Circulating Cell-Free DNA Assessment in Biofluids from Children with Neuroblastoma Demonstrates Feasibility and Potential for Minimally Invasive Molecular Diagnostics
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
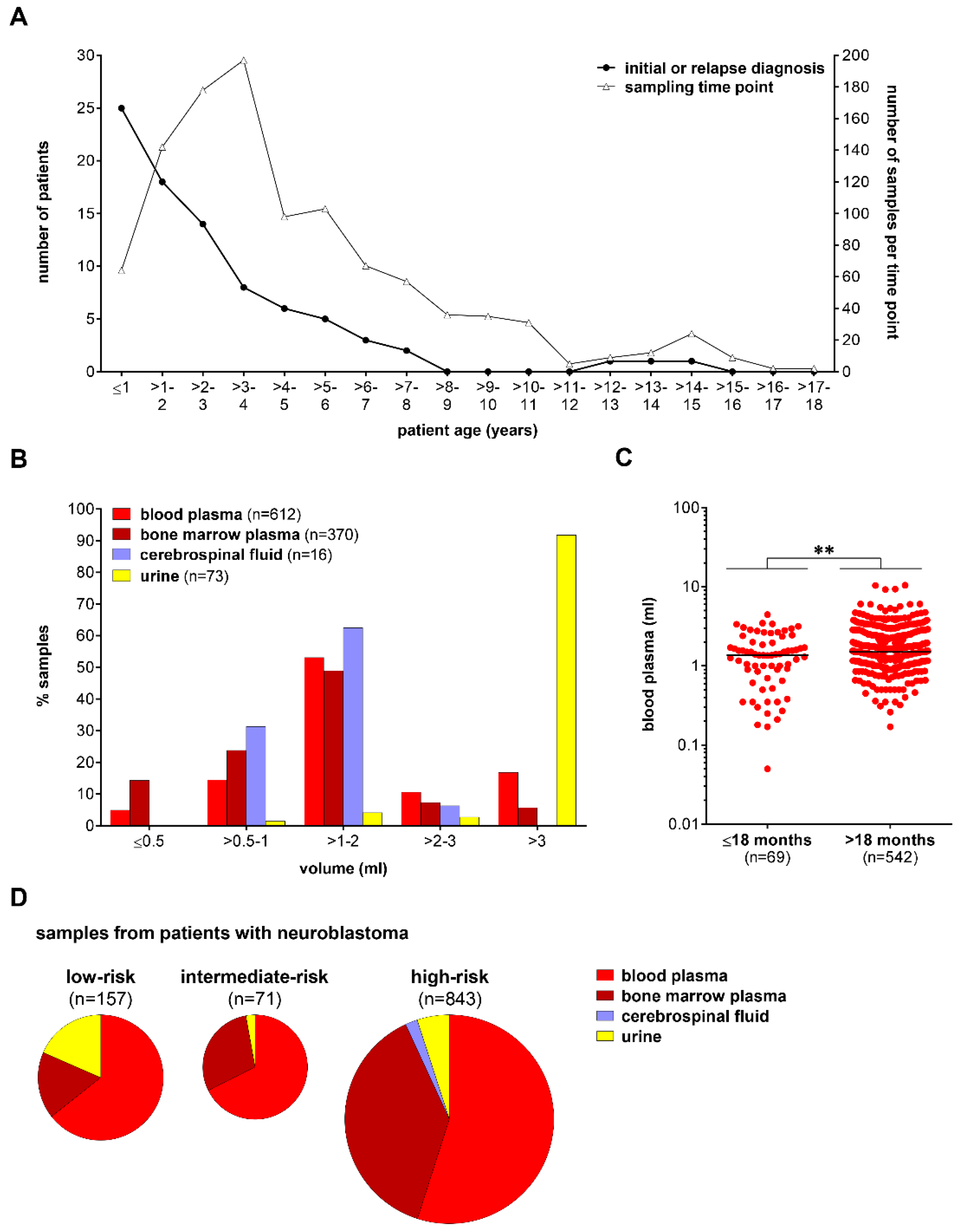
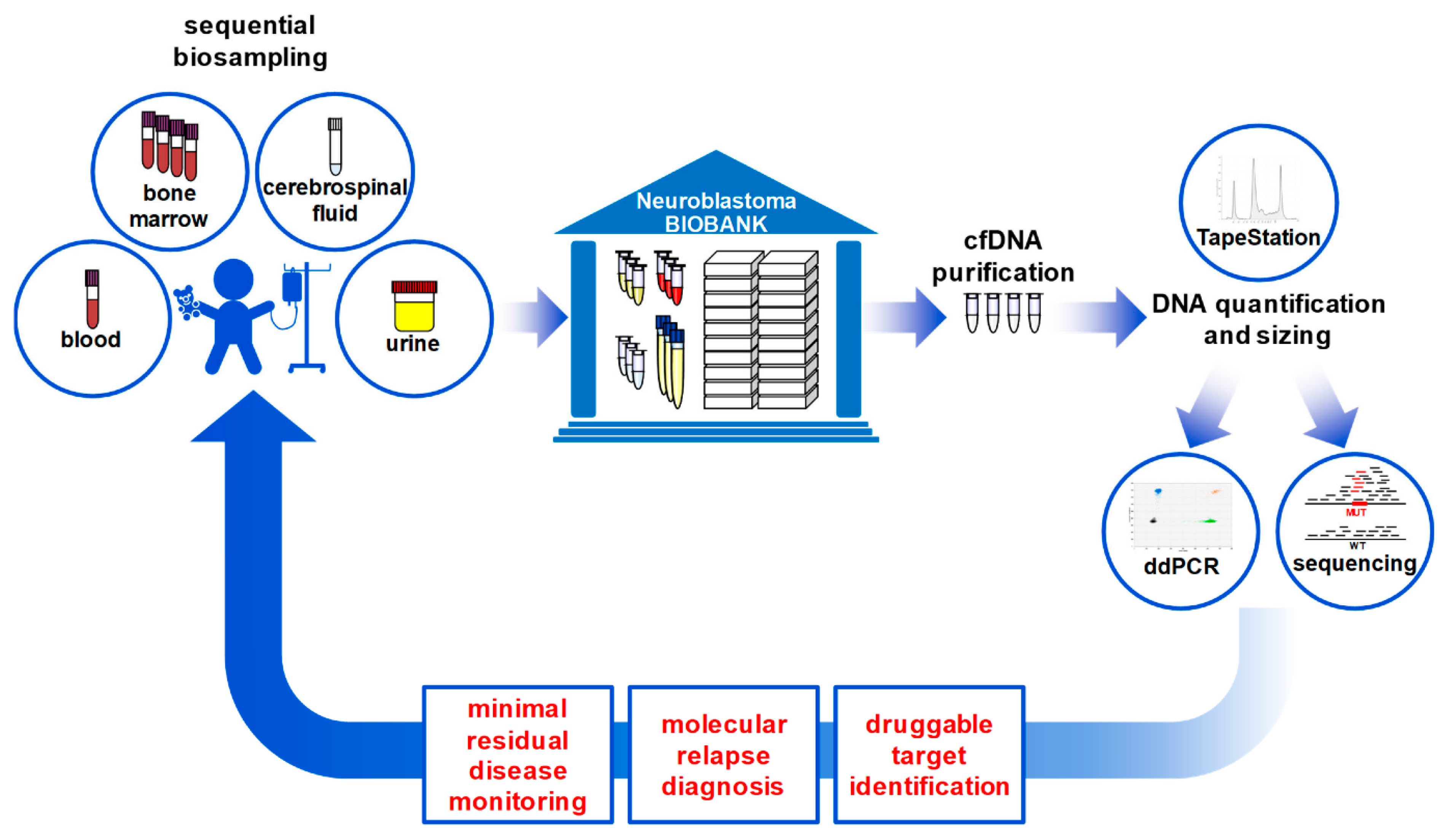
2.1. Patient Samples
2.2. Genomic and Cell-Free DNA Preparation
2.3. Droplet Digital PCR
2.4. Statistical Analysis
3. Results
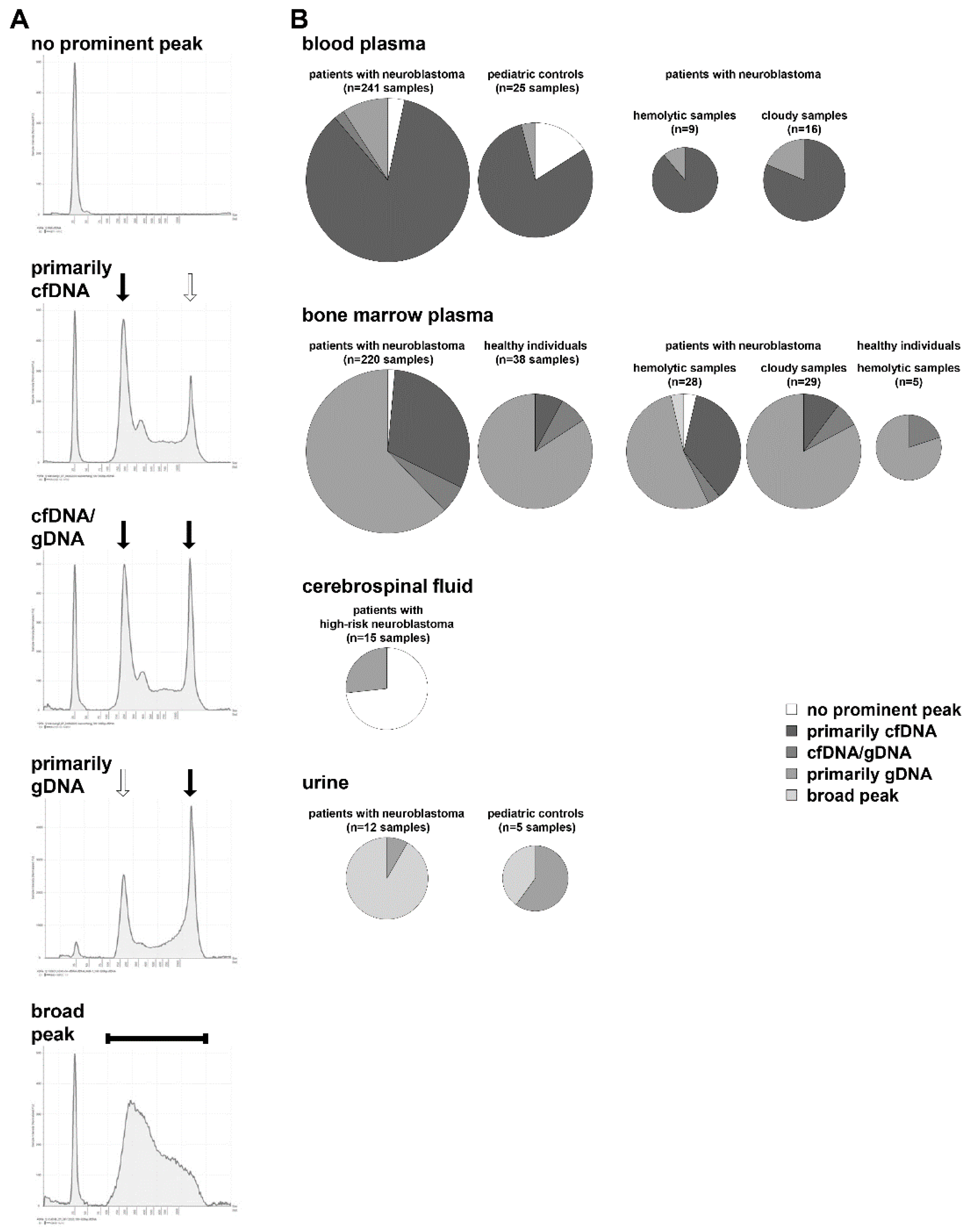
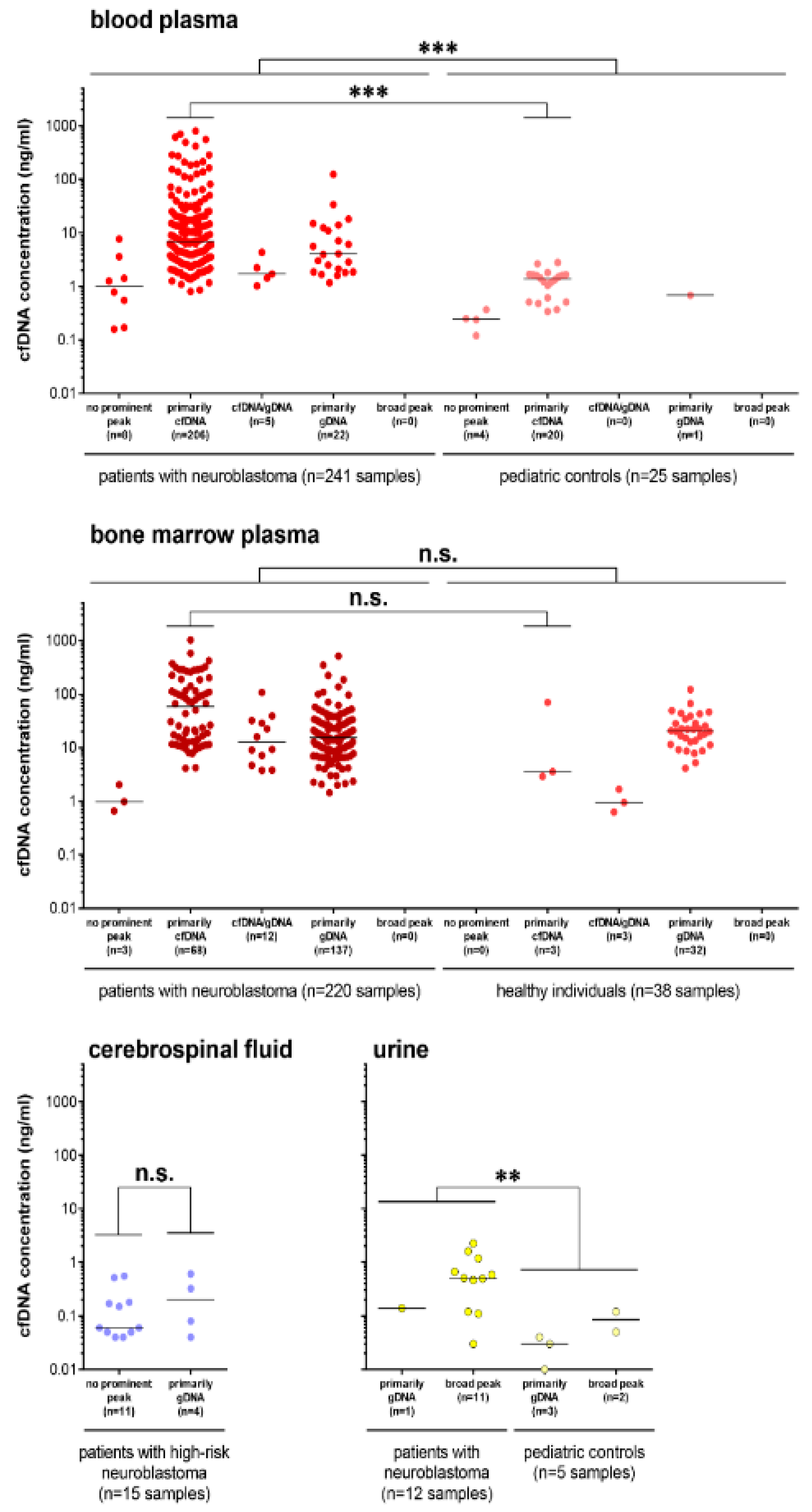
3.1. Cell-Free DNA Content Varies among Different Biofluids from Patients
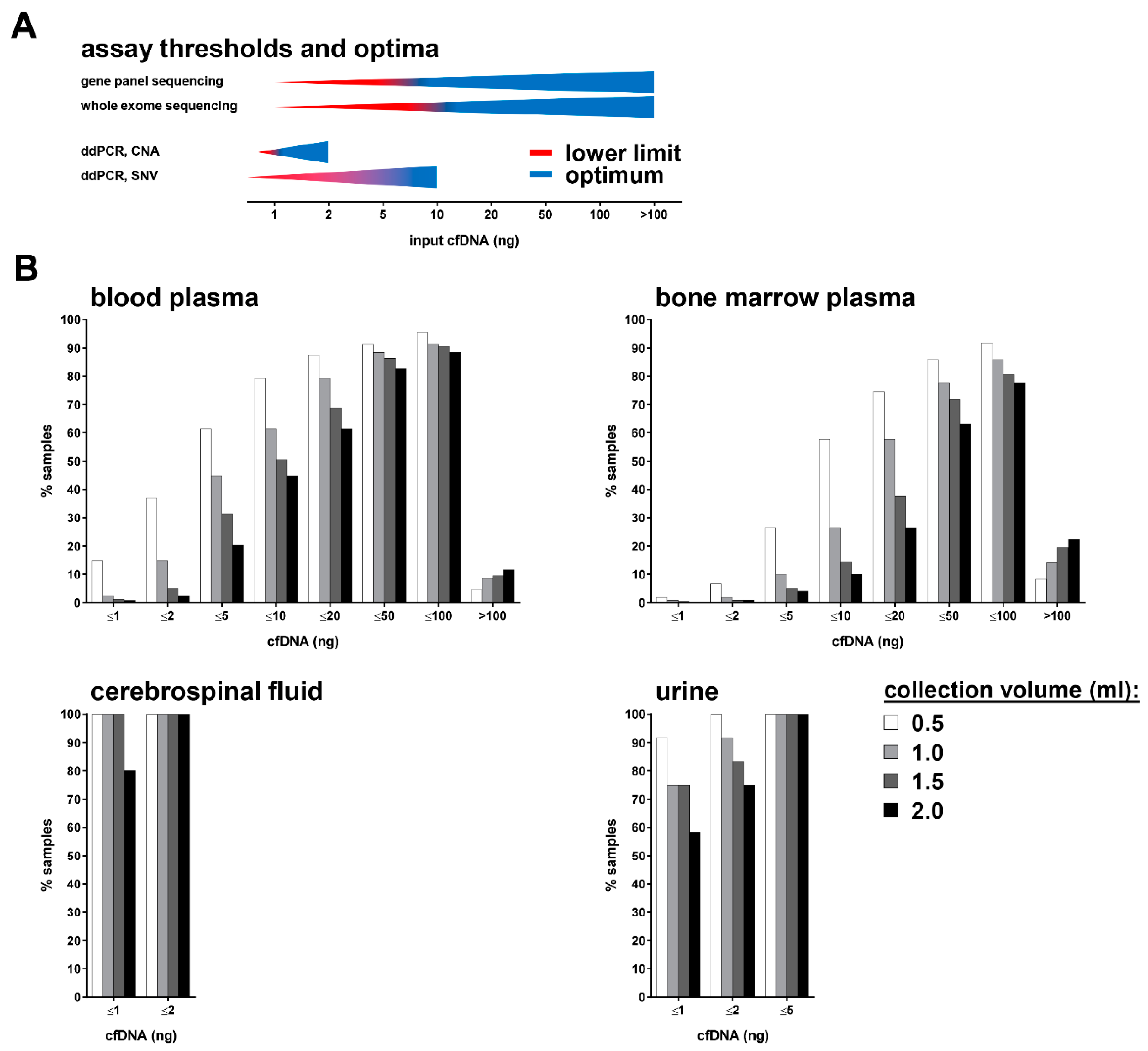
3.2. Multiple cfDNA Assays Can Be Conducted from 1 mL Collected Blood or Bone Marrow Plasma
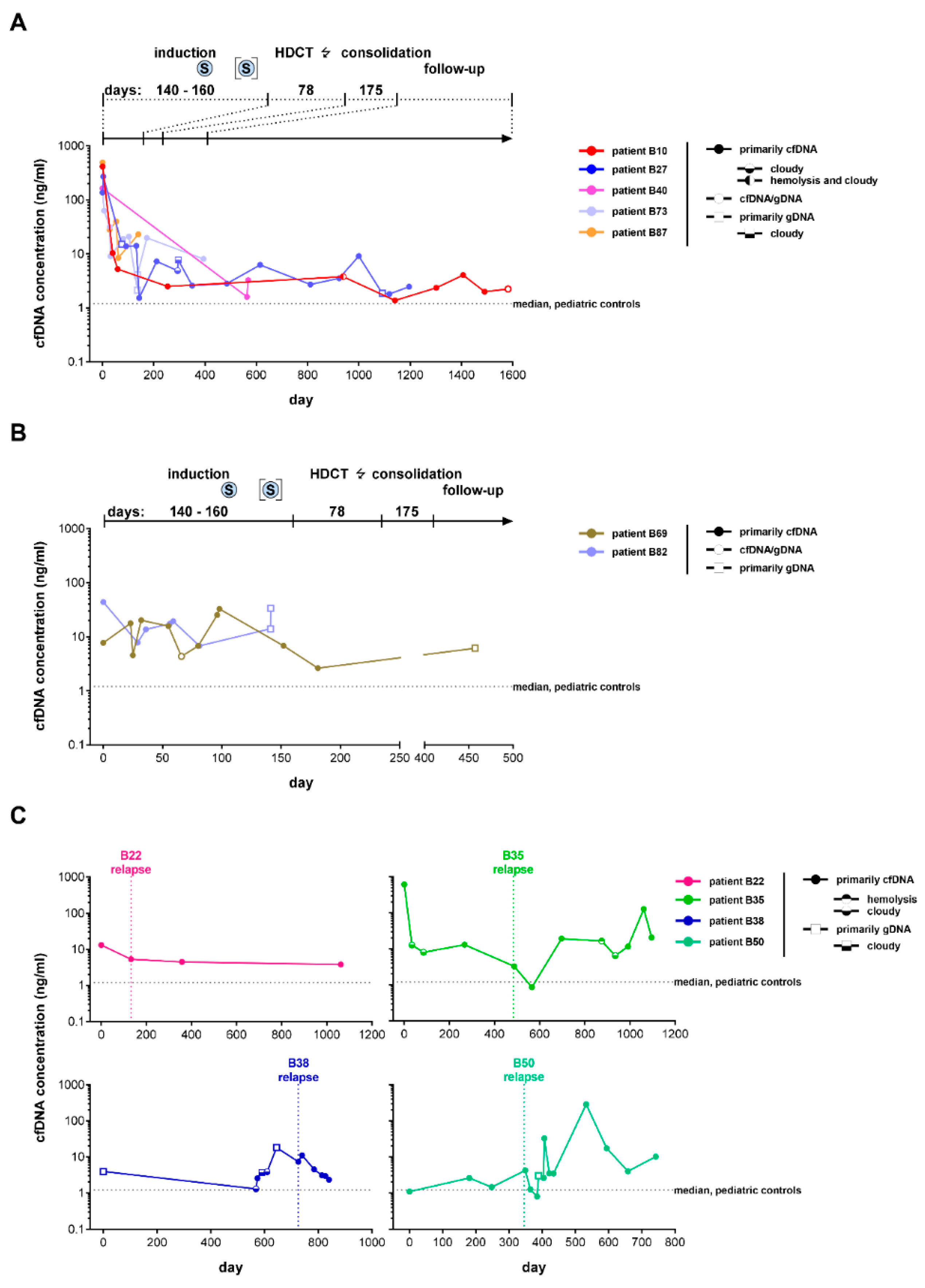
3.3. Favorable Treatment Response in Patients with High-Risk Neuroblastoma Is Associated with a Rapid Decrease in Peripheral Blood-Derived cfDNA
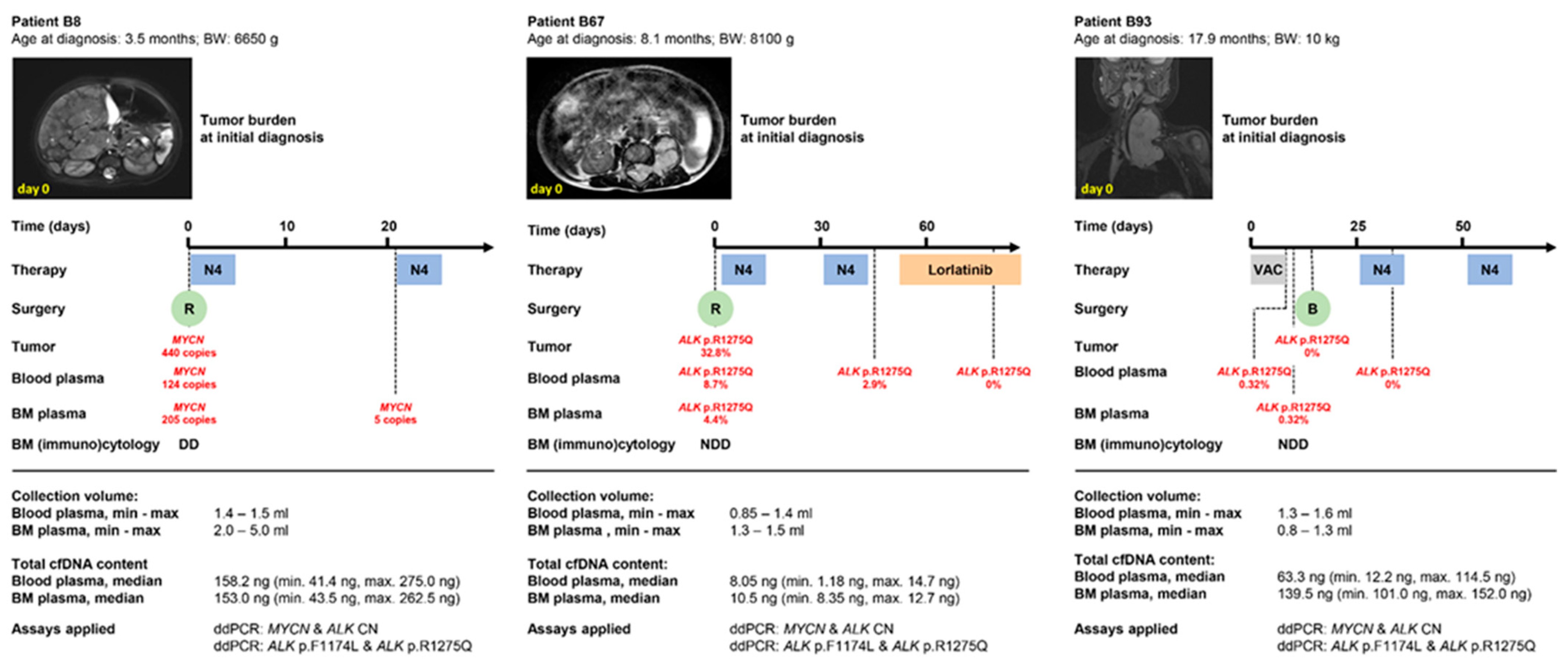
3.4. Peripheral Blood-Derived cfDNA Is Suitable for Molecular Neuroblastoma Profiling in Infants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, J.R.; Eggert, A.; Caron, H. Neuroblastoma: Biology, prognosis, and treatment. Hematol. Oncol. Clin. N. Am. 2010, 24, 65–86. [Google Scholar] [CrossRef] [PubMed]
- Matthay, K.K.; Maris, J.M.; Schleiermacher, G.; Nakagawara, A.; Mackall, C.L.; Diller, L.; Weiss, W.A. Neuroblastoma. Nat. Rev. Dis. Primers 2016, 2, 16078. [Google Scholar] [CrossRef] [PubMed]
- Maris, J.M.; Hogarty, M.D.; Bagatell, R.; Cohn, S.L. Neuroblastoma. Lancet 2007, 369, 2106–2120. [Google Scholar] [CrossRef]
- Berthold, F.; Spix, C.; Kaatsch, P.; Lampert, F. Incidence, Survival, and Treatment of Localized and Metastatic Neuroblastoma in Germany 1979–2015. Pediatric Drugs 2017, 19, 577–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, D.; Fagman, H.; Dalin, M.G.; Stahlberg, A. Circulating cell-free tumor DNA analysis in pediatric cancers. Mol. Asp. Med. 2020, 72, 100819. [Google Scholar] [CrossRef]
- Van Paemel, R.; Vlug, R.; de Preter, K.; van Roy, N.; Speleman, F.; Willems, L.; Lammens, T.; Laureys, G.; Schleiermacher, G.; Tytgat, G.A.M.; et al. The pitfalls and promise of liquid biopsies for diagnosing and treating solid tumors in children: A review. Eur. J. Pediatric 2020, 179, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Combaret, V.; Audoynaud, C.; Iacono, I.; Favrot, M.C.; Schell, M.; Bergeron, C.; Puisieux, A. Circulating MYCN DNA as a tumor-specific marker in neuroblastoma patients. Cancer Res. 2002, 62, 3646–3648. [Google Scholar]
- Gotoh, T.; Hosoi, H.; Iehara, T.; Kuwahara, Y.; Osone, S.; Tsuchiya, K.; Ohira, M.; Nakagawara, A.; Kuroda, H.; Sugimoto, T. Prediction of MYCN amplification in neuroblastoma using serum DNA and real-time quantitative polymerase chain reaction. J. Clin. Oncol. 2005, 23, 5205–5210. [Google Scholar] [CrossRef]
- Combaret, V.; Bergeron, C.; Noguera, R.; Iacono, I.; Puisieux, A. Circulating MYCN DNA predicts MYCN-amplification in neuroblastoma. J. Clin. Oncol. 2005, 23, 8919–8920. [Google Scholar] [CrossRef]
- Combaret, V.; Hogarty, M.D.; London, W.B.; McGrady, P.; Iacono, I.; Brejon, S.; Swerts, K.; Noguera, R.; Gross, N.; Rousseau, R.; et al. Influence of neuroblastoma stage on serum-based detection of MYCN amplification. Pediatric Blood Cancer 2009, 53, 329–331. [Google Scholar] [CrossRef] [Green Version]
- Kojima, M.; Hiyama, E.; Fukuba, I.; Yamaoka, E.; Ueda, Y.; Onitake, Y.; Kurihara, S.; Sueda, T. Detection of MYCN amplification using blood plasma: Noninvasive therapy evaluation and prediction of prognosis in neuroblastoma. Pediatric Surg. Int. 2013, 29, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Yagyu, S.; Iehara, T.; Tanaka, S.; Gotoh, T.; Misawa-Furihata, A.; Sugimoto, T.; London, W.B.; Hogarty, M.D.; Teramukai, S.; Nakagawara, A.; et al. Serum-Based Quantification of MYCN Gene Amplification in Young Patients with Neuroblastoma: Potential Utility as a Surrogate Biomarker for Neuroblastoma. PLoS ONE 2016, 11, e0161039. [Google Scholar] [CrossRef] [PubMed]
- Chicard, M.; Boyault, S.; Daage, L.C.; Richer, W.; Gentien, D.; Pierron, G.; Lapouble, E.; Bellini, A.; Clement, N.; Iacono, I.; et al. Genomic Copy Number Profiling Using Circulating Free Tumor DNA Highlights Heterogeneity in Neuroblastoma. Clin. Cancer Res. 2016, 22, 5564–5573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodrini, M.; Sprussel, A.; Astrahantseff, K.; Tiburtius, D.; Konschak, R.; Lode, H.N.; Fischer, M.; Keilholz, U.; Eggert, A.; Deubzer, H.E. Using droplet digital PCR to analyze MYCN and ALK copy number in plasma from patients with neuroblastoma. Oncotarget 2017, 8, 85234–85251. [Google Scholar] [CrossRef] [Green Version]
- Van Roy, N.; van der Linden, M.; Menten, B.; Dheedene, A.; Vandeputte, C.; van Dorpe, J.; Laureys, G.; Renard, M.; Sante, T.; Lammens, T.; et al. Shallow Whole Genome Sequencing on Circulating Cell-Free DNA Allows Reliable Noninvasive Copy-Number Profiling in Neuroblastoma Patients. Clin. Cancer Res. 2017, 23, 6305–6314. [Google Scholar] [CrossRef] [Green Version]
- Chicard, M.; Colmet-Daage, L.; Clement, N.; Danzon, A.; Bohec, M.; Bernard, V.; Baulande, S.; Bellini, A.; Deveau, P.; Pierron, G.; et al. Whole-Exome Sequencing of Cell-Free DNA Reveals Temporo-spatial Heterogeneity and Identifies Treatment-Resistant Clones in Neuroblastoma. Clin. Cancer Res. 2018, 24, 939–949. [Google Scholar] [CrossRef] [Green Version]
- Peitz, C.; Sprussel, A.; Linke, R.B.; Astrahantseff, K.; Grimaldi, M.; Schmelz, K.; Toedling, J.; Schulte, J.H.; Fischer, M.; Messerschmidt, C.; et al. Multiplexed Quantification of Four Neuroblastoma DNA Targets in a Single Droplet Digital PCR Reaction. J. Mol. Diagn. 2020, 22, 1309–1323. [Google Scholar] [CrossRef]
- van Paemel, R.; Vandeputte, C.; Raman, L.; van Thorre, J.; Willems, L.; van Dorpe, J.; van der Linden, M.; de Wilde, J.; de Koker, A.; Menten, B.; et al. The feasibility of using liquid biopsies as a complementary assay for copy number aberration profiling in routinely collected paediatric cancer patient samples. Eur. J. Cancer 2022, 160, 12–23. [Google Scholar] [CrossRef]
- Kobayashi, K.; Mizuta, S.; Yamane, N.; Hamabata, T.; Maihara, T.; Usami, I.; Heike, T. Cell-free DNA Oncogene Copy Number as a Surrogate Molecular Biomarker in ALK/MYCN-coamplified Neuroblastoma. J. Pediatric Hematol. Oncol. 2021, 43, e165–e168. [Google Scholar] [CrossRef]
- Kahana-Edwin, S.; Cain, L.E.; McCowage, G.; Darmanian, A.; Wright, D.; Mullins, A.; Saletta, F.; Karpelowsky, J. Neuroblastoma Molecular Risk-Stratification of DNA Copy Number and ALK Genotyping via Cell-Free Circulating Tumor DNA Profiling. Cancers 2021, 13, 3365. [Google Scholar] [CrossRef]
- Lodrini, M.; Graef, J.; Thole-Kliesch, T.M.; Astrahantseff, K.; Sprussel, A.; Grimaldi, M.; Peitz, C.; Linke, R.B.; Hollander, J.F.; Lankes, E.; et al. Targeted analysis of cell-free circulating tumor DNA is suitable for early relapse and actionable target detection in patients with neuroblastoma. Clin. Cancer Res. 2022; online ahead of print. [Google Scholar] [CrossRef]
- Combaret, V.; Iacono, I.; Bellini, A.; Brejon, S.; Bernard, V.; Marabelle, A.; Coze, C.; Pierron, G.; Lapouble, E.; Schleiermacher, G.; et al. Detection of tumor ALK status in neuroblastoma patients using peripheral blood. Cancer Med. 2015, 4, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Wang, H.; Chen, Y.; Chu, P.; Xing, T.; Gao, C.; Yue, Z.; Zheng, J.; Jin, M.; Gu, W.; et al. Whole exome sequencing reveals novel somatic alterations in neuroblastoma patients with chemotherapy. Cancer Cell Int. 2018, 18, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sausen, M.; Leary, R.J.; Jones, S.; Wu, J.; Reynolds, C.P.; Liu, X.; Blackford, A.; Parmigiani, G.; Diaz, L.A., Jr.; Papadopoulos, N.; et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat. Genet. 2013, 45, 12–17. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra224. [Google Scholar] [CrossRef] [Green Version]
- Klega, K.; Imamovic-Tuco, A.; Ha, G.; Clapp, A.N.; Meyer, S.; Ward, A.; Clinton, C.; Nag, A.; van Allen, E.; Mullen, E.; et al. Detection of Somatic Structural Variants Enables Quantification and Characterization of Circulating Tumor DNA in Children With Solid Tumors. JCO Precis. Oncol. 2018, 2, 1–13. [Google Scholar] [CrossRef]
- Kurihara, S.; Ueda, Y.; Onitake, Y.; Sueda, T.; Ohta, E.; Morihara, N.; Hirano, S.; Irisuna, F.; Hiyama, E. Circulating free DNA as non-invasive diagnostic biomarker for childhood solid tumors. J. Pediatric Surg. 2015, 50, 2094–2097. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, L.; Su, Y.; Yue, Z.; Xing, T.; Zhao, W.; Zhao, Q.; Duan, C.; Huang, C.; Zhang, D.; et al. Plasma cell-free DNA quantification is highly correlated to tumor burden in children with neuroblastoma. Cancer Med. 2018, 7, 3022–3030. [Google Scholar] [CrossRef]
- Su, Y.; Wang, L.; Wang, X.; Yue, Z.; Xing, T.; Zhao, W.; Zhao, Q.; Duan, C.; Huang, C.; Han, Y.; et al. Dynamic alterations of plasma cell free DNA in response to chemotherapy in children with neuroblastoma. Cancer Med. 2019, 8, 1558–1566. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Wang, L.; Jiang, C.; Yue, Z.; Fan, H.; Hong, H.; Duan, C.; Jin, M.; Zhang, D.; Qiu, L.; et al. Increased plasma concentration of cell-free DNA precedes disease recurrence in children with high-risk neuroblastoma. BMC Cancer 2020, 20, 102. [Google Scholar] [CrossRef] [Green Version]
- Yagyu, S.; Gotoh, T.; Iehara, T.; Miyachi, M.; Katsumi, Y.; Tsubai-Shimizu, S.; Kikuchi, K.; Tamura, S.; Tsuchiya, K.; Imamura, T.; et al. Circulating methylated-DCR2 gene in serum as an indicator of prognosis and therapeutic efficacy in patients with MYCN nonamplified neuroblastoma. Clin. Cancer Res. 2008, 14, 7011–7019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Zogchel, L.M.J.; van Wezel, E.M.; van Wijk, J.; Stutterheim, J.; Bruins, W.S.C.; Zappeij-Kannegieter, L.; Slager, T.J.E.; Schumacher-Kuckelkorn, R.; Verly, I.R.N.; van der Schoot, C.E.; et al. Hypermethylated RASSF1A as Circulating Tumor DNA Marker for Disease Monitoring in Neuroblastoma. JCO Precis. Oncol. 2020, 4, 291–306. [Google Scholar] [CrossRef] [PubMed]
- Van Zogchel, L.M.J.; Lak, N.S.M.; Verhagen, O.; Tissoudali, A.; Nuru, M.G.; Gelineau, N.U.; Zappeij-Kannengieter, L.; Javadi, A.; Zijtregtop, E.A.M.; Merks, J.H.M.; et al. Novel Circulating Hypermethylated RASSF1A ddPCR for Liquid Biopsies in Patients with Pediatric Solid Tumors. JCO Precis. Oncol. 2021, 5, 1738–1748. [Google Scholar] [CrossRef] [PubMed]
- Applebaum, M.A.; Barr, E.K.; Karpus, J.; West-Szymanski, D.C.; Oliva, M.; Sokol, E.A.; Zhang, S.; Zhang, Z.; Zhang, W.; Chlenski, A.; et al. 5-Hydroxymethylcytosine Profiles in Circulating Cell-Free DNA Associate with Disease Burden in Children with Neuroblastoma. Clin. Cancer Res. 2020, 26, 1309–1317. [Google Scholar] [CrossRef]
- Combaret, V.; Brejon, S.; Iacono, I.; Schleiermacher, G.; Pierron, G.; Ribeiro, A.; Bergeron, C.; Marabelle, A.; Puisieux, A. Determination of 17q gain in patients with neuroblastoma by analysis of circulating DNA. Pediatric Blood Cancer 2011, 56, 757–761. [Google Scholar] [CrossRef]
- Yagyu, S.; Iehara, T.; Gotoh, T.; Miyachi, M.; Katsumi, Y.; Kikuchi, K.; Tsuchiya, K.; Osone, S.; Kuroda, H.; Sugimoto, T.; et al. Preoperative analysis of 11q loss using circulating tumor-released DNA in serum: A novel diagnostic tool for therapy stratification of neuroblastoma. Cancer Lett. 2011, 309, 185–189. [Google Scholar] [CrossRef]
- Gerber, T.; Taschner-Mandl, S.; Saloberger-Sindhoringer, L.; Popitsch, N.; Heitzer, E.; Witt, V.; Geyeregger, R.; Hutter, C.; Schwentner, R.; Ambros, I.M.; et al. Assessment of Pre-Analytical Sample Handling Conditions for Comprehensive Liquid Biopsy Analysis. J. Mol. Diagn. 2020, 22, 1070–1086. [Google Scholar] [CrossRef]
- Simon, T.; Hero, B.; Schulte, J.H.; Deubzer, H.; Hundsdoerfer, P.; von Schweinitz, D.; Fuchs, J.; Schmidt, M.; Prasad, V.; Krug, B.; et al. 2017 GPOH Guidelines for Diagnosis and Treatment of Patients with Neuroblastic Tumors. Klin. Pediatric 2017, 229, 147–167. [Google Scholar] [CrossRef]
- Fan, H.C.; Blumenfeld, Y.J.; Chitkara, U.; Hudgins, L.; Quake, S.R. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. USA 2008, 105, 16266–16271. [Google Scholar] [CrossRef] [Green Version]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29 (Suppl. 1), S49–S52. [Google Scholar]
- Bennett, C.W.; Berchem, G.; Kim, Y.J.; El-Khoury, V. Cell-free DNA and next-generation sequencing in the service of personalized medicine for lung cancer. Oncotarget 2016, 7, 71013–71035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koeppel, F.; Blanchard, S.; Jovelet, C.; Genin, B.; Marcaillou, C.; Martin, E.; Rouleau, E.; Solary, E.; Soria, J.C.; Andre, F.; et al. Whole exome sequencing for determination of tumor mutation load in liquid biopsy from advanced cancer patients. PLoS ONE 2017, 12, e0188174. [Google Scholar]
- Bos, M.K.; Angus, L.; Nasserinejad, K.; Jager, A.; Jansen, M.; Martens, J.W.M.; Sleijfer, S. Whole exome sequencing of cell-free DNA—A systematic review and Bayesian individual patient data meta-analysis. Cancer Treat. Rev. 2020, 83, 101951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Brodeur, G.M. Spontaneous regression of neuroblastoma. Cell Tissue Res. 2018, 372, 277–286. [Google Scholar] [CrossRef]
- Liebs, S.; Eder, T.; Klauschen, F.; Schutte, M.; Yaspo, M.L.; Keilholz, U.; Tinhofer, I.; Kidess-Sigal, E.; Braunholz, D. Applicability of liquid biopsies to represent the mutational profile of tumor tissue from different cancer entities. Oncogene 2021, 40, 5204–5212. [Google Scholar] [CrossRef]
- Schmelz, K.; Toedling, J.; Huska, M.; Cwikla, M.C.; Kruetzfeldt, L.M.; Proba, J.; Ambros, P.F.; Ambros, I.M.; Boral, S.; Lodrini, M.; et al. Spatial and temporal intratumour heterogeneity has potential consequences for single biopsy-based neuroblastoma treatment decisions. Nat. Commun. 2021, 12, 6804. [Google Scholar] [CrossRef]
- Fischer, M.; Moreno, L.; Ziegler, D.S.; Marshall, L.V.; Zwaan, C.M.; Irwin, M.S.; Casanova, M.; Sabado, C.; Wulff, B.; Stegert, M.; et al. Ceritinib in paediatric patients with anaplastic lymphoma kinase-positive malignancies: An open-label, multicentre, phase 1, dose-escalation and dose-expansion study. Lancet Oncol. 2021, 22, 1764–1776. [Google Scholar] [CrossRef]
- Mosse, Y.P. Anaplastic Lymphoma Kinase as a Cancer Target in Pediatric Malignancies. Clin. Cancer Res. 2016, 22, 546–552. [Google Scholar] [CrossRef] [Green Version]
- Triana, S.; Vonficht, D.; Jopp-Saile, L.; Raffel, S.; Lutz, R.; Leonce, D.; Antes, M.; Hernandez-Malmierca, P.; Ordonez-Rueda, D.; Ramasz, B.; et al. Single-cell proteo-genomic reference maps of the hematopoietic system enable the purification and massive profiling of precisely defined cell states. Nat. Immunol. 2021, 22, 1577–1589. [Google Scholar] [CrossRef] [PubMed]
- McEwen, A.E.; Leary, S.E.S.; Lockwood, C.M. Beyond the Blood: CSF-Derived cfDNA for Diagnosis and Characterization of CNS Tumors. Front. Cell Dev. Biol. 2020, 8, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimoto, T.; Inoue, M.; Tokimasa, S.; Yagyu, S.; Iehara, T.; Hosoi, H.; Kawa, K. Detection of MYCN DNA in the cerebrospinal fluid for diagnosing isolated central nervous system relapse in neuroblastoma. Pediatric Blood Cancer 2011, 56, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Streleckiene, G.; Reid, H.M.; Arnold, N.; Bauerschlag, D.; Forster, M. Quantifying cell free DNA in urine: Comparison between commercial kits, impact of gender and inter-individual variation. Biotechniques 2018, 64, 225–230. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lodrini, M.; Wünschel, J.; Thole-Kliesch, T.M.; Grimaldi, M.; Sprüssel, A.; Linke, R.B.; Hollander, J.F.; Tiburtius, D.; Künkele, A.; Schulte, J.H.; et al. Circulating Cell-Free DNA Assessment in Biofluids from Children with Neuroblastoma Demonstrates Feasibility and Potential for Minimally Invasive Molecular Diagnostics. Cancers 2022, 14, 2080. https://doi.org/10.3390/cancers14092080
Lodrini M, Wünschel J, Thole-Kliesch TM, Grimaldi M, Sprüssel A, Linke RB, Hollander JF, Tiburtius D, Künkele A, Schulte JH, et al. Circulating Cell-Free DNA Assessment in Biofluids from Children with Neuroblastoma Demonstrates Feasibility and Potential for Minimally Invasive Molecular Diagnostics. Cancers. 2022; 14(9):2080. https://doi.org/10.3390/cancers14092080
Chicago/Turabian StyleLodrini, Marco, Jasmin Wünschel, Theresa M. Thole-Kliesch, Maddalena Grimaldi, Annika Sprüssel, Rasmus B. Linke, Jan F. Hollander, Daniela Tiburtius, Annette Künkele, Johannes H. Schulte, and et al. 2022. "Circulating Cell-Free DNA Assessment in Biofluids from Children with Neuroblastoma Demonstrates Feasibility and Potential for Minimally Invasive Molecular Diagnostics" Cancers 14, no. 9: 2080. https://doi.org/10.3390/cancers14092080
APA StyleLodrini, M., Wünschel, J., Thole-Kliesch, T. M., Grimaldi, M., Sprüssel, A., Linke, R. B., Hollander, J. F., Tiburtius, D., Künkele, A., Schulte, J. H., Lankes, E., Elgeti, T., Hundsdörfer, P., Astrahantseff, K., Simon, T., Eggert, A., & Deubzer, H. E. (2022). Circulating Cell-Free DNA Assessment in Biofluids from Children with Neuroblastoma Demonstrates Feasibility and Potential for Minimally Invasive Molecular Diagnostics. Cancers, 14(9), 2080. https://doi.org/10.3390/cancers14092080






