal Carcinoma. J. Pers. Med. 2021, 11, 31. [Google Scholar] [CrossRef]
- Khodadadi-Jamayran, A.; Akgol-Oksuz, B.; Afanasyeva, Y.; Heguy, A.; Thompson, M.; Ray, K.; Giro-Perafita, A.; Sánchez, I.; Wu, X.; Tripathy, D.; et al. Prognostic role of elevated mir-24-3p in breast cancer and its association with the metastatic process. Oncotarget 2018, 9, 12868–12878. [Google Scholar] [CrossRef]
- Qattan, A.; Al-Tweigeri, T.; Alkhayal, W.; Suleman, K.; Tulbah, A.; Amer, S. Clinical Identification of Dysregulated Circulating microRNAs and Their Implication in Drug Response in Triple Negative Breast Cancer (TNBC) by Target Gene Network and Meta-Analysis. Genes 2021, 12, 549. [Google Scholar] [CrossRef] [PubMed]
- Hannafon, B.N.; Trigoso, Y.D.; Calloway, C.L.; Zhao, Y.D.; Lum, D.H.; Welm, A.L.; Zhao, Z.J.; Blick, K.E.; Dooley, W.C.; Ding, W.Q. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016, 18, 90. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.H.; Espinoza-Sánchez, N.A.; El-Damen, A.; Fahim, S.A.; Badawy, M.A.; Greve, B.; El-Shinawi, M.; Götte, M.; Ibrahim, S.A. Small extracellular vesicle-encapsulated miR-181b-5p, miR-222-3p and let-7a-5p: Next generation plasma biopsy-based diagnostic biomarkers for inflammatory breast cancer. PLoS ONE 2021, 16, e0250642. [Google Scholar] [CrossRef] [PubMed]
- Sueta, A.; Yamamoto, Y.; Tomiguchi, M.; Takeshita, T.; Yamamoto-Ibusuki, M.; Iwase, H. Differential expression of exosomal miRNAs between breast cancer patients with and without recurrence. Oncotarget 2017, 8, 69934–69944. [Google Scholar] [CrossRef]
- Todorova, V.K.; Byrum, S.D.; Gies, A.J.; Haynie, C.; Smith, H.; Reyna, N.S.; Makhoul, I. Circulating Exosomal microRNAs as Predictive Biomarkers of Neoadjuvant Chemotherapy Response in Breast Cancer. Curr. Oncol. 2022, 29, 613–630. [Google Scholar] [CrossRef]
- Sueta, A.; Fujiki, Y.; Goto-Yamaguchi, L.; Tomiguchi, M.; Yamamoto-Ibusuki, M.; Iwase, H.; Yamamoto, Y. Exosomal miRNA profiles of triple-negative breast cancer in neoadjuvant treatment. Oncol. Lett. 2021, 22, 819. [Google Scholar] [CrossRef]
- Wu, H.; Wang, Q.; Zhong, H.; Li, L.; Zhang, Q.; Huang, Q.; Yu, Z. Differentially expressed microRNAs in exosomes of patients with breast cancer revealed by next-generation sequencing. Oncol. Rep. 2020, 43, 240–250. [Google Scholar] [CrossRef]
- Ni, Q.; Stevic, I.; Pan, C.; Müller, V.; Oliviera-Ferrer, L.; Pantel, K.; Schwarzenbach, H. Different signatures of miR-16, miR-30b and miR-93 in exosomes from breast cancer and DCIS patients. Sci. Rep. 2018, 8, 12974. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, A.; De Miguel-Pérez, D.; Ortega, F.G.; García-Puche, J.L.; Robles-Fernández, I.; Exposito, J.; Martorell-Marugan, J.; Carmona-Sáez, P.; Garrido-Navas, M.D.C.; Rolfo, C.; et al. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Res. 2019, 21, 21. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, W.; Li, B.; Stringer-Reasor, E.; Chu, C.; Sun, L.; Bae, S.; Chen, D.; Wei, S.; Jiao, K.; et al. MicroRNA-200c and microRNA-141 are regulated by a FOXP3-KAT2B axis and associated with tumor metastasis in breast cancer. Breast Cancer Res. 2017, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, P.M.M.; Vieira, E.; Lemos, D.S.; Souza, I.L.M.; Zanata, S.M.; Pankievicz, V.C.; Tuleski, T.R.; Souza, E.M.; Wowk, P.F.; Urban, C.D.A.; et al. Identification of miRNAs Enriched in Extracellular Vesicles Derived from Serum Samples of Breast Cancer Patients. Biomolecules 2020, 10, 150. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.X.; Cai, Y.Q.; Lv, M.M.; Chen, L.; Zhong, S.L.; Ma, T.F.; Zhao, J.H.; Tang, J.H. Exosomes from docetaxel-resistant breast cancer cells alter chemosensitivity by delivering microRNAs. Tumour Biol. 2014, 35, 9649–9659. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Lai, X.; Yu, S.; Chen, S.; Ma, Y.; Zhang, Y.; Li, H.; Zhu, X.; Yao, L.; Zhang, J. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res. Treat. 2014, 147, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Chen, X.; Wang, D.; Zhang, X.; Shen, H.; Yang, S.; Lv, M.; Tang, J.; Zhao, J. MicroRNA expression profiles of drug-resistance breast cancer cells and their exosomes. Oncotarget 2016, 7, 19601–19609. [Google Scholar] [CrossRef] [PubMed]
- Stevic, I.; Müller, V.; Weber, K.; Fasching, P.A.; Karn, T.; Marmé, F.; Schem, C.; Stickeler, E.; Denkert, C.; Van Mackelenbergh, M.; et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med. 2018, 16, 179. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, M.; Iinuma, H.; Umemoto, Y.; Yanagisawa, T.; Matsumoto, A.; Jinno, H. Exosome-encapsulated microRNA-223-3p as a minimally invasive biomarker for the early detection of invasive breast cancer. Oncol. Lett. 2018, 15, 9584–9592. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, L.; Yu, G.; Sun, Z.; Wang, T.; Tian, X.; Duan, X.; Zhang, C. Exosomal miR-1246 and miR-155 as predictive and prognostic biomarkers for trastuzumab-based therapy resistance in HER2-positive breast cancer. Cancer Chemother. Pharmacol. 2020, 86, 761–772. [Google Scholar] [CrossRef]
- Salvador-Coloma, C.; Santaballa, A.; Sanmartín, E.; Calvo, D.; García, A.; Hervás, D.; Cordón, L.; Quintas, G.; Ripoll, F.; Panadero, J.; et al. Immunosuppressive profiles in liquid biopsy at diagnosis predict response to neoadjuvant chemotherapy in triple-negative breast cancer. Eur. J. Cancer 2020, 139, 119–134. [Google Scholar] [CrossRef]
- Wang, K.; Yuan, Y.; Cho, J.H.; McClarty, S.; Baxter, D.; Galas, D.J. Comparing the MicroRNA spectrum between serum and plasma. PLoS ONE 2012, 7, e41561. [Google Scholar] [CrossRef]
- Hamam, R.; Ali, A.M.; Alsaleh, K.A.; Kassem, M.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. MicroRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci. Rep. 2016, 6, 25997. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, A.M.; Incoronato, M. Clinical Translatability of “Identified” Circulating miRNAs for Diagnosing Breast Cancer: Overview and Update. Cancers 2019, 11, 901. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, A.M.; Nuzzo, S.; Condorelli, G.; Salvatore, M.; Incoronato, M. Prognostic and Clinicopathological Significance of MiR-155 in Breast Cancer: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 5834. [Google Scholar] [CrossRef] [PubMed]
| miRNA (Expression) | Cohort | Sample | Clinical Significance | Ref |
|---|---|---|---|---|
| miR296-3p, miR-575, miR-3160-5p, miR-4483, miR-4710, miR-4755-3p, miR-5698, miR-8089 (new MBC) miR-8089 and miR-5698 (↓ OS) | MBC (n = 147) | Serum | Predictive, Prognostic (new distant metastasis, OS) | [104] |
| miR-331 (↑), mir-195 (↓ LA-MBC) miR-331 (↑), mir-181a (↓ BC) miR-181a (↑ HCs) | LA (n = 31), LA-MBC (n = 22), HCs (n = 21) | Whole blood | Diagnostic (MBC vs. luminal A) | [105] |
| miR-21, miR-181a, miR-10b (↑) miR-145 and let-7a (↓) | BC (n = 30), HCs (n = 20) | Plasma | Diagnostic, Predictive: (initial LABC, PFS) | [73] |
| miR-21, miR-23b, miR-200b, miR-200c (↑ MBC) miR-21, miR-190, miR-200b, miR-200c (↑ MBC) | Early BC (n = 133), MBC (n = 110) | Plasma | Diagnostic, Prognostic (MBC vs. Early BC; miR-200b ↑ → ↓ OS; miR23b and miR-190 ↑ → ↓PFS) | [106] |
| miR-1246, miR-206, miR-24, miR-373 (↑ BC) | BC (n = 226), HCs (n = 146) | Plasma (Serum) | Diagnostic (early-stage BC detection) | [107] |
| miR-106a-3p, miR-106a-5p *, miR-20b-5p *, miR-92a-2-5p miR-106a-5p *, miR-20b-5p *, miR-19b-3p, miR-92a-3p miR-106a-5p *, miR-20b-5p * (↑) | BC (n = 200), HCs (n = 200) ** BC (n = 204), HCs (n = 202) ** Plasma: BC (n = 32), HCs (n = 32) ***; serum: BC (n = 32), HCs (n = 32), ***; tissues: BC (n = 32), HCs (n = 32) *** | Plasma Serum Plasma, Serum, Tissues | Diagnostic, prognostic (BC vs. HCs; association with clinical/pathological featuresDiagnostic (BC vs. HCs) | [108] |
| miR-210 (↑ BC) miR-152 (↑ BC) | BC (n = 30), BBC (n = 5), HCs (n = 5) | Plasma | Diagnostic, prognostic: early diagnosis, staging | [109] |
| miR-24-3p (↑ MBC) | BC (n = 115); MBC (n = 115) | Plasma | Prognostic, predictive (metastasis, OS) | [110] |
| miR-19a/b-3p; miR-25–3p; miR-22-3p; miR-93-5p; miR-210-3p (↑), miR-199a-3p (↓) | TNBC (n = 36), LA (n = 16), LB (n = 41), HCs (n = 34) | Plasma | Predictive, prognostic (drug-resistance in TNBC; OS) | [111] |
| Lapatinib: miR-376c-3p, miR-874-3p; miR-197-3p, miR-320c, miR-100-5p (pre); miR-144-3p, miR-362-3p, miR-100-5p (post) Trastuzumab: miR-374a-5p, miR-574-3p, miR-140-5p, miR-328-3p, miR-145-5p (post) Lapatinib+Trastuzumab: miR-34a-5p, miR-98-5p, miR-100-5p (post) | HER2+ BC (n = 429) | Plasma (pre/2 wks post NAC) | Predictive, prognostic (pCR-NAC response) | [97] |
| miRNA (Expression) | Cohort | Sample | Clinical Significance | Ref |
|---|---|---|---|---|
| miR-21-3p, miR 105-5p (↑ MBC pre-NAC); miR-222-3p (↑ basal-like, LB pre-NAC); | 47 LBC (n = 47), MBC (n = 6), HCs (n = 8) | Serum | Diagnostic, predictive, prognostic (trastuzumab resistance) | [119] |
| miR-16 (↑BC, DCIS) miR-93 ((↑DCIS) miR-30b (↓ recurrence) | BC (n = 111), DCIS (n = 42), HCs (n = 39) | Plasma | Diagnostic, predictive (associated with clinical/pathological features, recurrence) | [118] |
| (a#) miR-142-5p, miR-320a, miR-4433b-5p (↑BC vs. HCs, LA vs. HCs); (b#) miR-142-5p and miR-320a (↑BC vs. HCs, LA vs. HCs); (c#) miR142-5p, miR-150-5p (↑LA vs. TNBC) | LA (n = 26), TNBC (n = 23), HCs (n = 26) | Serum | Diagnostic, prognostic: (BC vs. HCs, LA vs. TNBC; associated with clinical/pathological features) | [121] |
| miR-181b-5p, miR-222-3p (↑ IBC) let-7a-5p (↓ IBC) | IBC (n = 23), non-IBC (n = 34), HCs (n = 20) | Plasma | Diagnostic | [113] |
| miR-16, miR-27a, miR-27b, miR-143, miR-365 (↓ TNBC vs. Her2+) let-7g, miR-148a, miR-202, miR-335, miR-370, miR-376c, miR-382, miR-422a, miR-433, miR-489, miR-628, miR-652, miR-891a (↑ TNBC vs. Her2+) miR155, miR301 (best predictor pCR) | Her2+ BC (n = 211), TNBC (n = 224), HCs (n = 20) | Plasma | Diagnostic, predictive (associated with clinical/pathological features, pCR) | [125] |
| (a) miR-4448, miR-2392, miR-2467-3p and miR-4800-3p (↑ pCR+) (b) recurrence: 15 miRs ↑ (miR-195-5p: 4.43 fc, p = 0.02); 28 miRs ↓ (miR-548ab: 0.23 fc, p < 0.001) | TNBC: (a) pCR (n = 12), w/o pCR (n = 12) (b) w/o pCR: recurrence (n = 8), w/o recurrence (n = 8) | Serum pre a/post b NAC | Predictive, prognostic (pCR+/pCR−; higher expression → longer OS) | [116] |
| Recurrence: miR-338-3p, miR-340-5p, miR-124-3p (↑ serum); miR-340, 195-5p, miR-17-5p, miR-93-5p, miR-130a-3p (↑ tumor) miR-340, 195-5p, miR-17-5p, miR-93-5p, miR-130a-3p (↑ tumor) | BC: recurrence (n = 16), w/o recurrence (n = 16) Tissues: recurrence (n = 35), w/o recurrence (n = 39) | Serum, tumor tissue | Predictive, prognostic (associated with recurrence) | [114] |
| miR-223-3p (↑IDC) | BC (n = 185), HCs (n = 146) | Plasma | Diagnostic (IDC vs. DCIS; associated with clinical/pathological features) | [126] |
| miR-1246 and miR-155 (↑Trastuzumab-resistant) | Early BC (n = 107), MBC (n = 68) | Plasma | Predictive, prognostic (trastuzumab: resistant vs. sensitive pt; EFS, PFS) | [127] |
| (a) miR-30b (↑), miR-328, miR-423 (↓pCR+) (b) exo-miR-141 (↓pCR+); miR-34a, miR183 (↑), miR-182 (↓pCR−) | IDC (n = 20) | Plasma (pre/2 wks post NAC) | Predictive (pCR to NAC) | [115] |
| miR-185, miR-4283, miR-5008 and miR-3613 (↓) miR-1302, miR-4715 and miR-3144 (↑) | TNBC (n = 34) | Plasma | Predictive (poor NAC responders) | [128] |
| NCT Number | Title | Study Type and Design | Brief Description | Enrolled Patients | Start–End Status | Country |
|---|---|---|---|---|---|---|
| NCT01612871 | Circulating miRNAs as Biomarker of Hormone Sensitivity in Breast Cancer (MIRHO) | Interventional, Phase 4; Single group assignment; Prospective | Test in the advanced setting of the role of circulating MiRNA in comparison with tissue | 39 | June 2012 Jan 2016 Completed | France |
| NCT05151224 | Circulating microRNA 21 Expression Level Before and After Neoadjuvant Systemic Therapy in Breast Carcinoma | Observational cohort; Prospective Diagnostic Test: microRNA21 | miR-21 expression in early setting (before and after NAC) | 40 | Dec 2021 Jan 2024 Not yet recruiting | Egypt |
| NCT02065908 | Circulating miRNAs as Biomarkers of Cardiotoxicity in Breast Cancer | Observational Cohort; Prospective | Circulating microRNA in serum as sensitive and specific biomarker of cardiotoxicity in cancer patients treated with anthracycline-based chemotherapy. | 128 | Jan 2014 Dec 2016 Completed | Poland |
| NCT01722851 | Circulating miRNAs | Observational: Cohort Prospective | Circulating miRNA markers as predictive of neoadjuvant and adjuvant chemotherapy | 255 | May 2011 Dec 2021 Completed | Ireland |
| NCT04720508 | Aberrant Expression of Micro RNA for Diagnosis of Breast Cancer | Observational: Case-control; Retrospective Diagnostic test: blood sample collection | Assess serum miRNA-373 and miRNA 425-5p, correlate with clinicopathological features | 50 | Dec 2021 Dec 2023 Not yet recruiting | Egypt |
| NCT03779022 | miRNA and Relevant Biomarkers of BC Patients Undergoing Neoadjuvant Treatment | Observational cohort; Prospective Genetic: microRNA | Investigate miRNAs as predictive biomarkers of neoadjuvant therapy | 100 | Nov 2015 Dec 2019 Unknown | China |
| NCT03528473 | Adapted Physical Activity (APA) in a Breast Cancer Population | Interventional; Randomized | Investigate miRNA levels in serum of post-menopausal, inactive patients in follow-up, w/wo HT hormone therapy after completed AR/AC therapy | 100 | Jan 2019 Dec 2022 Active, not recruiting | Italy |
| NCT04771871 | MicroRNA Profiles in Triple Negative Breast Cancer (TARMAC) | Interventional, Phase 2; Single group | Assess blood miRNA and ctDNA levels during and after standard chemotherapy in black TNBC patients | 42 | Nov 2021 Aug 2023 Recruiting | Nigeria |
| NCT02618538 | The Andromeda Study Predictive Value of Combined Criteria to Tailor BC Screening | Observational: Cross-Sectional | Investigate variance of plasma miRNAs associated with BC risk in HCs and BC patients | 26,600 | Jul 2015 Mar 2018 Completed | Italy |
| NCT03118882 | STI.VI Study: How to improve Lifestyle in Screening Context (STIVI) | Interventional: Randomized; Prevention Biological sampling (blood and saliva) | Investigate deregulation of plasma miRNAs associated with breast/colorectal risk in HCs and cancer patients: screening as potential biomarkers (follow up to 10 years after study end) | 1270 | May 2020 June 2014 Completed | Italy |
| NCT03255486 | Identification and Evaluation of Biomarkers of Resistance to Neoadjuvant Chemotherapy (IDEASEIN) | Interventional: Single group Assignment; Prevention; Blood Sample | Biomarkers of resistance to neoadjuvant chemotherapy in locally advanced breast cancer | 164 | Jul 2013 Dec 2016 Completed | France |
| NCT02605512 | BreAst Cancer and Cardiotoxicity Induced by RAdioTherapy: the BACCARAT Study (BACCARAT) | Interventional: Single Group Assignment; Screening | Evaluate circulating biomarkers as predictive of whether 3DCRT induces cardiac toxicity | 120 | Oct 2015 Sept 2020 Unknown | France |
| NCT04781062 | Development of a Horizontal Data Integration Classifier for Noninvasive Early Diagnosis of Breast Cancer (RENOVATE) | Interventional: non Randomized; Sequential Assignment; Blood and urine (T0-T1) | Develop a non-invasive molecular profile for early diagnosis, correlating blood (ctDNA, proteins, exosomes) and urine (ctDNA) biomarkers | 750 | Jan 2021 Dec 2024 Recruiting | Italy |
| NCT04530890 | Interest of Circulating Tumor DNA in Digestive and Gynecologic/Breast Cancer | Interventional: Single Group Assignment; Basic Science; Blood Sample | Assess diagnostic, prognostic and predictive impact of ctDNA and exosomes in digestive and gynecological/breast cancers (pre and during treatment, progression and/or relapse, monitoring or treatment break) | 1000 | Jan 2021 Sept 2030 Recruiting | France |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardinali, B.; Tasso, R.; Piccioli, P.; Ciferri, M.C.; Quarto, R.; Del Mastro, L. Circulating miRNAs in Breast Cancer Diagnosis and Prognosis. Cancers 2022, 14, 2317. https://doi.org/10.3390/cancers14092317
Cardinali B, Tasso R, Piccioli P, Ciferri MC, Quarto R, Del Mastro L. Circulating miRNAs in Breast Cancer Diagnosis and Prognosis. Cancers. 2022; 14(9):2317. https://doi.org/10.3390/cancers14092317
Chicago/Turabian StyleCardinali, Barbara, Roberta Tasso, Patrizia Piccioli, Maria Chiara Ciferri, Rodolfo Quarto, and Lucia Del Mastro. 2022. "Circulating miRNAs in Breast Cancer Diagnosis and Prognosis" Cancers 14, no. 9: 2317. https://doi.org/10.3390/cancers14092317
APA StyleCardinali, B., Tasso, R., Piccioli, P., Ciferri, M. C., Quarto, R., & Del Mastro, L. (2022). Circulating miRNAs in Breast Cancer Diagnosis and Prognosis. Cancers, 14(9), 2317. https://doi.org/10.3390/cancers14092317








