Phenotypic and Histological Distribution Analysis Identify Mast Cell Heterogeneity in Non-Small Cell Lung Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Patients Samples
2.2. Histology
2.3. Flow Cytometry
2.4. Statistical Analysis
2.5. Key Resources Table
3. Results
3.1. NSCLC Patient Cohort
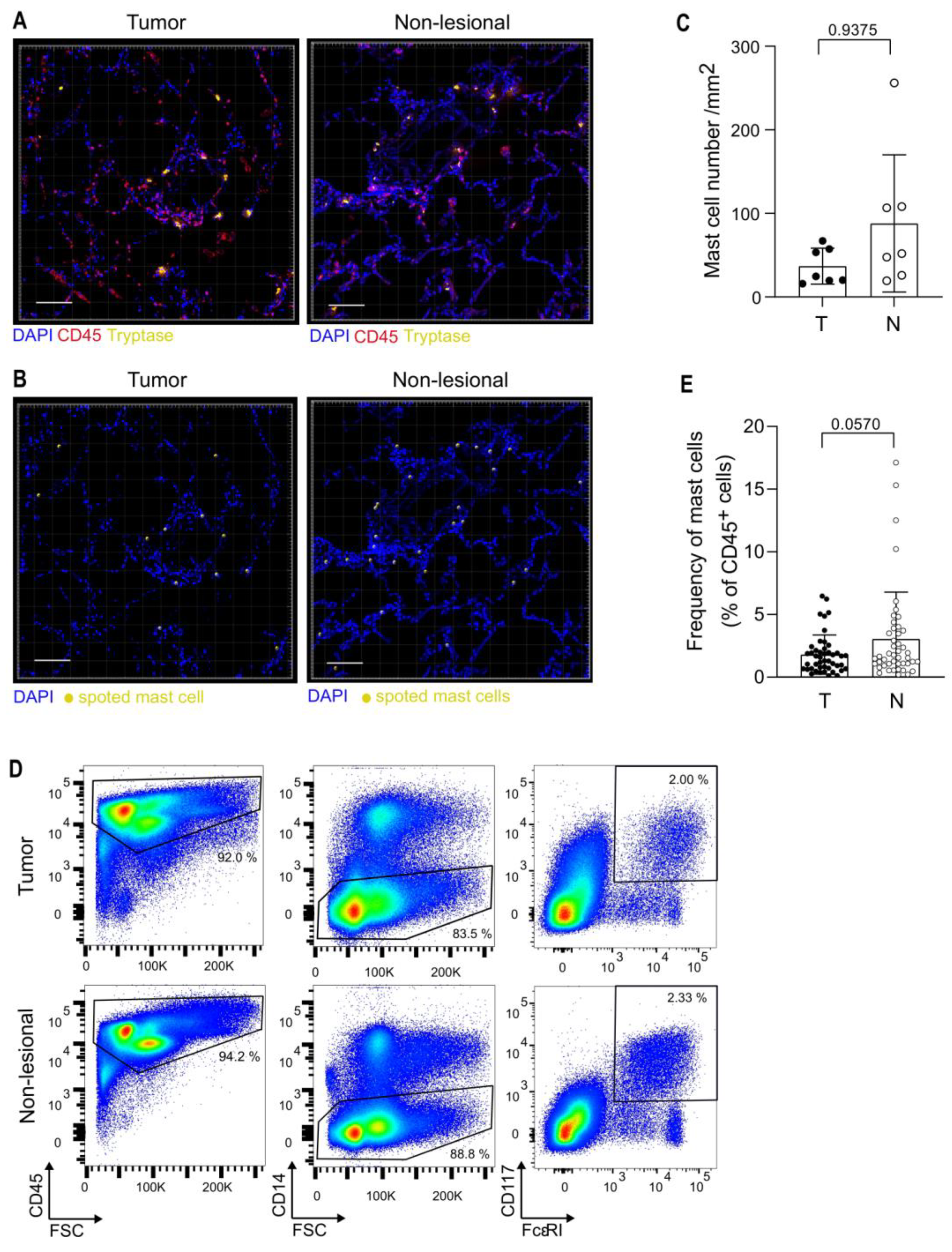
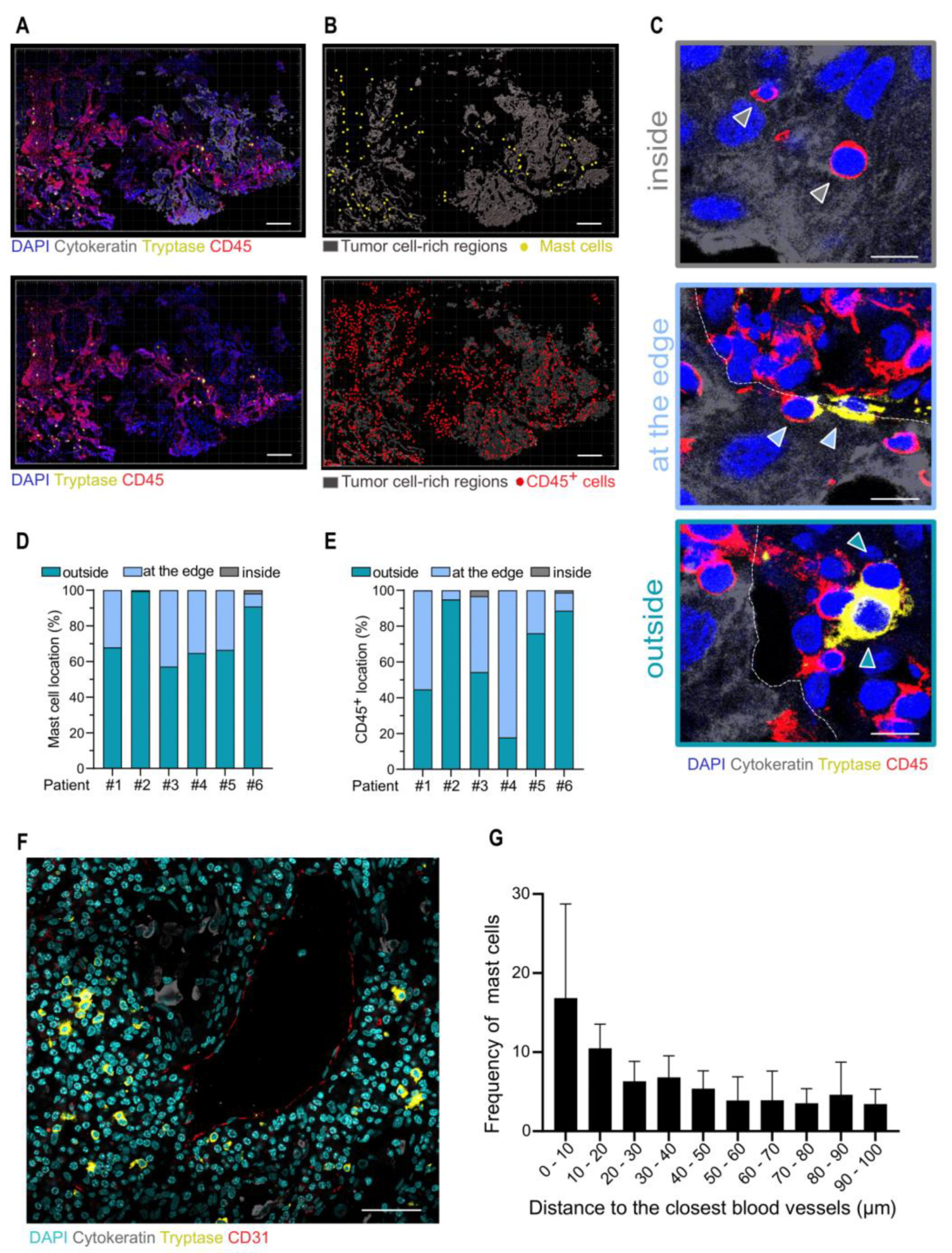
3.2. Analysis of Mast Cells in the TME
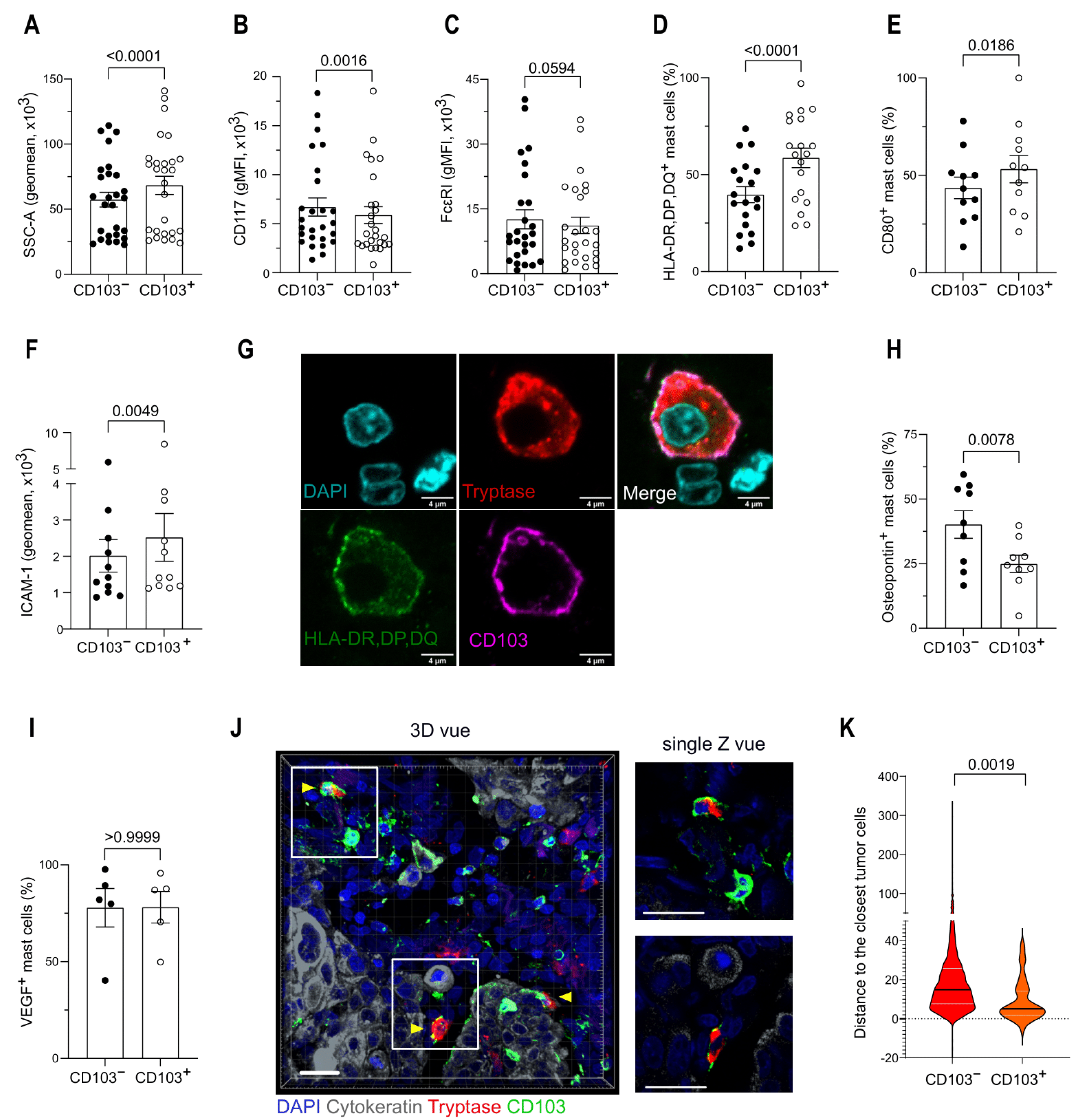
3.3. Mast Cell Phenotype Is Impacted by the Tumor Microenvironment
3.4. CD103 Identifies Two Distinct Mast Cell Populations in the TME
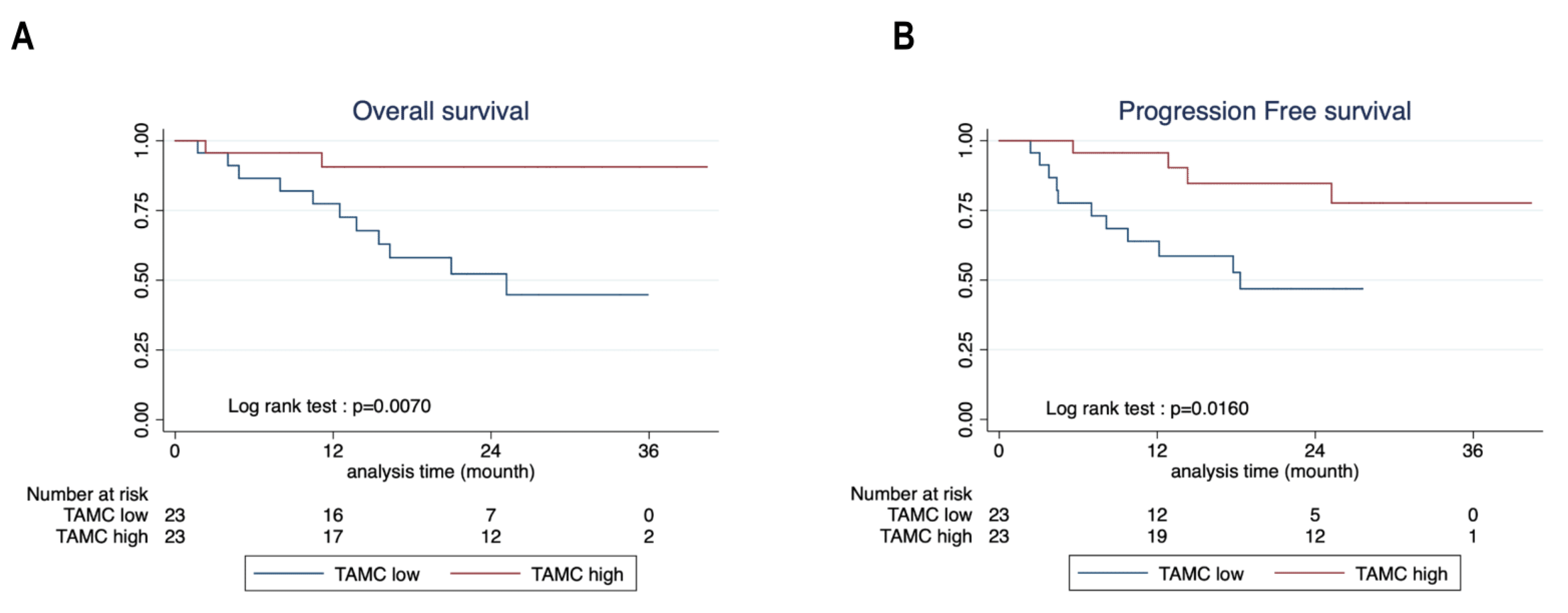
3.5. Mast Cells Are Associated with Better Survival in NSCLC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malvezzi, M.; Carioli, G.; Bertuccio, P.; Boffetta, P.; Levi, F.; La Vecchia, C.; Negri, E. European Cancer Mortality Predictions for the Year 2017, with Focus on Lung Cancer. Ann. Oncol. 2017, 28, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V.; et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina, J.R.; Yang, P.; Cassivi, S.D.; Schild, S.E.; Adjei, A.A. Non-Small Cell Lung Cancer: Epidemiology, Risk Factors, Treatment, and Survivorship. Mayo Clin. Proc. 2008, 83, 584–594. [Google Scholar] [CrossRef]
- Suresh, K.; Naidoo, J.; Lin, C.T.; Danoff, S. Immune Checkpoint Immunotherapy for Non-Small Cell Lung Cancer. Chest 2018, 154, 1416–1423. [Google Scholar] [CrossRef] [PubMed]
- Provencio, M.; Nadal, E.; Insa, A.; García-Campelo, M.R.; Casal-Rubio, J.; Dómine, M.; Majem, M.; Rodríguez-Abreu, D.; Martínez-Martí, A.; De Castro Carpeño, J.; et al. Neoadjuvant Chemotherapy and Nivolumab in Resectable Non-Small-Cell Lung Cancer (NADIM): An Open-Label, Multicentre, Single-Arm, Phase 2 Trial. Lancet Oncol. 2020, 21, 1413–1422. [Google Scholar] [CrossRef]
- Stankovic, B.; Bjørhovde, H.A.K.; Skarshaug, R.; Aamodt, H.; Frafjord, A.; Müller, E.; Hammarström, C.; Beraki, K.; Bækkevold, E.S.; Woldbæk, P.R.; et al. Immune Cell Composition in Human Non-Small Cell Lung Cancer. Front. Immunol. 2019, 9, 3101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, S.; Wei, M.; Wang, S.; Dong, J.; Wei, J. Pan-Cancer Analysis of Immune Cell Infiltration Identifies a Prognostic Immune-Cell Characteristic Score (ICCS) in Lung Adenocarcinoma. Front. Immunol. 2020, 11, 1218. [Google Scholar] [CrossRef]
- Banat, G.-A.; Tretyn, A.; Pullamsetti, S.S.; Wilhelm, J.; Weigert, A.; Olesch, C.; Ebel, K.; Stiewe, T.; Grimminger, F.; Seeger, W.; et al. Immune and Inflammatory Cell Composition of Human Lung Cancer Stroma. PLoS ONE 2015, 10, e0139073. [Google Scholar] [CrossRef]
- Gurish, M.F.; Austen, K.F. Developmental Origin and Functional Specialization of Mast Cell Subsets. Immunity 2012, 37, 25–33. [Google Scholar] [CrossRef] [Green Version]
- Andersson, C.K.; Mori, M.; Bjermer, L.; Lofdahl, C.-G.; Erjefalt, J.S. Novel Site-Specific Mast Cell Subpopulations in the Human Lung. Thorax 2009, 64, 297–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahlin, J.S.; Hallgren, J. Mast Cell Progenitors: Origin, Development and Migration to Tissues. Mol. Immunol. 2015, 63, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Abonia, J.P.; Hallgren, J.; Jones, T.; Shi, T.; Xu, Y.; Koni, P.; Flavell, R.A.; Boyce, J.A.; Austen, K.F.; Gurish, M.F. Alpha-4 Integrins and VCAM-1, but Not MAdCAM-1, Are Essential for Recruitment of Mast Cell Progenitors to the Inflamed Lung. Blood 2006, 108, 1588–1594. [Google Scholar] [CrossRef] [PubMed]
- Komi, D.E.A.; Redegeld, F.A. Role of Mast Cells in Shaping the Tumor Microenvironment. Clin. Rev. Allerg Immunol. 2020, 58, 313–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domagala, M.; Laplagne, C.; Leveque, E.; Laurent, C.; Fournié, J.-J.; Espinosa, E.; Poupot, M. Cancer Cells Resistance Shaping by Tumor Infiltrating Myeloid Cells. Cancers 2021, 13, 165. [Google Scholar] [CrossRef]
- Varricchi, G.; Galdiero, M.R.; Loffredo, S.; Marone, G.; Iannone, R.; Marone, G.; Granata, F. Are Mast Cells MASTers in Cancer? Front. Immunol. 2017, 8, 424. [Google Scholar] [CrossRef] [Green Version]
- Aponte-López, A.; Fuentes-Pananá, E.M.; Cortes-Muñoz, D.; Muñoz-Cruz, S. Mast Cell, the Neglected Member of the Tumor Microenvironment: Role in Breast Cancer. J. Immunol. Res. 2018, 2018, 2584243. [Google Scholar] [CrossRef] [Green Version]
- Imada, A.; Shijubo, N.; Kojima, H.; Abe, S. Mast Cells Correlate with Angiogenesis and Poor Outcome in Stage I Lung Adenocarcinoma. Eur. Respir. J. 2000, 15, 1087. [Google Scholar] [CrossRef] [Green Version]
- Welsh, T.J.; Green, R.H.; Richardson, D.; Waller, D.A.; O’Byrne, K.J.; Bradding, P. Macrophage and Mast-Cell Invasion of Tumor Cell Islets Confers a Marked Survival Advantage in Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2005, 23, 8959–8967. [Google Scholar] [CrossRef]
- Tomita, M.; Matsuzaki, Y.; Onitsuka, T. Correlation between Mast Cells and Survival Rates in Patients with Pulmonary Adenocarcinoma. Lung Cancer 1999, 26, 103–108. [Google Scholar] [CrossRef]
- Tataroğlu, C.; Kargı, A.; Özkal, S.; Eşrefoğlu, N.; Akkoçlu, A. Association of Macrophages, Mast Cells and Eosinophil Leukocytes with Angiogenesis and Tumor Stage in Non-Small Cell Lung Carcinomas (NSCLC). Lung Cancer 2004, 43, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Dou, A.; Fang, J. Heterogeneous Myeloid Cells in Tumors. Cancers 2021, 13, 3772. [Google Scholar] [CrossRef] [PubMed]
- Campillo-Navarro, M.; Chávez-Blanco, A.D.; Wong-Baeza, I.; Serafín-López, J.; Flores-Mejía, R.; Estrada-Parra, S.; Estrada-García, I.; Chacón-Salinas, R. Mast Cells in Lung Homeostasis: Beyond Type I Hypersensitivity. Curr. Respir. Med. Rev. 2014, 10, 115–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankova, L.G.; Dwyer, D.F.; Liu, A.Y.; Austen, K.F.; Gurish, M.F. Maturation of Mast Cell Progenitors to Mucosal Mast Cells during Allergic Pulmonary Inflammation in Mice. Mucosal. Immunol. 2015, 8, 596–606. [Google Scholar] [CrossRef]
- Dahlin, J.S.; Malinovschi, A.; Öhrvik, H.; Sandelin, M.; Janson, C.; Alving, K.; Hallgren, J. Lin−CD34hi CD117int/Hi FcεRI+ Cells in Human Blood Constitute a Rare Population of Mast Cell Progenitors. Blood 2016, 127, 383–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derakhshan, T.; Samuchiwal, S.K.; Hallen, N.; Bankova, L.G.; Boyce, J.A.; Barrett, N.A.; Austen, K.F.; Dwyer, D.F. Lineage-Specific Regulation of Inducible and Constitutive Mast Cells in Allergic Airway Inflammation. J. Exp. Med. 2021, 218, e20200321. [Google Scholar] [CrossRef] [PubMed]
- Plum, T.; Wang, X.; Rettel, M.; Krijgsveld, J.; Feyerabend, T.B.; Rodewald, H.-R. Human Mast Cell Proteome Reveals Unique Lineage, Putative Functions, and Structural Basis for Cell Ablation. Immunity 2020, 52, 404–416. [Google Scholar] [CrossRef]
- Dwyer, D.F.; Barrett, N.A.; Austen, K.F. Expression Profiling of Constitutive Mast Cells Reveals a Unique Identity within the Immune System. Nat. Immunol. 2016, 17, 878–887. [Google Scholar] [CrossRef]
- Rönnberg, E.; Boey, D.Z.H.; Ravindran, A.; Säfholm, J.; Orre, A.-C.; Al-Ameri, M.; Adner, M.; Dahlén, S.-E.; Dahlin, J.S.; Nilsson, G. Immunoprofiling Reveals Novel Mast Cell Receptors and the Continuous Nature of Human Lung Mast Cell Heterogeneity. Front. Immunol. 2021, 12, 804812. [Google Scholar] [CrossRef]
- Atiakshin, D.; Samoilova, V.; Buchwalow, I.; Tiemann, M. Expression of CD38 in Mast Cells: Cytological and Histotopographic Features. Cells 2021, 10, 2511. [Google Scholar] [CrossRef]
- Lamort, A.-S.; Giopanou, I.; Psallidas, I.; Stathopoulos, G.T. Osteopontin as a Link between Inflammation and Cancer: The Thorax in the Spotlight. Cells 2019, 8, 815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dwyer, D.F.; Ordovas-Montanes, J.; Allon, S.J.; Buchheit, K.M.; Vukovic, M.; Derakhshan, T.; Feng, C.; Lai, J.; Hughes, T.K.; Nyquist, S.K.; et al. Human Airway Mast Cells Proliferate and Acquire Distinct Inflammation-Driven Phenotypes during Type 2 Inflammation. Sci. Immunol. 2021, 6, eabb7221. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J.C.; Schön, M.P. Integrin AE(CD103)Β7 in Epithelial Cancer. Cancers 2021, 13, 6211. [Google Scholar] [CrossRef]
- Corgnac, S.; Boutet, M.; Kfoury, M.; Naltet, C.; Mami-Chouaib, F. The Emerging Role of CD8+ Tissue Resident Memory T (TRM) Cells in Antitumor Immunity: A Unique Functional Contribution of the CD103 Integrin. Front. Immunol. 2018, 9, 1904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Floc’h, A.; Jalil, A.; Vergnon, I.; Chansac, B.L.M.; Lazar, V.; Bismuth, G.; Chouaib, S.; Mami-Chouaib, F. AEβ7 Integrin Interaction with E-Cadherin Promotes Antitumor CTL Activity by Triggering Lytic Granule Polarization and Exocytosis. J. Exp. Med. 2007, 204, 559–570. [Google Scholar] [CrossRef]
- Wright, S.H.; Brown, J.; Knight, P.A.; Thornton, E.M.; Kilshaw, P.J.; Miller, H.R.P. Transforming Growth Factor-Bβ 1 Mediates Coexpression of the Integrin Subunit Aα E and the Chymase Mouse Mast Cell Protease-1 during the Early Differentiation of Bone Marrow-Derived Mucosal Mast Cell Homologues: Integrin and Chymase Expression. Clin. Exp. Allergy 2002, 32, 315–324. [Google Scholar] [CrossRef]
- Brown, J.K.; Knight, P.A.; Pemberton, A.D.; Wright, S.H.; Pate, J.A.; Thornton, E.M.; Miller, H.R.P. Expression of Integrin-AE by Mucosal Mast Cells in the Intestinal Epithelium and Its Absence in Nematode-Infected Mice Lacking the Transforming Growth Factor-Β1-Activating Integrin Avβ6. Am. J. Pathol. 2004, 165, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Gaudenzio, N.; Espagnolle, N.; Mars, L.T.; Liblau, R.; Valitutti, S.; Espinosa, E. Cell-Cell Cooperation at the T Helper Cell/Mast Cell Immunological Synapse. Blood 2009, 114, 4979–4988. [Google Scholar] [CrossRef] [Green Version]
- Gaudenzio, N.; Laurent, C.; Valitutti, S.; Espinosa, E. Human Mast Cells Drive Memory CD4(+) T Cells toward an Inflammatory IL-22(+) Phenotype. J. Allergy Clin. Immunol. 2013, 131, 1400–1407. [Google Scholar] [CrossRef]
- Moorman, H.R.; Poschel, D.; Klement, J.D.; Lu, C.; Redd, P.S.; Liu, K. Osteopontin: A Key Regulator of Tumor Progression and Immunomodulation. Cancers 2020, 12, 3379. [Google Scholar] [CrossRef]
- Nagasaka, A.; Matsue, H.; Matsushima, H.; Aoki, R.; Nakamura, Y.; Kambe, N.; Kon, S.; Uede, T.; Shimada, S. Osteopontin Is Produced by Mast Cells and Affects IgE-Mediated Degranulation and Migration of Mast Cells. Eur. J. Immunol. 2008, 38, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wang, X. Role of Osteopontin in Lung Cancer Evolution and Heterogeneity. Semin. Cell Dev. Biol. 2017, 64, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Castello, L.M.; Raineri, D.; Salmi, L.; Clemente, N.; Vaschetto, R.; Quaglia, M.; Garzaro, M.; Gentilli, S.; Navalesi, P.; Cantaluppi, V.; et al. Osteopontin at the Crossroads of Inflammation and Tumor Progression. Mediat. Inflamm. 2017, 2017, 4049098. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D.; Crivellato, E. Mast Cells, Angiogenesis, and Tumour Growth. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2012, 1822, 2–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shikotra, A.; Ohri, C.M.; Green, R.H.; Waller, D.A.; Bradding, P. Mast Cell Phenotype, TNFα Expression and Degranulation Status in Non-Small Cell Lung Cancer. Sci. Rep. 2016, 6, 38352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grujic, M.; Hellman, L.; Gustafson, A.-M.; Akula, S.; Melo, F.R.; Pejler, G. Protective Role of Mouse Mast Cell Tryptase Mcpt6 in Melanoma. Pigment. Cell Melanoma Res. 2020, 33, 579–590. [Google Scholar] [CrossRef]
- Rabelo Melo, F.; Santosh Martin, S.; Sommerhoff, C.P.; Pejler, G. Exosome-Mediated Uptake of Mast Cell Tryptase into the Nucleus of Melanoma Cells: A Novel Axis for Regulating Tumor Cell Proliferation and Gene Expression. Cell Death Dis. 2019, 10, 659. [Google Scholar] [CrossRef] [Green Version]


| Characteristics | Patients n = 48 |
|---|---|
| Sex | |
| Male | 28 (58.33) |
| Female | 20 (41.67) |
| Age, year | 64.02 (61.39–66.65) |
| Smoking history | 44 (91.67) |
| Histology | |
| LUAD | 35 (72.92) |
| LUSC | 9 (18.75) |
| Other | 4 (8.33) |
| pTNM staging | |
| I | 9 (18.75) |
| II | 12 (25.00) |
| III | 27 (56.25) |
| IV | 0 |
| Reagent or Resource | Source | Identifier | Dilution |
|---|---|---|---|
| Antibodies—Flow Cytometry | |||
| Mouse Anti-Human CD117 (c-Kit) BUV395 | BD Biosciences | Cat# 745733 RRID: AB_2743207 | 1/50 |
| Mouse anti-Human FcεRI APC | eBiosciences | Cat# 17-5899-42 RRID: AB_10671394 | 1/50 |
| Mouse anti-Human HLA-DR,DP,DQ BV650 | BD Biosciences | Cat# 740582 RRID:AB_2740041 | 1/100 |
| Mouse anti-Human CD54 BV786 | BD Biosciences | Cat# 740978 RRID: AB_2740602 | 1/50 |
| Mouse anti-Human CD80 PE-Cy5 | BD Biosciences | Cat# 559370 RRID: AB_397239 | 1/25 |
| Mouse anti-Human CD38 Pe-vio 770 | Miltenyi Biotec | Cat# 130-118-982 RRID: AB_2751601 | 1/50 |
| Mouse anti-Human CD45 Amcyan | Miltenyi Biotec | Cat# 130-110-638 RRID: AB_2658245 | 1/100 |
| Mouse anti-Human CD103 BV 421 | BD Biosciences | Cat# 563882 RRID: AB_2738464 | 1/50 |
| Mouse anti-Human CD14 PECF-594 | BD Biosciences | Cat# 562335 RRID: AB_11153663 | 1/100 |
| Mouse anti-Human MRGX2 PE | Biolegend | Cat# 359003 RRID: AB_2562300 | 1/25 |
| Rat anti-Integrin b7 BV650 | BD Biosciences | Cat# 564284 RRID:AB_2738729 | 1/50 |
| Mouse anti-Human osteopontin FITC | R and D systems | Cat# IC14331F RRID: AB_10888852 | 1/25 |
| Mouse anti-Human VEGF PE | R and D systems | Cat# IC2931P RRID: AB_357311 | 1/50 |
| Antibodies—Histology | |||
| Mouse (IgG1) anti Human-Tryptase | Agilent | Cat# M7052 RRID: AB_2206478 | 1/200 |
| Rabbit anti-Human CD103 | Abcam | Cat# ab224202 RRID: AB_2891141 | 1/100 |
| Rabbit anti-Human CD103 | Diagomics | Cat# BSB2864 | 1/100 |
| Rabbit anti-Human pan-cytokeratin | Thermo Fisher Scientific | Cat# PA127114 RRID: AB_780038 | 1/100 |
| Mouse (IgG2a) anti-Human CD31 | Thermo Fisher Scientific | Cat# MA3100 RRID: AB_223516 | 1/100 |
| Rat anti-Human CD45 | Thermo Fisher Scientific | Cat# MA517687 RRID: AB_2539077 | 1/100 |
| Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 555 | Thermo Fisher Scientific | Cat# A32732 RRID: AB_2633281 | 1/200 |
| Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 647 | Thermo Fisher Scientific | Cat# A32733 RRID: AB_2633282 | 1/200 |
| Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 660 | Thermo Fisher Scientific | Cat# A-21074 RRID: AB_2535735 | 1/200 |
| Goat anti-mouse IgG2a Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Thermo Fisher Scientific | Cat# A21135 RRID: AB_1500827 | 1/200 |
| Goat anti-mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A21121 RRID: AB_2535764 | 1/200 |
| Goat anti-Rat IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 546 | Thermo Fisher Scientific | Cat# A-11081 RRID: AB_141738 | 1/200 |
| Chemicals, Peptides, and Recombinant Proteins | |||
| eBioscience™ Fixable Viability Dye eFluor™ 780 | Thermo Fisher Scientific | 65-0865-18 | 1/500 |
| Citrate Buffer, pH 6.0, 10×, Antigen Retriever | Sigma-Aldrich | Cat# C9999 | |
| ACK lysing buffer | Gibco | Cat# A1049201 | |
| DAPI | Thermofisher | Cat# D1306 | 1 μg/mL |
| Critical Commercial Assays | |||
| Tumor Dissociation Kit, human | Miltenyi Biotec | Cat# 130-095-929 | |
| Software and Algorithms | |||
| GraphPad Prism 9 | graphpad.com | N/A | |
| Flowjo 10.0 | Flowjo.com | ||
| Zen | Carl Zeiss Microscopy | ||
| ImageJ | ImageJ | ||
| Imaris | Imaris | ||
| Characteristics | TAMC Low n = 23 (%) | TAMC High n = 23 (%) | p Value |
|---|---|---|---|
| Sex | 0.765 | ||
| Male | 14 (60.87) | 13 (56.52) | |
| Female | 9 (39.13) | 10 (43.48) | |
| Age, year | 65.39 (62.48–68.29) | 62.52 (57.75–67.29) | 0.292 |
| Smoking history | 22 (95.65) | 20 (86.96) | 0.295 |
| Histology | |||
| LUAD | 19 (82.61) | 14 (60.87) | 0.102 |
| LUSC | 3 (13.04) | 6 (26.09) | 0.265 |
| Other | 1 (4.35) | 3 (13.04) | 0.129 |
| pTNM staging | |||
| I | 5 (21.74) | 4 (17.39) | 0.710 |
| II | 3 (13.04) | 8 (34.78) | 0.223 |
| III | 15 (65.22) | 11 (47.83) | 0.429 |
| IV | 0 (0) | 0 (0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leveque, E.; Rouch, A.; Syrykh, C.; Mazières, J.; Brouchet, L.; Valitutti, S.; Espinosa, E.; Lafouresse, F. Phenotypic and Histological Distribution Analysis Identify Mast Cell Heterogeneity in Non-Small Cell Lung Cancer. Cancers 2022, 14, 1394. https://doi.org/10.3390/cancers14061394
Leveque E, Rouch A, Syrykh C, Mazières J, Brouchet L, Valitutti S, Espinosa E, Lafouresse F. Phenotypic and Histological Distribution Analysis Identify Mast Cell Heterogeneity in Non-Small Cell Lung Cancer. Cancers. 2022; 14(6):1394. https://doi.org/10.3390/cancers14061394
Chicago/Turabian StyleLeveque, Edouard, Axel Rouch, Charlotte Syrykh, Julien Mazières, Laurent Brouchet, Salvatore Valitutti, Eric Espinosa, and Fanny Lafouresse. 2022. "Phenotypic and Histological Distribution Analysis Identify Mast Cell Heterogeneity in Non-Small Cell Lung Cancer" Cancers 14, no. 6: 1394. https://doi.org/10.3390/cancers14061394
APA StyleLeveque, E., Rouch, A., Syrykh, C., Mazières, J., Brouchet, L., Valitutti, S., Espinosa, E., & Lafouresse, F. (2022). Phenotypic and Histological Distribution Analysis Identify Mast Cell Heterogeneity in Non-Small Cell Lung Cancer. Cancers, 14(6), 1394. https://doi.org/10.3390/cancers14061394






