The Role of Artificial Intelligence in Early Cancer Diagnosis
Abstract
Simple Summary
Abstract
1. Introduction
2. An Overview of Artificial Intelligence in Oncology
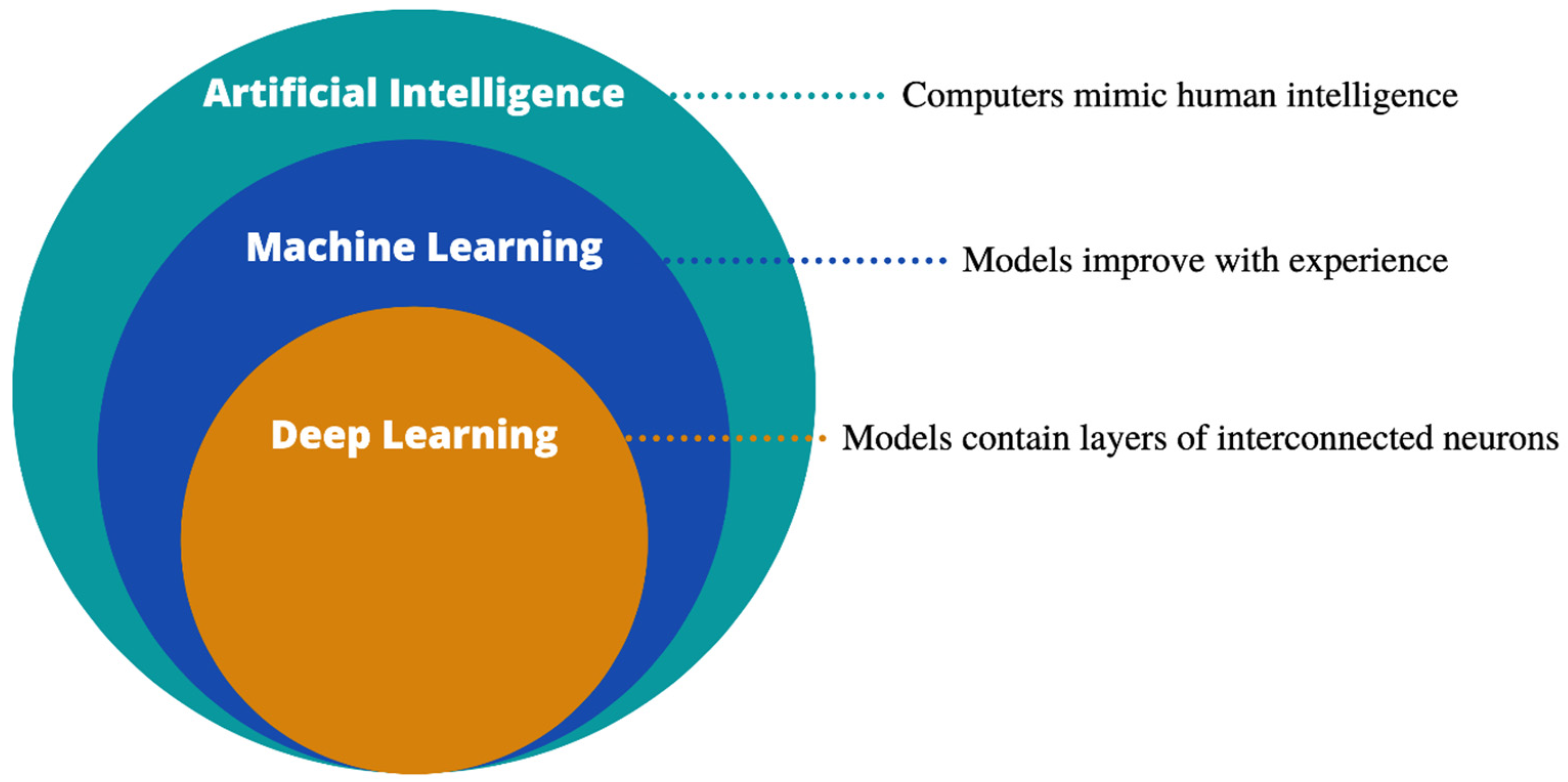
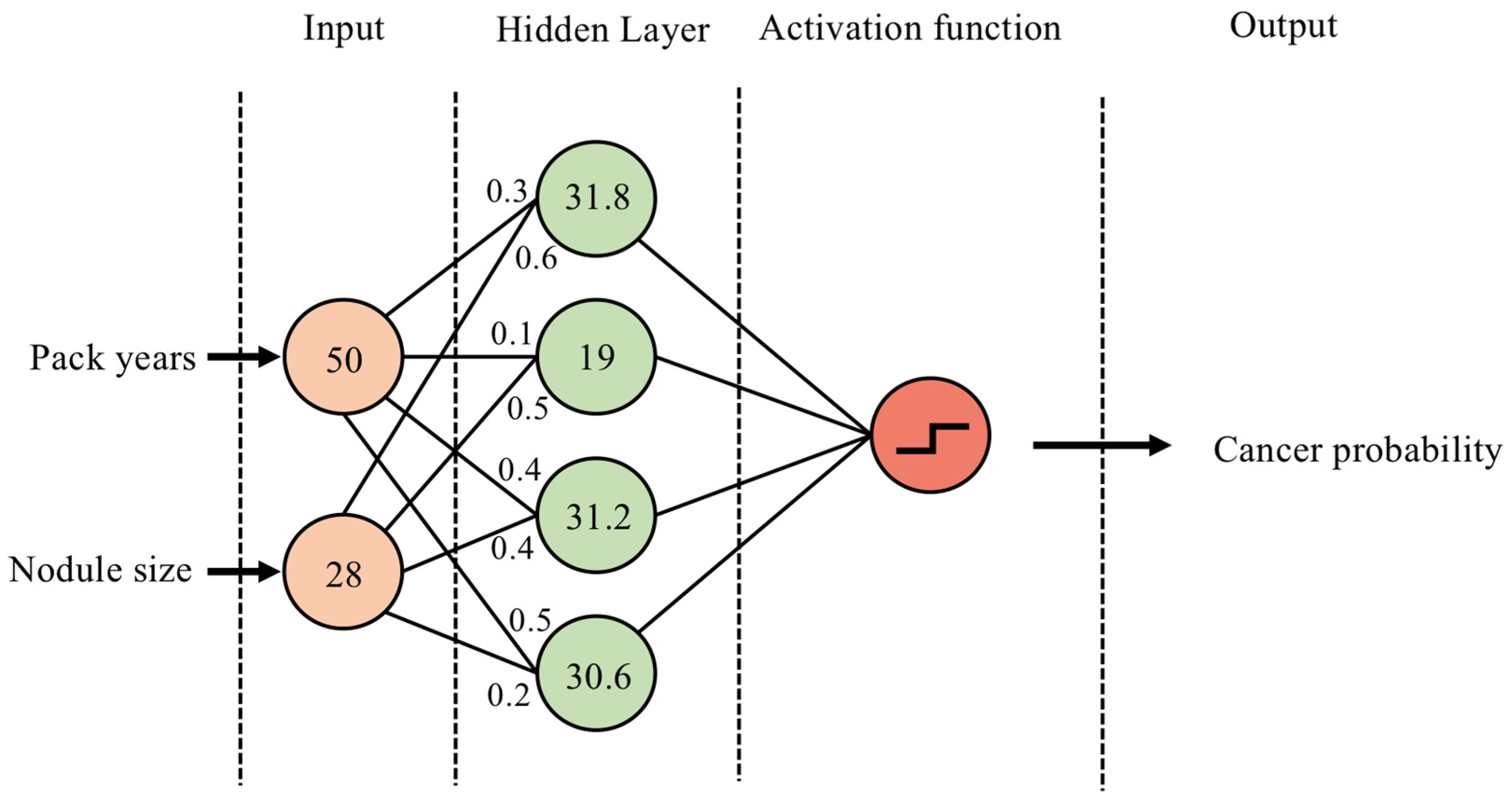
2.1. Definitions and Model Architectures
2.2. Data Types: Electronic Healthcare Records
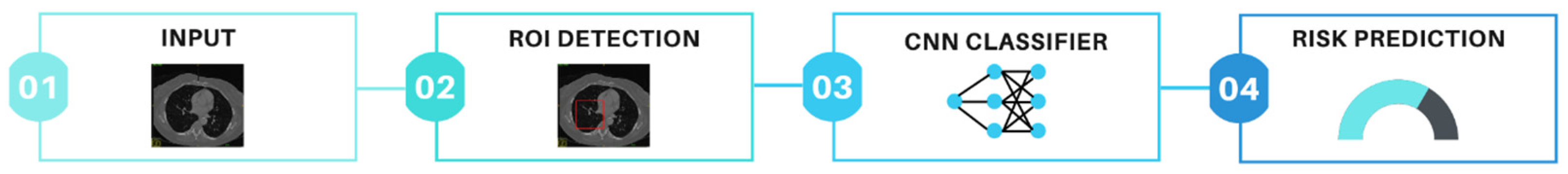
2.3. Data Types: Radiology
2.4. Data Types: Digital Pathology
2.5. Data Types: Multi-Omic Data
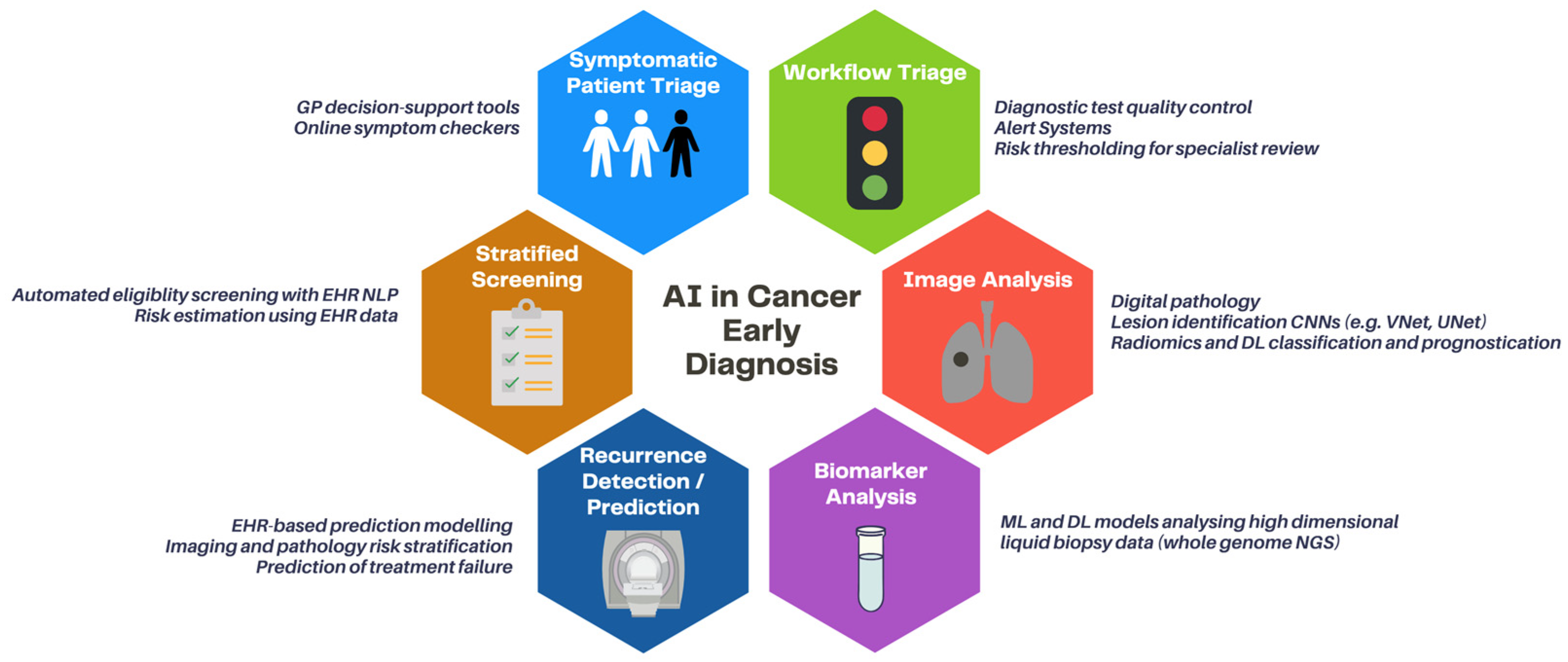
3. Clinical Applications
3.1. Risk-Stratified Screening of Asymptomatic Patients
3.2. Symptomatic Patient Triage
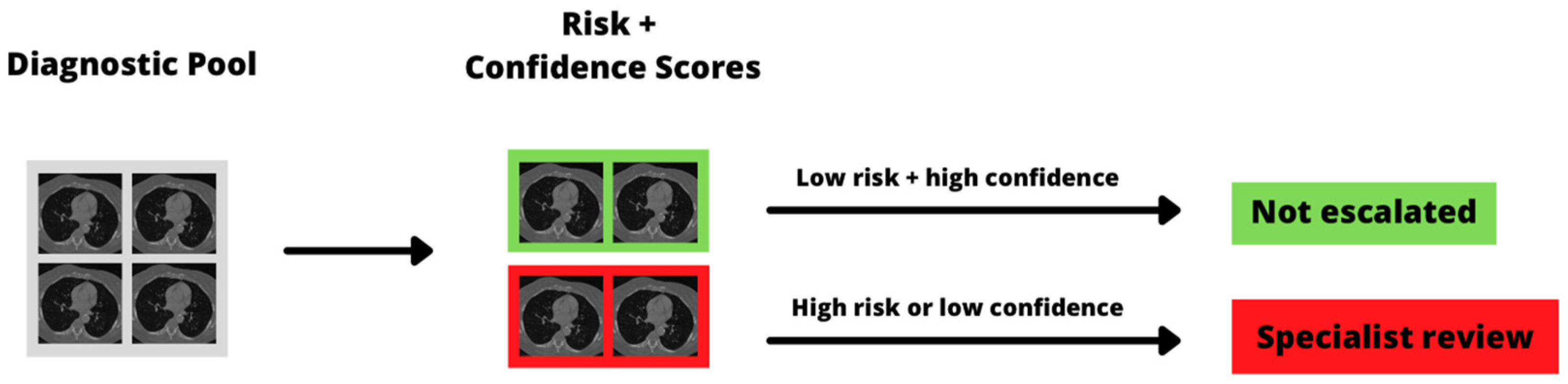
3.3. Diagnostic Workflow Triage
3.4. Early Detection
3.5. Early Detection of Recurrence
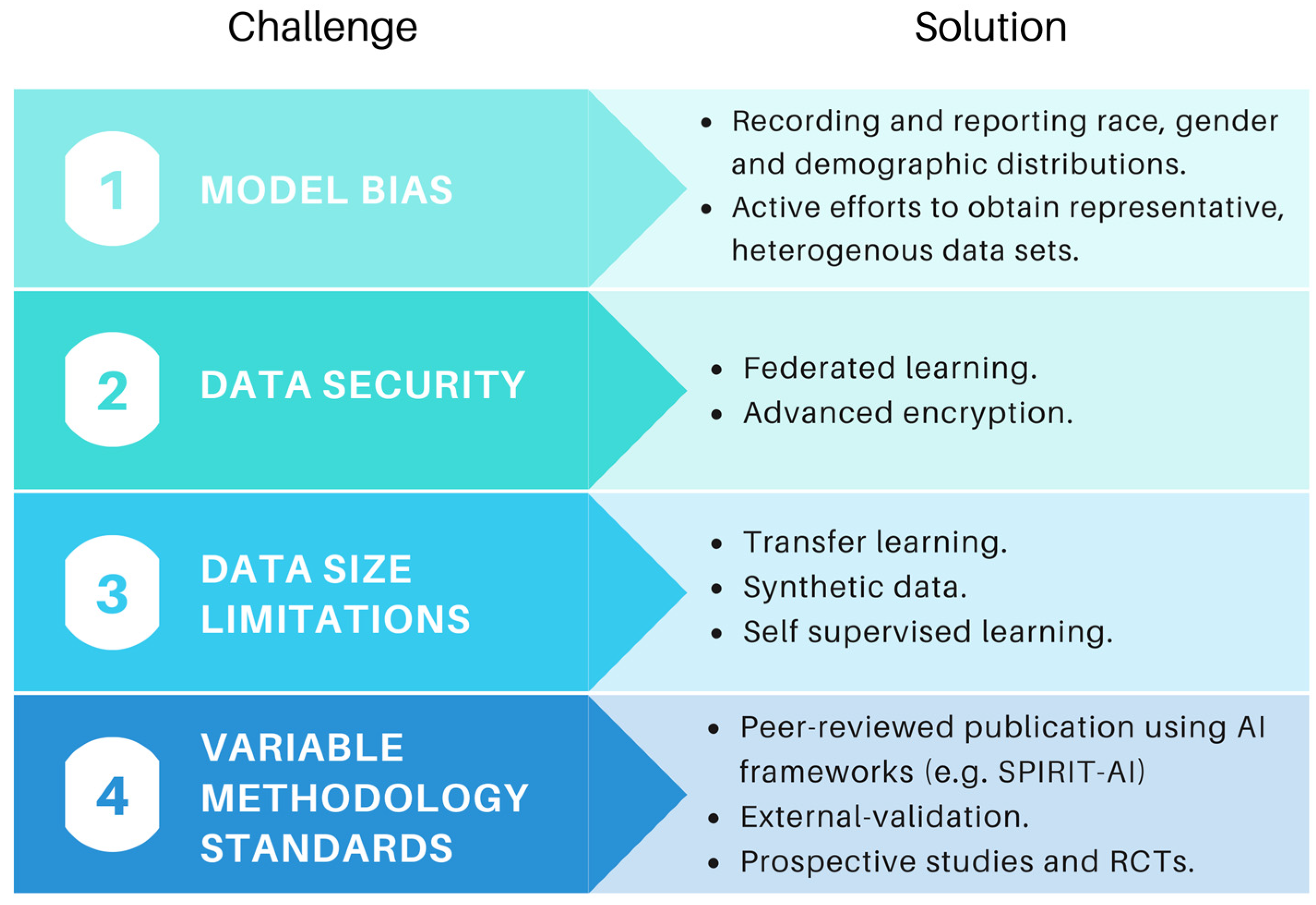
4. Challenges and Future Directions
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McPhail, S.; Johnson, S.; Greenberg, D.; Peake, M.; Rous, B. Stage at diagnosis and early mortality from cancer in England. Br. J. Cancer 2015, 112, S108–S115. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.B.; Crosbie, P.A.; Balata, H.; Chudziak, J.; Hussell, T.; Dive, C. Progress and prospects of early detection in lung cancer. Open Biol. 2017, 7, 170070. [Google Scholar] [CrossRef]
- National Cancer Registration and Analysis Service: Staging Data in England. Available online: https://www.cancerdata.nhs.uk/stage_at_diagnosis (accessed on 14 October 2021).
- NHS NHS Long Term Plan: Cancer. Available online: https://www.longtermplan.nhs.uk/areas-of-work/cancer/ (accessed on 30 March 2021).
- Sasieni, P. Evaluation of the UK breast screening programmes. Ann. Oncol. 2003, 14, 1206–1208. [Google Scholar] [CrossRef] [PubMed]
- Maroni, R.; Massat, N.J.; Parmar, D.; Dibden, A.; Cuzick, J.; Sasieni, P.D.; Duffy, S.W. A case-control study to evaluate the impact of the breast screening programme on mortality in England. Br. J. Cancer 2020, 124, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Esserman, L.J. The WISDOM Study: Breaking the deadlock in the breast cancer screening debate. NPJ Breast Cancer 2017, 3, 34. [Google Scholar] [CrossRef] [PubMed]
- Dembrower, K.; Wåhlin, E.; Liu, Y.; Salim, M.; Smith, K.; Lindholm, P.; Eklund, M.; Strand, F. Effect of artificial intelligence-based triaging of breast cancer screening mammograms on cancer detection and radiologist workload: A retrospective simulation study. Lancet Digit. Health 2020, 2, e468–e474. [Google Scholar] [CrossRef]
- Meystre, S.M.; Heider, P.M.; Kim, Y.; Aruch, D.B.; Britten, C.D. Automatic trial eligibility surveillance based on unstructured clinical data. Int. J. Med. Inform. 2019, 129, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.T.; Rammage, M.; Jackson, G.P.; Preininger, A.M.; Dankwa-Mullan, I.; Roebuck, M.C.; Torres, A.; Holtzen, H.; Coverdill, S.E.; Williamson, M.P.; et al. Artificial Intelligence Tool for Optimizing Eligibility Screening for Clinical Trials in a Large Community Cancer Center. JCO Clin. Cancer Inform. 2020, 4, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Yang, J.; Fong, S.; Zhao, Q. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett. 2020, 471, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Yim, W.; Yetisgen, M.; Harris, W.P.; Kwan, S.W. Natural Language Processing in Oncology: A Review. JAMA Oncol. 2016, 2, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Chhatwal, J.; Alagoz, O.; Lindstrom, M.J.; Kahn, C.E.; Shaffer, K.A.; Burnside, E.S. A logistic regression model based on the national mammography database format to aid breast cancer diagnosis. Am. J. Roentgenol. 2009, 192, 1117–1127. [Google Scholar] [CrossRef]
- Zhang, F.; Kaufman, H.L.; Deng, Y.; Drabier, R. Recursive SVM biomarker selection for early detection of breast cancer in peripheral blood. BMC Med. Genom. 2013, 6 (Suppl. 1), S4. [Google Scholar] [CrossRef] [PubMed]
- Olatunji, S.O.; Alotaibi, S.; Almutairi, E.; Alrabae, Z.; Almajid, Y.; Altabee, R.; Altassan, M.; Basheer Ahmed, M.I.; Farooqui, M.; Alhiyafi, J. Early diagnosis of thyroid cancer diseases using computational intelligence techniques: A case study of a Saudi Arabian dataset. Comput. Biol. Med. 2021, 131, 104267. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.H.; Chen, P.R.; Gou, Z.P.; Li, Y.Z.; Li, M.; Xiang, L.C.; Feng, P. Prostate cancer prediction using the random forest algorithm that takes into account transrectal ultrasound findings, age, and serum levels of prostate-specific antigen. Asian J. Androl. 2017, 19, 586. [Google Scholar] [CrossRef] [PubMed]
- Liew, X.Y.; Hameed, N.; Clos, J. An investigation of XGBoost-based algorithm for breast cancer classification. Mach. Learn. Appl. 2021, 6, 100154. [Google Scholar] [CrossRef]
- Muhammad, W.; Hart, G.R.; Nartowt, B.; Farrell, J.J.; Johung, K.; Liang, Y.; Deng, J. Pancreatic Cancer Prediction Through an Artificial Neural Network. Front. Artif. Intell. 2019, 2, 2. [Google Scholar] [CrossRef] [PubMed]
- Suh, Y.J.; Jung, J.; Cho, B.J. Automated Breast Cancer Detection in Digital Mammograms of Various Densities via Deep Learning. J. Personal. Med. 2020, 10, 211. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, ICML 2019, Long Beach, CA, USA, 9–15 June 2019; pp. 10691–10700. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition CVPR 2017, Honolulu, HI, USA, 21–26 July 2016; pp. 2261–2269. [Google Scholar] [CrossRef]
- Gillum, R.F. From papyrus to the electronic tablet: A brief history of the clinical medical record with lessons for the digital age. Am. J. Med. 2013, 126, 853–857. [Google Scholar] [CrossRef] [PubMed]
- DATA-CAN: Health Data Research Hub for Cancer|UCLPartners. Available online: https://uclpartners.com/work/data-can-health-data-research-hub-cancer/ (accessed on 18 October 2021).
- NHS Digital Cancer Waiting Times Data Collection (CWT)—NHS Digital. Available online: https://digital.nhs.uk/data-and-information/data-collections-and-data-sets/data-collections/cancerwaitingtimescwt#uk-cancer-tools-and-intelligence (accessed on 18 October 2021).
- Digital Transformation of Screening—NHSX. Available online: https://www.nhsx.nhs.uk/key-tools-and-info/digital-transformation-of-screening/ (accessed on 24 January 2022).
- Benefits of the new NHS Cervical Screening Management System—NHS Digital. Available online: https://digital.nhs.uk/services/screening-services/national-cervical-screening/new-cervical-screening-management-system/benefits (accessed on 24 January 2022).
- Benke, K.; Benke, G. Artificial intelligence and big data in public health. Int. J. Environ. Res. Public Health 2018, 15, 2796. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.R.; Farrag, A.; Ashkin, E. Using natural language processing to extract abnormal results from cancer screening reports. J. Patient Saf. 2017, 13, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Nayor, J.; Borges, L.F.; Goryachev, S.; Gainer, V.S.; Saltzman, J.R. Natural Language Processing Accurately Calculates Adenoma and Sessile Serrated Polyp Detection Rates. Dig. Dis. Sci. 2018, 63, 1794–1800. [Google Scholar] [CrossRef] [PubMed]
- Glaser, A.P.; Jordan, B.J.; Cohen, J.; Desai, A.; Silberman, P.; Meeks, J.J. Automated Extraction of Grade, Stage, and Quality Information From Transurethral Resection of Bladder Tumor Pathology Reports Using Natural Language Processing. JCO Clin. Cancer Inform. 2018, 2, 1–8. [Google Scholar] [CrossRef]
- Danforth, K.N.; Early, M.I.; Ngan, S.; Kosco, A.E.; Zheng, C.; Gould, M.K. Automated identification of patients with pulmonary nodules in an integrated health system using administrative health plan data, radiology reports, and natural language processing. J. Thorac. Oncol. 2012, 7, 1257–1262. [Google Scholar] [CrossRef] [PubMed]
- Farjah, F.; Halgrim, S.; Buist, D.S.M.; Gould, M.K.; Zeliadt, S.B.; Loggers, E.T.; Carrell, D.S. An Automated Method for Identifying Individuals with a Lung Nodule Can Be Feasibly Implemented Across Health Systems. eGEMs 2016, 4, 15. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Beyer, S.E.; McKee, B.J.; Regis, S.M.; McKee, A.B.; Flacke, S.; Saadawi, G.; El Wald, C. Automatic Lung-RADSTM classification with a natural language processing system. J. Thorac. Dis. 2017, 9, 3114. [Google Scholar] [CrossRef] [PubMed]
- Roch, A.M.; Mehrabi, S.; Krishnan, A.; Schmidt, H.E.; Kesterson, J.; Beesley, C.; Dexter, P.R.; Palakal, M.; Schmidt, C.M. Automated pancreatic cyst screening using natural language processing: A new tool in the early detection of pancreatic cancer. Hpb 2015, 17, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Hunter, B.; Reis, S.; Campbell, D.; Matharu, S.; Ratnakumar, P.; Mercuri, L.; Hindocha, S.; Kalsi, H.; Mayer, E.; Glampson, B.; et al. Development of a Structured Query Language and Natural Language Processing Algorithm to Identify Lung Nodules in a Cancer Centre. Front. Med. 2021, 8, 748168. [Google Scholar] [CrossRef]
- Ni, Y.; Wright, J.; Perentesis, J.; Lingren, T.; Deleger, L.; Kaiser, M.; Kohane, I.; Solti, I. Increasing the efficiency of trial-patient matching: Automated clinical trial eligibility Pre-screening for pediatric oncology patients Clinical decision-making, knowledge support systems, and theory. BMC Med. Inform. Decis. Mak. 2015, 15, 28. [Google Scholar] [CrossRef]
- Morin, O.; Vallières, M.; Braunstein, S.; Ginart, J.B.; Upadhaya, T.; Woodruff, H.C.; Zwanenburg, A.; Chatterjee, A.; Villanueva-Meyer, J.E.; Valdes, G.; et al. An artificial intelligence framework integrating longitudinal electronic health records with real-world data enables continuous pan-cancer prognostication. Nat. Cancer 2021, 2, 709–722. [Google Scholar] [CrossRef]
- Chen, X.; Feng, B.; Chen, Y.; Liu, K.; Li, K.; Duan, X.; Hao, Y.; Cui, E.; Liu, Z.; Zhang, C.; et al. A CT-based radiomics nomogram for prediction of lung adenocarcinomas and granulomatous lesions in patient with solitary sub-centimeter solid nodules. Cancer Imaging 2020, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Beig, N.; Khorrami, M.; Alilou, M.; Prasanna, P.; Braman, N.; Orooji, M.; Rakshit, S.; Bera, K.; Rajiah, P.; Ginsberg, J.; et al. Perinodular and Intranodular Radiomic Features on Lung CT Images Distinguish Adenocarcinomas from Granulomas. Radiology 2019, 290, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Shakir, H.; Deng, Y.; Rasheed, H.; Khan, T.M.R. Radiomics based likelihood functions for cancer diagnosis. Sci. Rep. 2019, 9, 9501. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Arshad, M.; Thornton, A.; Avesani, G.; Cunnea, P.; Curry, E.; Kanavati, F.; Liang, J.; Nixon, K.; Williams, S.T.; et al. A mathematical-descriptor of tumor-mesoscopic-structure from computed-tomography images annotates prognostic- and molecular-phenotypes of epithelial ovarian cancer. Nat. Commun. 2019, 10, 764. [Google Scholar] [CrossRef] [PubMed]
- Astaraki, M.; Zakko, Y.; Toma Dasu, I.; Smedby, Ö.; Wang, C. Benign-malignant pulmonary nodule classification in low-dose CT with convolutional features. Phys. Medica 2021, 83, 146–153. [Google Scholar] [CrossRef]
- Guan, Y.; Aamir, M.; Rahman, Z.; Ali, A.; Abro, W.A.; Dayo, Z.A.; Bhutta, M.S.; Hu, Z.; Guan, Y.; Aamir, M.; et al. A framework for efficient brain tumor classification using MRI images. Math. Biosci. Eng. 2021, 18, 5790–5815. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Lect. Notes Comput. Sci. 2015, 9351, 234–241. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 4th International Conference on 3D Vision, 3DV, Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Baccouche, A.; Garcia-Zapirain, B.; Castillo Olea, C.; Elmaghraby, A.S. Connected-UNets: A deep learning architecture for breast mass segmentation. NPJ Breast Cancer 2021, 7, 1–12. [Google Scholar] [CrossRef]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Baldwin, D.R.; Gustafson, J.; Pickup, L.; Arteta, C.; Novotny, P.; Declerck, J.; Kadir, T.; Figueiras, C.; Sterba, A.; Exell, A.; et al. External validation of a convolutional neural network artificial intelligence tool to predict malignancy in pulmonary nodules. Thorax 2020, 75, 306–312. [Google Scholar] [CrossRef]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef]
- Papadimitroulas, P.; Brocki, L.; Christopher Chung, N.; Marchadour, W.; Vermet, F.; Gaubert, L.; Eleftheriadis, V.; Plachouris, D.; Visvikis, D.; Kagadis, G.C.; et al. Artificial intelligence: Deep learning in oncological radiomics and challenges of interpretability and data harmonization. Phys. Medica 2021, 83, 108–121. [Google Scholar] [CrossRef]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.J.; Lee, J.; Oien, K.A.; Treanor, D. Digital pathology access and usage in the UK: Results from a national survey on behalf of the National Cancer Research Institute’s CM-Path initiative. J. Clin. Pathol. 2018, 71, 463. [Google Scholar] [CrossRef] [PubMed]
- Schüffler, P.J.; Geneslaw, L.; Yarlagadda, D.V.K.; Hanna, M.G.; Samboy, J.; Stamelos, E.; Vanderbilt, C.; Philip, J.; Jean, M.-H.; Corsale, L.; et al. Integrated digital pathology at scale: A solution for clinical diagnostics and cancer research at a large academic medical center. J. Am. Med. Inform. Assoc. 2021, 28, 1874. [Google Scholar] [CrossRef] [PubMed]
- Browning, L.; Colling, R.; Rakha, E.; Rajpoot, N.; Rittscher, J.; James, J.A.; Salto-Tellez, M.; Snead, D.R.J.; Verrill, C. Digital pathology and artificial intelligence will be key to supporting clinical and academic cellular pathology through COVID-19 and future crises: The PathLAKE consortium perspective. J. Clin. Pathol. 2021, 74, 443–447. [Google Scholar] [CrossRef]
- Dash, R.C.; Jones, N.; Merrick, R.; Haroske, G.; Harrison, J.; Sayers, C.; Haarselhorst, N.; Wintell, M.; Herrmann, M.D.; Macary, F. Integrating the Health-care Enterprise Pathology and Laboratory Medicine Guideline for Digital Pathology Interoperability. J. Pathol. Inform. 2021, 12, 16. [Google Scholar] [CrossRef]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef]
- Sui, D.; Liu, W.; Chen, J.; Zhao, C.; Ma, X.; Guo, M.; Tian, Z. A Pyramid Architecture-Based Deep Learning Framework for Breast Cancer Detection. Biomed Res. Int. 2021, 2021, 1–10. [Google Scholar] [CrossRef]
- Ehteshami Bejnordi, B.; Mullooly, M.; Pfeiffer, R.M.; Fan, S.; Vacek, P.M.; Weaver, D.L.; Herschorn, S.; Brinton, L.A.; van Ginneken, B.; Karssemeijer, N.; et al. Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod. Pathol. 2018, 31, 1502–1512. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Werneck Krauss Silva, V.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Woerl, A.C.; Eckstein, M.; Geiger, J.; Wagner, D.C.; Daher, T.; Stenzel, P.; Fernandez, A.; Hartmann, A.; Wand, M.; Roth, W.; et al. Deep Learning Predicts Molecular Subtype of Muscle-invasive Bladder Cancer from Conventional Histopathological Slides. Eur. Urol. 2020, 78, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Naik, N.; Madani, A.; Esteva, A.; Keskar, N.S.; Press, M.F.; Ruderman, D.; Agus, D.B.; Socher, R. Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains. Nat. Commun. 2020, 11, 5727. [Google Scholar] [CrossRef] [PubMed]
- Ström, P.; Kartasalo, K.; Olsson, H.; Solorzano, L.; Delahunt, B.; Berney, D.M.; Bostwick, D.G.; Evans, A.J.; Grignon, D.J.; Humphrey, P.A.; et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: A population-based, diagnostic study. Lancet Oncol. 2020, 21, 222–232. [Google Scholar] [CrossRef]
- Madabhushi, A.; Feldman, M.D.; Leo, P. Deep-learning approaches for Gleason grading of prostate biopsies. Lancet Oncol. 2020, 21, 187–189. [Google Scholar] [CrossRef]
- Lokhande, A.; Bonthu, S.; Singhal, N. Carcino-Net: A Deep Learning Framework for Automated Gleason Grading of Prostate Biopsies. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, Montreal, QC, Canada, 20–24 July 2020; pp. 1380–1383. [Google Scholar]
- Bera, K.; Katz, I.; Madabhushi, A. Reimagining T Staging Through Artificial Intelligence and Machine Learning Image Processing Approaches in Digital Pathology. JCO Clin. Cancer Inform. 2020, 4, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- da Silva, L.M.; Pereira, E.M.; Salles, P.G.O.; Godrich, R.; Ceballos, R.; Kunz, J.D.; Casson, A.; Viret, J.; Chandarlapaty, S.; Ferreira, C.G.; et al. Independent real-world application of a clinical-grade automated prostate cancer detection system. J. Pathol. 2021, 254, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Halse, H.; Colebatch, A.J.; Petrone, P.; Henderson, M.A.; Mills, J.K.; Snow, H.; Westwood, J.A.; Sandhu, S.; Raleigh, J.M.; Behren, A.; et al. Multiplex immunohistochemistry accurately defines the immune context of metastatic melanoma. Sci. Rep. 2018, 8, 11158. [Google Scholar] [CrossRef] [PubMed]
- Fassler, D.J.; Abousamra, S.; Gupta, R.; Chen, C.; Zhao, M.; Paredes, D.; Batool, S.A.; Knudsen, B.S.; Escobar-Hoyos, L.; Shroyer, K.R.; et al. Deep learning-based image analysis methods for brightfield-acquired multiplex immunohistochemistry images. Diagn. Pathol. 2020, 15, 1–11. [Google Scholar] [CrossRef]
- Kawakami, E.; Tabata, J.; Yanaihara, N.; Ishikawa, T.; Koseki, K.; Iida, Y.; Saito, M.; Komazaki, H.; Shapiro, J.S.; Goto, C.; et al. Application of artificial intelligence for preoperative diagnostic and prognostic prediction in epithelial ovarian cancer based on blood biomarkers. Clin. Cancer Res. 2019, 25, 3006–3015. [Google Scholar] [CrossRef] [PubMed]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, Y.; Pan, X.; Li, M.; Yang, S.; Li, S.C. DNA methylation markers for pan-cancer prediction by deep learning. Genes 2019, 10, 778. [Google Scholar] [CrossRef]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xia, C.; Wang, G. Multi-Omics Analysis in Initiation and Progression of Meningiomas: From Pathogenesis to Diagnosis. Front. Oncol. 2020, 10, 1491. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Takahashi, M.; Tanaka, S.; Takayanagi, S.; Takami, H.; Yamazawa, E.; Nambu, S.; Miyake, M.; Satomi, K.; Ichimura, K.; et al. A new era of neuro-oncology research pioneered by multi-omics analysis and machine learning. Biomolecules 2021, 11, 565. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, A.; Idbaih, A.; Dehais, C.; Elarouci, N.; Carpentier, C.; Letouzé, E.; Colin, C.; Mokhtari, K.; Jouvet, A.; Uro-Coste, E.; et al. Integrated multi-omics analysis of oligodendroglial tumours identifies three subgroups of 1p/19q co-deleted gliomas. Nat. Commun. 2016, 7, 11263. [Google Scholar] [CrossRef] [PubMed]
- Franco, E.F.; Rana, P.; Cruz, A.; Calderón, V.V.; Azevedo, V.; Ramos, R.T.J.; Ghosh, P. Performance comparison of deep learning autoencoders for cancer subtype detection using multi-omics data. Cancers 2021, 13, 2013. [Google Scholar] [CrossRef] [PubMed]
- Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; Sicks, J.R.D. Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef] [PubMed]
- de Koning, H.J.; van der Aalst, C.M.; de Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.-W.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N.; et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Dyer, O. US task force recommends extending lung cancer screenings to over 50s. BMJ 2021, 372, n698. [Google Scholar] [CrossRef] [PubMed]
- Krist, A.H.; Davidson, K.W.; Mangione, C.M.; Barry, M.J.; Cabana, M.; Caughey, A.B.; Davis, E.M.; Donahue, K.E.; Doubeni, C.A.; Kubik, M.; et al. Screening for Lung Cancer: US Preventive Services Task Force Recommendation Statement. JAMA-J. Am. Med. Assoc. 2021, 325, 962–970. [Google Scholar] [CrossRef]
- Richards, T.B.; Doria-Rose, V.P.; Soman, A.; Klabunde, C.N.; Caraballo, R.S.; Gray, S.C.; Houston, K.A.; White, M.C. Lung Cancer Screening Inconsistent With U.S. Preventive Services Task Force Recommendations. Am. J. Prev. Med. 2019, 56, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.X.; Baggett, T.P.; Pandharipande, P.V.; Park, E.R.; Fintelmann, F.J.; Percac-Lima, S.; Shepard, J.A.O.; Flores, E.J. Barriers to Lung Cancer Screening Engagement from the Patient and Provider Perspective. Radiology 2019, 290, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.T.; Raghu, V.K.; Mayrhofer, T.; Aerts, H.J.W.L.; Hoffmann, U. Deep Learning Using Chest Radiographs to Identify High-Risk Smokers for Lung Cancer Screening Computed Tomography: Development and Validation of a Prediction Model. Ann. Intern. Med. 2020, 173, 704–713. [Google Scholar] [CrossRef] [PubMed]
- Gould, M.K.; Tang, T.; Liu, I.L.A.; Lee, J.; Zheng, C.; Danforth, K.N.; Kosco, A.E.; Di Fiore, J.L.; Suh, D.E. Recent trends in the identification of incidental pulmonary nodules. Am. J. Respir. Crit. Care Med. 2015, 192, 1208–1214. [Google Scholar] [CrossRef] [PubMed]
- Green, T.; Atkin, K.; Macleod, U. Cancer detection in primary care: Insights from general practitioners. Br. J. Cancer 2015, 112, S41–S49. [Google Scholar] [CrossRef]
- Hamilton, W.; Green, T.; Martins, T.; Elliott, K.; Rubin, G.; Macleod, U. Evaluation of risk assessment tools for suspected cancer in general practice: A cohort study. Br. J. Gen. Pract. 2013, 63, e30. [Google Scholar] [CrossRef] [PubMed]
- C the Signs|Find Cancer Earlier. Available online: https://cthesigns.co.uk/ (accessed on 18 November 2021).
- An AI Support Tool to Help Healthcare Professionals in Primary Care to Identify Patients at Risk of Cancer Earlier—NHSX. Available online: https://www.nhsx.nhs.uk/key-tools-and-info/digital-playbooks/cancer-digital-playbook/an-AI-support-tool-to-help-healthcare-professionals-in-primary-care-to-identify-patients-at-risk-of-cancer-earlier/ (accessed on 9 November 2021).
- Babylon Health UK—The Online Doctor and…|Babylon Health. Available online: https://www.babylonhealth.com/ (accessed on 18 November 2021).
- Baker, A.; Perov, Y.; Middleton, K.; Baxter, J.; Mullarkey, D.; Sangar, D.; Butt, M.; DoRosario, A.; Johri, S. A Comparison of Artificial Intelligence and Human Doctors for the Purpose of Triage and Diagnosis. Front. Artif. Intell. 2020, 3, 100. [Google Scholar] [CrossRef] [PubMed]
- UK’s MHRA Says It Has ‘Concerns’ about Babylon Health—And Flags Legal Gap around Triage Chatbots|TechCrunch. Available online: https://techcrunch.com/2021/03/05/uks-mhra-says-it-has-concerns-about-babylon-health-and-flags-legal-gap-around-triage-chatbots/?guccounter=1&guce_referrer=aHR0cHM6Ly93d3cuZ29vZ2xlLmNvbS8&guce_referrer_sig=AQAAAJ2qLhRLfYrjSPpC_FG85UfLrUX2HsTyVUXcolTGJngMUtHeaXEGQZ2chY8JI7KXbe3ZJYFx6sdH4o3YQFd_3QQnYQkmr7F5qw_AkShAdghtDIMvSt3L7rZfxGxWSl4LmzoaTdI-5O3WKmlGslD2V3FCugaQcV6MCwrIOr4Tfhwb (accessed on 18 November 2021).
- Anderson, M.; O’Neill, C.; Macleod Clark, J.; Street, A.; Woods, M.; Johnston-Webber, C.; Charlesworth, A.; Whyte, M.; Foster, M.; Majeed, A.; et al. Securing a sustainable and fit-for-purpose UK health and care workforce. Lancet 2021, 397, 1992–2011. [Google Scholar] [CrossRef]
- Van Haren, R.M.; Delman, A.M.; Turner, K.M.; Waits, B.; Hemingway, M.; Shah, S.A.; Starnes, S.L. Impact of the COVID-19 Pandemic on Lung Cancer Screening Program and Subsequent Lung Cancer. J. Am. Coll. Surg. 2021, 232, 600. [Google Scholar] [CrossRef] [PubMed]
- Armato, S.G.; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A Completed Reference Database of Lung Nodules on CT Scans. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef] [PubMed]
- Gehrung, M.; Crispin-Ortuzar, M.; Berman, A.G.; O’Donovan, M.; Fitzgerald, R.C.; Markowetz, F. Triage-driven diagnosis of Barrett’s esophagus for early detection of esophageal adenocarcinoma using deep learning. Nat. Med. 2021, 27, 833–841. [Google Scholar] [CrossRef]
- Shaheen, N.J.; Falk, G.W.; Iyer, P.G.; Gerson, L.B. ACG Clinical Guideline: Diagnosis and Management of Barrett’s Esophagus. Am. J. Gastroenterol. 2016, 111, 30–50. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, R.C.; di Pietro, M.; O’Donovan, M.; Maroni, R.; Muldrew, B.; Debiram-Beecham, I.; Gehrung, M.; Offman, J.; Tripathi, M.; Smith, S.G.; et al. Cytosponge-trefoil factor 3 versus usual care to identify Barrett’s oesophagus in a primary care setting: A multicentre, pragmatic, randomised controlled trial. Lancet 2020, 396, 333–344. [Google Scholar] [CrossRef]
- Schaffter, T.; Buist, D.S.M.; Lee, C.I.; Nikulin, Y.; Ribli, D.; Guan, Y.; Lotter, W.; Jie, Z.; Du, H.; Wang, S.; et al. Evaluation of Combined Artificial Intelligence and Radiologist Assessment to Interpret Screening Mammograms. JAMA Netw. Open 2020, 3, e200265. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Ruiz, A.; Lång, K.; Gubern-Merida, A.; Broeders, M.; Gennaro, G.; Clauser, P.; Helbich, T.H.; Chevalier, M.; Tan, T.; Mertelmeier, T.; et al. Stand-Alone Artificial Intelligence for Breast Cancer Detection in Mammography: Comparison with 101 Radiologists. JNCI J. Natl. Cancer Inst. 2019, 111, 916. [Google Scholar] [CrossRef]
- Kim, H.E.; Kim, H.H.; Han, B.K.; Kim, K.H.; Han, K.; Nam, H.; Lee, E.H.; Kim, E.K. Changes in cancer detection and false-positive recall in mammography using artificial intelligence: A retrospective, multireader study. Lancet Digit. Health 2020, 2, e138–e148. [Google Scholar] [CrossRef]
- The Technologies|Artificial Intelligence in Mammography|Advice|NICE. Available online: https://www.nice.org.uk/advice/mib242/chapter/The-technologies (accessed on 24 November 2021).
- Yi, P.H.; Singh, D.; Harvey, S.C.; Hager, G.D.; Mullen, L.A. DeepCAT: Deep Computer-Aided Triage of Screening Mammography. J. Digit. Imaging 2021, 34, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Oke, J.L.; Pickup, L.C.; Declerck, J.; Callister, M.E.; Baldwin, D.; Gustafson, J.; Peschl, H.; Ather, S.; Tsakok, M.; Exell, A.; et al. Development and validation of clinical prediction models to risk stratify patients presenting with small pulmonary nodules: A research protocol. Diagn. Progn. Res. 2018, 2, e28110. [Google Scholar] [CrossRef] [PubMed]
- Schelb, P.; Kohl, S.; Radtke, J.P.; Wiesenfarth, M.; Kickingereder, P.; Bickelhaupt, S.; Kuder, T.A.; Stenzinger, A.; Hohenfellner, M.; Schlemmer, H.P.; et al. Classification of Cancer at Prostate MRI: Deep Learning versus Clinical PI-RADS Assessment. Radiology 2019, 293, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Mehralivand, S.; Harmon, S.A.; Shih, J.H.; Smith, C.P.; Lay, N.; Argun, B.; Bednarova, S.; Baroni, R.H.; Canda, A.E.; Ercan, K.; et al. Multicenter Multireader Evaluation of an Artificial Intelligence-Based Attention Mapping System for the Detection of Prostate Cancer With Multiparametric MRI. AJR. Am. J. Roentgenol. 2020, 215, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Wildeboer, R.R.; van Sloun, R.J.G.; Wijkstra, H.; Mischi, M. Artificial intelligence in multiparametric prostate cancer imaging with focus on deep-learning methods. Comput. Methods Programs Biomed. 2020, 189, 105316. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genom. 2019, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Galleri Blood Test for Cancer|Tests and Scans|Cancer Research UK. Available online: https://www.cancerresearchuk.org/about-cancer/cancer-in-general/tests/blood-tests/Galleri-blood-test (accessed on 7 December 2021).
- Tao, K.; Bian, Z.; Zhang, Q.; Guo, X.; Yin, C.; Wang, Y.; Zhou, K.; Wan, S.; Shi, M.; Bao, D.; et al. Machine learning-based genome-wide interrogation of somatic copy number aberrations in circulating tumor DNA for early detection of hepatocellular carcinoma-NC-ND license. EBioMedicine 2020, 56, 102811. [Google Scholar] [CrossRef]
- Shin, H.; Oh, S.; Hong, S.; Kang, M.; Kang, D.; Ji, Y.G.; Choi, B.H.; Kang, K.W.; Jeong, H.; Park, Y.; et al. Early-Stage Lung Cancer Diagnosis by Deep Learning-Based Spectroscopic Analysis of Circulating Exosomes. ACS Nano 2020, 14, 5435–5444. [Google Scholar] [CrossRef]
- Best, M.G.; In ’t Veld, S.G.J.G.; Sol, N.; Wurdinger, T. RNA sequencing and swarm intelligence–enhanced classification algorithm development for blood-based disease diagnostics using spliced blood platelet RNA. Nat. Protoc. 2019, 14, 1206–1234. [Google Scholar] [CrossRef]
- Cucchiara, F.; Del Re, M.; Valleggi, S.; Romei, C.; Petrini, I.; Lucchesi, M.; Crucitta, S.; Rofi, E.; De Liperi, A.; Chella, A.; et al. Integrating Liquid Biopsy and Radiomics to Monitor Clonal Heterogeneity of EGFR-Positive Non-Small Cell Lung Cancer. Front. Oncol. 2020, 10, 2664. [Google Scholar] [CrossRef]
- Roth, P.; Wischhusen, J.; Happold, C.; Chandran, P.A.; Hofer, S.; Eisele, G.; Weller, M.; Keller, A. A specific miRNA signature in the peripheral blood of glioblastoma patients. J. Neurochem. 2011, 118, 449–457. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Wong, C.W.; Chaudhry, A. Radiogenomics of lung cancer. J. Thorac. Dis. 2020, 12, 5104–5109. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.; Baldassano, S.N.; Loh, P.L.; Kording, K.; Litt, B.; Issadore, D. Machine Learning To Detect Signatures of Disease in Liquid Biopsies—A User’s Guide. Lab Chip 2018, 18, 395. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Liu, T.; Cui, J.; Borole, P.; Benjamin, A.; Kording, K.; Kording, K.; Issadore, D.; Issadore, D. A web-based automated machine learning platform to analyze liquid biopsy data. Lab Chip 2020, 20, 2166–2174. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.C.; Kim, K.H.; Woo, S.M.; Myung, J.K. Clinical multi-omics strategies for the effective cancer management. J. Proteom. 2018, 188, 97–106. [Google Scholar] [CrossRef]
- Alonzi, R. Functional Radiotherapy Targeting using Focused Dose Escalation. Clin. Oncol. 2015, 27, 601–617. [Google Scholar] [CrossRef]
- Rezaeijo, S.M.; Hashemi, B.; Mofid, B.; Bakhshandeh, M.; Mahdavi, A.; Hashemi, M.S. The feasibility of a dose painting procedure to treat prostate cancer based on mpMR images and hierarchical clustering. Radiat. Oncol. 2021, 16, 182. [Google Scholar] [CrossRef]
- Zhou, G.Q.; Wu, C.F.; Deng, B.; Gao, T.S.; Lv, J.W.; Lin, L.; Chen, F.P.; Kou, J.; Zhang, Z.X.; Huang, X.D.; et al. An optimal posttreatment surveillance strategy for cancer survivors based on an individualized risk-based approach. Nat. Commun. 2020, 11, 3872. [Google Scholar] [CrossRef]
- Ting, W.C.; Lu, Y.C.A.; Ho, W.C.; Cheewakriangkrai, C.; Chang, H.R.; Lin, C.L. Machine Learning in Prediction of Second Primary Cancer and Recurrence in Colorectal Cancer. Int. J. Med. Sci. 2020, 17, 280–291. [Google Scholar] [CrossRef]
- Chang, C.C.; Chen, S.H. Developing a Novel Machine Learning-Based Classification Scheme for Predicting SPCs in Breast Cancer Survivors. Front. Genet. 2019, 10, 848. [Google Scholar] [CrossRef]
- Thomas, C.; Mandrik, O.; Saunders, C.L.; Thompson, D.; Whyte, S.; Griffin, S.; Usher-Smith, J.A. The Costs and Benefits of Risk Stratification for Colorectal Cancer Screening Based On Phenotypic and Genetic Risk: A Health Economic Analysis. Cancer Prev. Res. 2021, 14, 811–822. [Google Scholar] [CrossRef]
- Hasnain, Z.; Mason, J.; Gill, K.; Miranda, G.; Gill, I.S.; Kuhn, P.; Newton, P.K. Machine learning models for predicting post-cystectomy recurrence and survival in bladder cancer patients. PLoS ONE 2019, 14, e0210976. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, P.M.; Robinson, E.J.; Pradhan, J.S.; Gartrell-Corrado, R.D.; Rohr, B.R.; Trager, M.H.; Geskin, L.J.; Kluger, H.M.; Wong, P.F.; Acs, B.; et al. Deep Learning Based on Standard H&E Images of Primary Melanoma Tumors Identifies Patients at Risk for Visceral Recurrence and Death. Clin. Cancer Res. 2020, 26, 1126–1134. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, R.; Long, J.; Saleem, A.; Rubin, D.L.; Shen, J. Deep learning predicts postsurgical recurrence of hepatocellular carcinoma from digital histopathologic images. Sci. Rep. 2021, 11, 2047. [Google Scholar] [CrossRef]
- Tokuyama, N.; Saito, A.; Muraoka, R.; Matsubara, S.; Hashimoto, T.; Satake, N.; Matsubayashi, J.; Nagao, T.; Mirza, A.H.; Graf, H.-P.; et al. Prediction of non-muscle invasive bladder cancer recurrence using machine learning of quantitative nuclear features. Mod. Pathol. 2021, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Jones, H.J.S.; Cunningham, C.; Askautrud, H.A.; Danielsen, H.E.; Kerr, D.J.; Domingo, E.; Maughan, T.; Leedham, S.J.; Koelzer, V.H. Stromal composition predicts recurrence of early rectal cancer after local excision. Histopathology 2021, 79, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Tang, X. The role of artificial intelligence in medical imaging research. BJR Open 2019, 2, 20190031. [Google Scholar] [CrossRef]
- Shen, W.C.; Chen, S.W.; Wu, K.C.; Hsieh, T.C.; Liang, J.A.; Hung, Y.C.; Yeh, L.S.; Chang, W.C.; Lin, W.C.; Yen, K.Y.; et al. Prediction of local relapse and distant metastasis in patients with definitive chemoradiotherapy-treated cervical cancer by deep learning from [18F]-fluorodeoxyglucose positron emission tomography/computed tomography. Eur. Radiol. 2019, 29, 6741–6749. [Google Scholar] [CrossRef]
- Zhang, W.; Fang, M.; Dong, D.; Wang, X.; Ke, X.; Zhang, L.; Hu, C.; Guo, L.; Guan, X.; Zhou, J.; et al. Development and validation of a CT-based radiomic nomogram for preoperative prediction of early recurrence in advanced gastric cancer. Radiother. Oncol. 2020, 145, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Lou, B.; Doken, S.; Zhuang, T.; Wingerter, D.; Gidwani, M.; Mistry, N.; Ladic, L.; Kamen, A.; Abazeed, M.E. An image-based deep learning framework for individualizing radiotherapy dose. Lancet. Digit. Health 2019, 1, e136–e147. [Google Scholar] [CrossRef]
- Cirillo, D.; Catuara-Solarz, S.; Morey, C.; Guney, E.; Subirats, L.; Mellino, S.; Gigante, A.; Valencia, A.; Rementeria, M.J.; Chadha, A.S.; et al. Sex and gender differences and biases in artificial intelligence for biomedicine and healthcare. NPJ Digit. Med. 2020, 3, 81. [Google Scholar] [CrossRef]
- Mhasawade, V.; Zhao, Y.; Chunara, R. Machine learning and algorithmic fairness in public and population health. Nat. Mach. Intell. 2021, 3, 659–666. [Google Scholar] [CrossRef]
- Winter, J.S. AI in healthcare: Data governance challenges. J. Hosp. Manag. Health Policy 2021, 5. [Google Scholar] [CrossRef]
- Morley, J.; Machado, C.C.V.; Burr, C.; Cowls, J.; Joshi, I.; Taddeo, M.; Floridi, L. The ethics of AI in health care: A mapping review. Soc. Sci. Med. 2020, 260, 113172. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Ethics and Governance of Artificial Intelligence for Health: WHO Guidance; World Health Organization: Geneva, Switzerland, 2021; pp. 1–148.
- Hindocha, S.; Badea, C. Moral exemplars for the virtuous machine: The clinician’s role in ethical artificial intelligence for healthcare. AI Ethics 2021, 1, 1–9. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Zou, J.; Schiebinger, L. AI can be sexist and racist—It’s time to make it fair. Nature 2018, 559, 324–326. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Khan, S.M.; Xu, A.J.; Ibrahim, H.; Smith, L.; Caballero, J.; Zepeda, L.; de Blas Perez, C.; Denniston, A.K.; Liu, X.; et al. Characteristics of publicly available skin cancer image datasets: A systematic review. Lancet Digit. Health 2021, 4, e64–e74. [Google Scholar] [CrossRef]
- Robinson, W.R.; Renson, A.; Naimi, A.I. Teaching yourself about structural racism will improve your machine learning. Biostatistics 2020, 21, 339. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 1–9. [Google Scholar] [CrossRef]
- Kim, Y.G.; Kim, S.; Cho, C.E.; Song, I.H.; Lee, H.J.; Ahn, S.; Park, S.Y.; Gong, G.; Kim, N. Effectiveness of transfer learning for enhancing tumor classification with a convolutional neural network on frozen sections. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Wang, X.; Chen, H.; Xiang, H.; Lin, H.; Lin, X.; Heng, P.A. Deep virtual adversarial self-training with consistency regularization for semi-supervised medical image classification. Med. Image Anal. 2021, 70, 102010. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Bentley, P.; Mori, K.; Misawa, K.; Fujiwara, M.; Rueckert, D. Self-supervised learning for medical image analysis using image context restoration. Med. Image Anal. 2019, 58, 101539. [Google Scholar] [CrossRef] [PubMed]
- Azizi, S.; Mustafa, B.; Ryan, F.; Beaver, Z.; Freyberg, J.; Deaton, J.; Loh, A.; Karthikesalingam, A.; Kornblith, S.; Chen, T.; et al. Big Self-Supervised Models Advance Medical Image Classification. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 3478–3488. [Google Scholar]
- Liu, Y.; Zhou, Y.; Liu, X.; Dong, F.; Wang, C.; Wang, Z. Wasserstein GAN-Based Small-Sample Augmentation for New-Generation Artificial Intelligence: A Case Study of Cancer-Staging Data in Biology. Engineering 2019, 5, 156–163. [Google Scholar] [CrossRef]
- Vaidya, J.; Shafiq, B.; Jiang, X.; Ohno-Machado, L. Identifying inference attacks against healthcare data repositories. AMIA Summits Transl. Sci. Proc. 2013, 2013, 262–266. [Google Scholar]
- NHS Data Breach Affects 150,000 Patients in England—BBC News. Available online: https://www.bbc.co.uk/news/technology-44682369 (accessed on 14 December 2021).
- Kairouz, P.; McMahan, H.B.; Avent, B.; Bellet, A.; Bennis, M.; Bhagoji, A.N.; Bonawitz, K.; Charles, Z.; Cormode, G.; Cummings, R.; et al. Advances and Open Problems in Federated Learning. Found. Trends Mach. Learn. 2019, 14, 1–210. [Google Scholar] [CrossRef]
- Kaissis, G.; Ziller, A.; Passerat-Palmbach, J.; Ryffel, T.; Usynin, D.; Trask, A.; Lima, I.; Mancuso, J.; Jungmann, F.; Steinborn, M.M.; et al. End-to-end privacy preserving deep learning on multi-institutional medical imaging. Nat. Mach. Intell. 2021, 3, 473–484. [Google Scholar] [CrossRef]
- Baker, M. 1500 scientists lift the lid on reproducibility. Nature 2016, 533, 452–454. [Google Scholar] [CrossRef]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019, 17, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Li, R.; Liu, Z.; Chen, J.; Yang, Y.; Chen, H.; Lin, Z.; Lai, W.; Long, E.; Wu, X.; et al. Diagnostic Efficacy and Therapeutic Decision-making Capacity of an Artificial Intelligence Platform for Childhood Cataracts in Eye Clinics: A Multicentre Randomized Controlled Trial. EClinicalMedicine 2019, 9, 52–59. [Google Scholar] [CrossRef]
- Freeman, K.; Geppert, J.; Stinton, C.; Todkill, D.; Johnson, S.; Clarke, A.; Taylor-Phillips, S. Use of artificial intelligence for image analysis in breast cancer screening programmes: Systematic review of test accuracy. BMJ 2021, 374, n1872. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.S.; De Groot, J.A.; Dutton, S.; Omar, O.; Shanyinde, M.; Tajar, A.; Voysey, M.; Wharton, R.; Yu, L.M.; Moons, K.G.; et al. External validation of multivariable prediction models: A systematic review of methodological conduct and reporting. BMC Med. Res. Methodol. 2014, 14, 40. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Faes, L.; Calvert, M.J.; Denniston, A.K. Extension of the CONSORT and SPIRIT statements. Lancet 2019, 394, 1225. [Google Scholar] [CrossRef]
- Faes, L.; Liu, X.; Wagner, S.K.; Fu, D.J.; Balaskas, K.; Sim, D.; Bachmann, L.M.; Keane, P.A.; Denniston, A.K. A clinician’s guide to artificial intelligence: How to critically appraise machine learning studies. Transl. Vis. Sci. Technol. 2020, 9, 7. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed]
| Model | Type | Description | Example |
|---|---|---|---|
| LR | R | Uses logistic function to predict categorical outcomes | Chhatwal et al. [13] |
| SVM | R, C | Constructs hyperplanes to maximise data separation | Zhang et al. [14] |
| NB | C | Utilises Bayesian probability including priors for classification | Olatunji et al. [15] |
| RF | R, C | Ensembles predictions of random decision trees | Xiao et al. [16] |
| XGB | R, C | As RF, but sequential errors minimised by gradient descent | Liew et al. [17] |
| ANN | R, C | Multiplies input by weights and biases to predict outcome | Muhammad [18] |
| CNN | R, C | Uses kernels to detect image features | Suh [19] |
| Traditional Machine Learning | Deep Learning |
|---|---|
| Requires ROI segmentation | ROI segmentation optional |
| Features are pre-specified | Features generated by model |
| Features are easily quantified | Features difficult to quantify |
| Computationally less intensive | Computationally more intensive |
| May perform better on small datasets | May perform better on large datasets |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hunter, B.; Hindocha, S.; Lee, R.W. The Role of Artificial Intelligence in Early Cancer Diagnosis. Cancers 2022, 14, 1524. https://doi.org/10.3390/cancers14061524
Hunter B, Hindocha S, Lee RW. The Role of Artificial Intelligence in Early Cancer Diagnosis. Cancers. 2022; 14(6):1524. https://doi.org/10.3390/cancers14061524
Chicago/Turabian StyleHunter, Benjamin, Sumeet Hindocha, and Richard W. Lee. 2022. "The Role of Artificial Intelligence in Early Cancer Diagnosis" Cancers 14, no. 6: 1524. https://doi.org/10.3390/cancers14061524
APA StyleHunter, B., Hindocha, S., & Lee, R. W. (2022). The Role of Artificial Intelligence in Early Cancer Diagnosis. Cancers, 14(6), 1524. https://doi.org/10.3390/cancers14061524






