Molecular Composition of Serum Exosomes Could Discriminate Rectal Cancer Patients with Different Responses to Neoadjuvant Radiotherapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
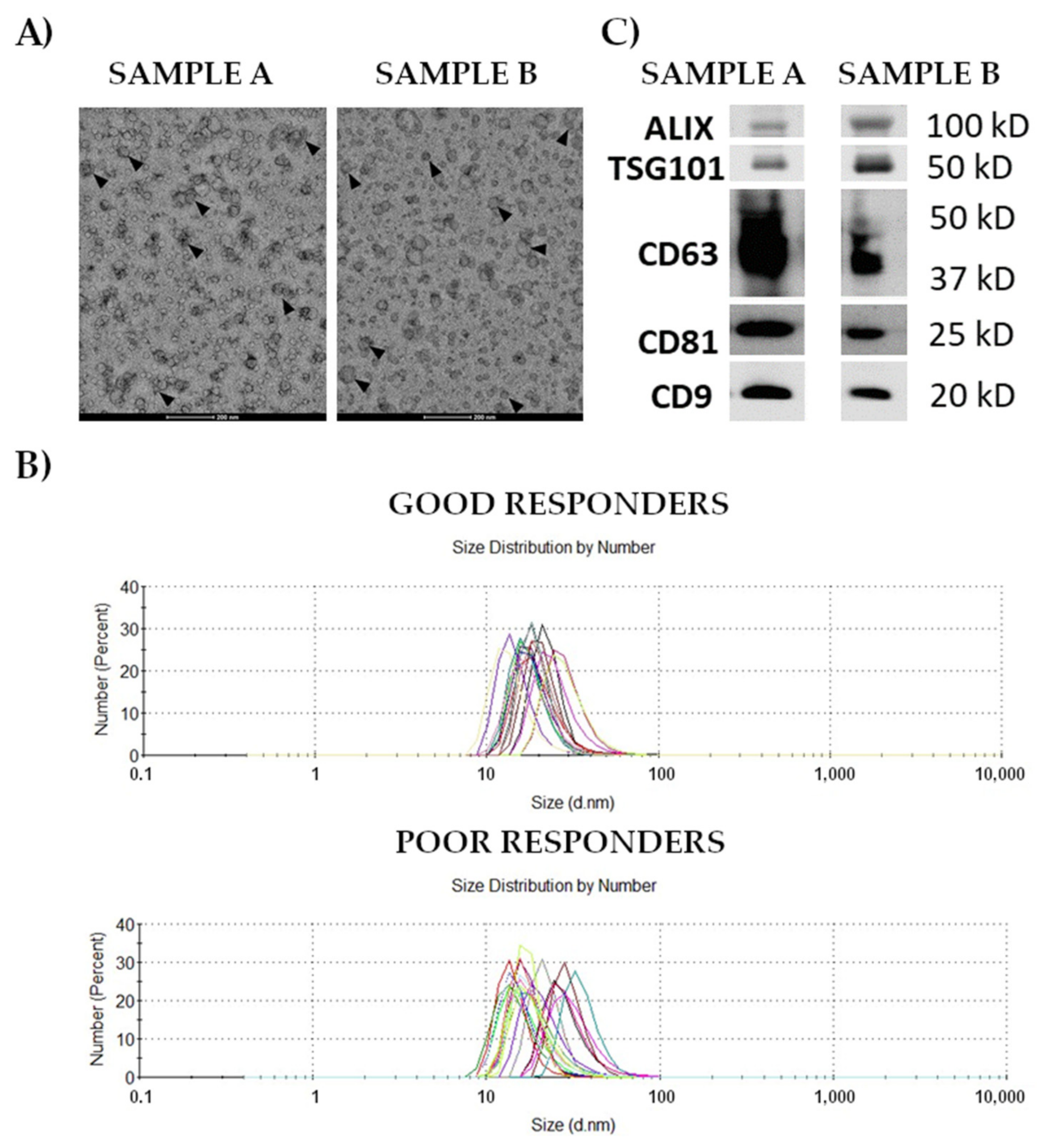
2.2. Isolation and Characterization of Small Extracellular Vesicles (Exosomes)
2.3. Protein Extraction, Peptide Generation, and LC-MS/MS Analysis
2.4. Metabolite Extraction, Derivatization, and GC/MS Analysis
2.5. Lipid Extraction and Mass Spectrometry Analysis
2.6. Statistical Analyses
2.7. Bioinformatics Analyses
3. Results
3.1. Characteristics and Composition of Serum-Derived Exosomes
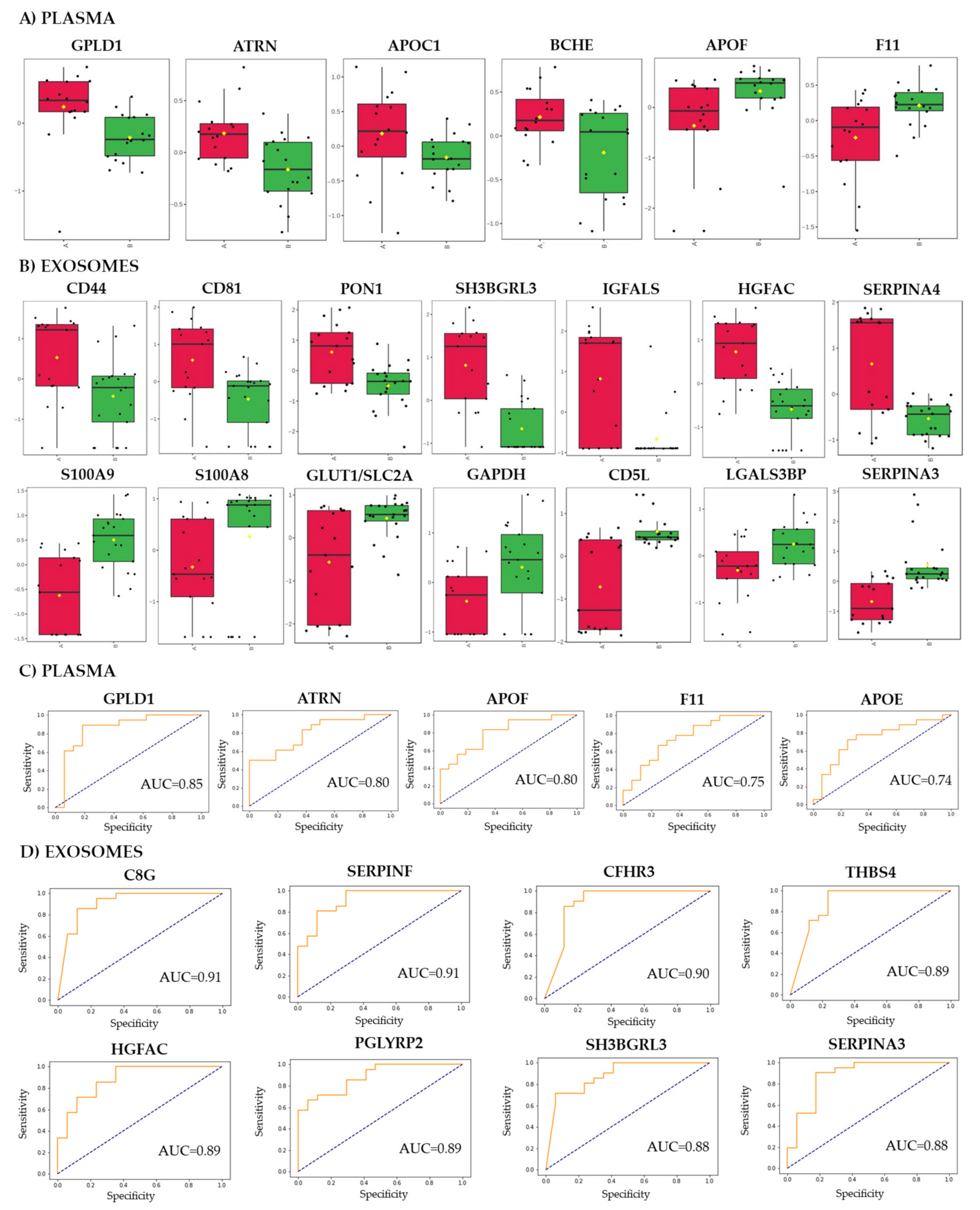
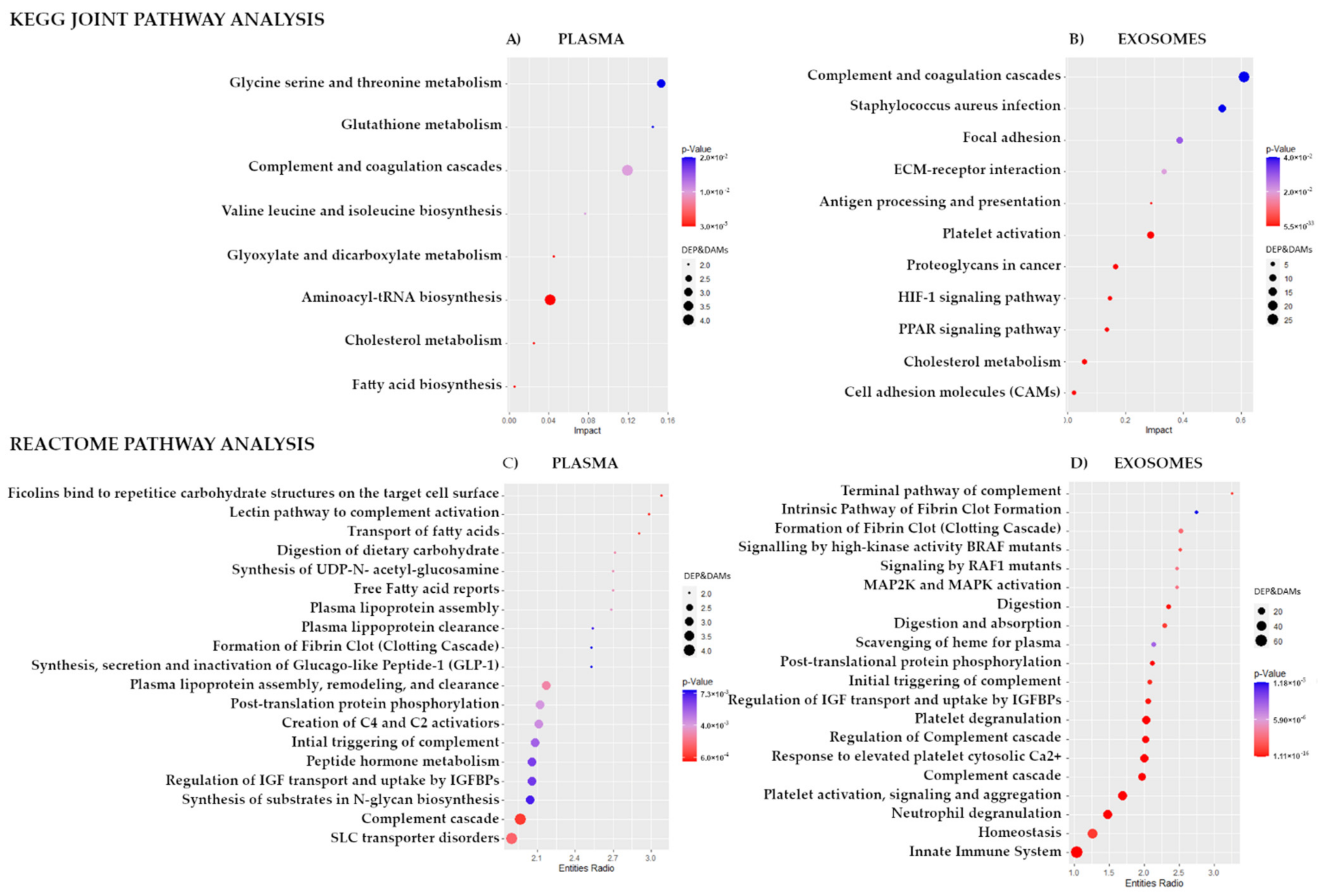
3.2. Proteins That Discriminated Patients with Different Responses to the Treatment
3.3. Metabolites That Discriminated Patients with Different Responses to the Treatment
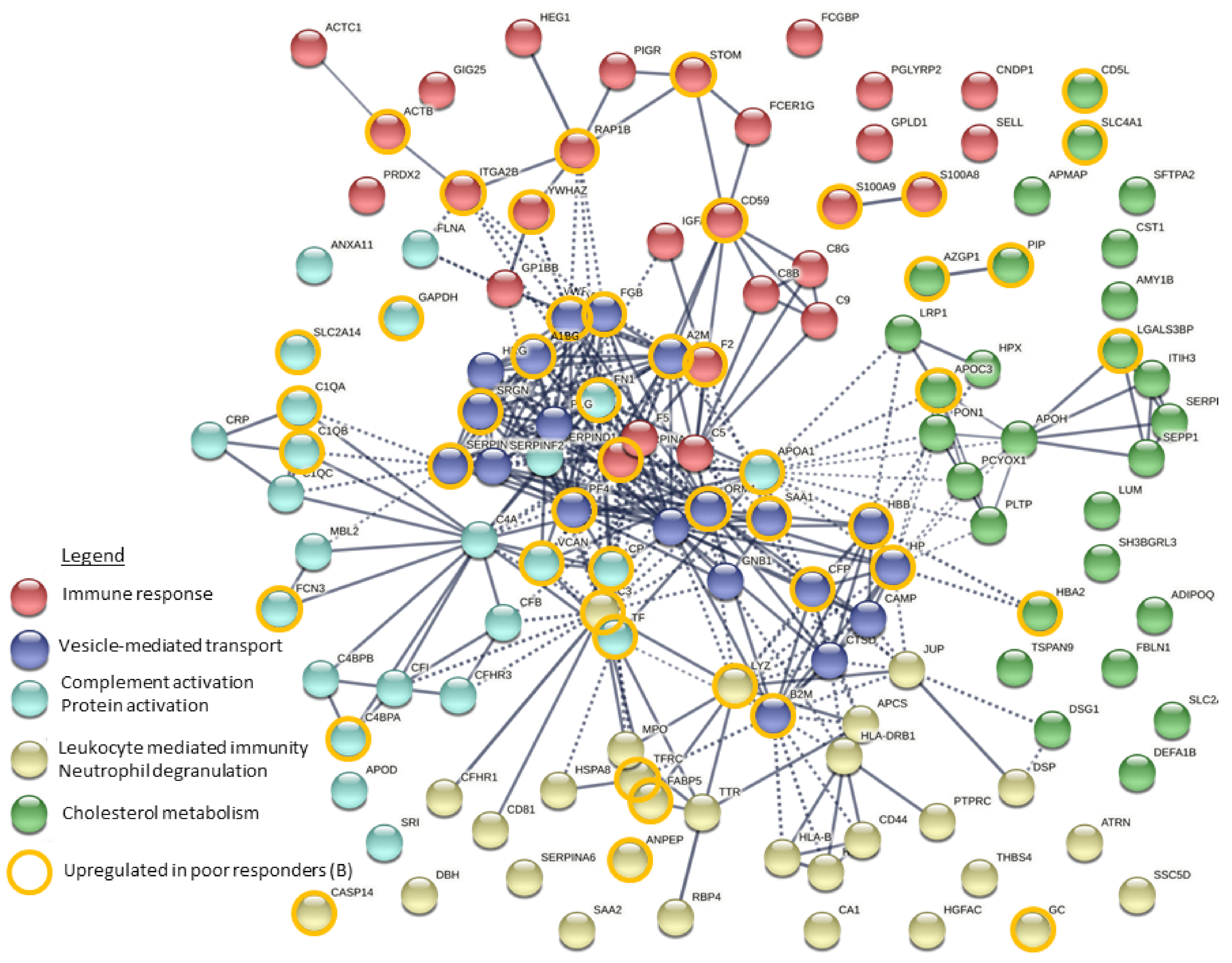
3.4. Integration of Data for Differently Expresses/Accumulated Proteins and Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Y.; Wang, J.; Ma, X.; Tan, L.; Yan, Y.; Xue, C.; Hui, B.; Liu, R.; Ma, H.; Ren, J. A Review of Neoadjuvant Chemoradiotherapy for Locally Advanced Rectal Cancer. Int. J. Biol. Sci. 2016, 12, 1022–1031. [Google Scholar] [CrossRef] [PubMed]
- Millan, M.; Merino, S.; Caro, A.; Feliu, F.; Escuder, J.; Francesh, T. Treatment of colorectal cancer in the elderly. World J. Gastrointest. Oncol. 2015, 7, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.-F.; Yu, W.D.; Pan, H.D.; Wang, L.; Li, W.; Yao, Y.-F.; Zhao, J.; Gu, J. Tumor regression grades: Potential outcome predictor of locally advanced rectal adenocarcinoma after preoperative radiotherapy. World J. Gastroenterol. 2015, 21, 1851. [Google Scholar] [CrossRef] [PubMed]
- Roeder, F.; Meldolesi, E.; Gerum, S.; Rödel, C. Recent advances in (chemo-)radiation therapy for rectal cancer: A comprehensive review. Radiat. Oncol. 2020, 15, 262. [Google Scholar] [CrossRef]
- Koncina, E.; Haan, S.; Rauh, S.; Letellier, E. Prognostic and Predictive Molecular Biomarkers for Colorectal Cancer: Updates and Challenges. Cancers 2020, 12, 319. [Google Scholar] [CrossRef]
- Lu, M.; Zhan, X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 2018, 9, 77–102. [Google Scholar] [CrossRef]
- Lacombe, J.; Azria, D.; Mange, A.; Solassol, J. Proteomic approaches to identify biomarkers predictive of radiotherapy outcomes. Expert Rev. Proteom. 2013, 10, 33–42. [Google Scholar] [CrossRef]
- Chang, L.; Graham, P.; Hao, J.; Bucci, J.; Malouf, D.; Gillatt, D.; Li, Y. Proteomics discovery of radioresistant cancer biomarkers for radiotherapy. Cancer Lett. 2015, 369, 289–297. [Google Scholar] [CrossRef]
- Jelonek, K.; Pietrowska, M.; Widlak, P. Systemic effects of ionizing radiation at the proteome and metabolome levels in the blood of cancer patients treated with radiotherapy: The influence of inflammation and radiationtoxicity. Int. J. Radiat. Biol. 2017, 93, 683–696. [Google Scholar] [CrossRef]
- Holm, M.; Joenväärä, S.; Saraswat, M.; Tohmola, T.; Ristimäki, A.; Renkonen, R.; Haglund, C. Preoperative Radiotherapy Leads to Significant Differences in the Plasma Protein Profile of Rectal Cancer Patients. Oncology 2020, 98, 493–500. [Google Scholar] [CrossRef]
- Allal, A.S.; Kähne, T.; Reverdin, A.K.; Lippert, H.; Schlegel, W.; Reymond, M.A. Radioresistance-related proteins in rectal cancer. Proteomics 2004, 4, 2261–2269. [Google Scholar] [CrossRef]
- Repetto, O.; De Re, V.; Paoli, A.; De Belluco, C.; Alessandrini, L.; Canzonieri, V.; Cannizzaro, R. Identification of protein clusters predictive of tumor response in rectal cancer patients receiving neoadjuvant chemo-radiotherapy. Oncotarget 2017, 8, 28328–28341. [Google Scholar] [CrossRef]
- Bowden, D.L.; Sutton, P.A.; Wall, M.A.; Jithesh, P.V.; Jenkins, R.E.; Palmer, D.H.; Goldring, C.E.; Parsons, J.L.; Park, B.K.; Kitteringham, N.R.; et al. Proteomic profiling of rectal cancer reveals acid ceramidase is implicated in radiation response. J. Proteom. 2018, 179, 53–60. [Google Scholar] [CrossRef]
- Jia, H.; Shen, X.; Guan, Y.; Xu, M.; Tu, J.; Mo, M.; Xie, L.; Yuan, J.; Zhang, Z.; Cai, S.; et al. Predicting the pathological response to neoadjuvant chemoradiation using untargeted metabolomics in locally advanced rectal cancer. Radiother. Oncol. 2018, 128, 548–556. [Google Scholar] [CrossRef]
- Rodríguez-Tomàs, E.; Arenas, M.; Gómez, J.; Acosta, J.; Trilla, J.; López, Y.; Árquez, M.; Torres, L.; Araguas, P.; Hernández-Aguilera, A.; et al. Identification of potential metabolic biomarkers of rectal cancer and of the effect of neoadjuvant radiochemotherapy. PLoS ONE 2021, 16. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Whiteside, T.L. The potential of tumor-derived exosomes for noninvasive cancer monitoring. Expert Rev. Mol. Diagn. 2015, 15, 1293–1310. [Google Scholar] [CrossRef]
- Lafitte, M.; Lecointre, C.; Roche, S. Roles of exosomes in metastatic colon cancer. Am. J. Physiol. Cell Physiol. 2019, 317, C869–C880. [Google Scholar] [CrossRef]
- Hon, K.W.; Abu, N.; Ab Mutalib, N.S.; Jamal, R. Exosomes as Potential Biomarkers and Targeted Therapy in Colorectal Cancer: A Mini-Review. Front. Pharmacol. 2017, 8, 583. [Google Scholar] [CrossRef]
- Mannavola, F.; Salerno, T.; Passarelli, A.; Tucci, M.; Internò, V.; Silvestris, F. Revisiting the Role of Exosomes in Colorectal Cancer: Where Are We Now? Front. Oncol. 2019, 9, 521. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhong, J.; Zhong, B.; Huang, J.; Jiang, L.; Jiang, Y.; Yuan, J.; Sun, J.; Dai, L.; Yang, C.; et al. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett. 2020, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-C.; Chan, M.-H.; Li, C.-H.; Fang, C.-Y.; Hsiao, M.; Chen, C.-L. Exosomal Components and Modulators in Colorectal Cancer: Novel Diagnosis and Prognosis Biomarkers. Biomedicines 2021, 9, 931. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Wang, X.; Gong, Z.; Yu, M.; Wu, H.; Zhang, D. Exosome-mediated metabolic reprogramming: The emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct. Target. Ther. 2020, 5, 242. [Google Scholar] [CrossRef] [PubMed]
- Eylem, C.C.; Yilmaz, M.; Derkus, B.; Nemutlu, E.; Camci, C.B.; Yilmaz, E.; Turkoglu, M.A.; Aytac, B.; Ozyurt, N.; Emregul, E. Untargeted multi-omic analysis of colorectal cancer-specific exosomes reveals joint pathways of colorectal cancer in both clinical samples and cell culture. Cancer Lett. 2019, 469, 186–194. [Google Scholar] [CrossRef]
- Jelonek, K.; Widlak, P.; Pietrowska, M. The Influence of Ionizing Radiation on Exosome Composition, Secretion and Intercellular Communication. Protein Pept. Lett. 2016, 23, 656–663. [Google Scholar] [CrossRef]
- Steinbichler, T.B.; Dudás, J.; Skvortsov, S.; Ganswindt, U.; Riechelmann, H.; Skvortsova, I.I. Therapy resistance mediated by exosomes. Mol. Cancer 2019, 18, 58. [Google Scholar] [CrossRef]
- Ni, J.; Bucci, J.; Malouf, D.; Knox, M.; Graham, P.; Li, Y. Exosomes in Cancer Radioresistance. Front. Oncol. 2019, 9, 869. [Google Scholar] [CrossRef]
- Smolarz, M.; Pietrowska, M.; Matysiak, N.; Mielanczyk, Ł.; Widlak, P. Proteome Profiling of Exosomes Purified from a Small Amount of Human Serum: The Problem of Co-Purified Serum Components. Proteomes 2019, 7, 18. [Google Scholar] [CrossRef]
- Ludwig, N.; Hong, C.S.; Ludwig, S.; Azambuja, J.H.; Sharma, P.; Theodoraki, M.-N.; Whiteside, T.L. Isolation and Analysis of Tumor-Derived Exosomes. Curr. Protoc. Immunol. 2019, 127, e91. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef]
- Herzog, R.; Schuhmann, K.; Schwudke, D.; Sampaio, J.L.; Bornstein, S.R.; Schroeder, M.; Shevchenko, A. Lipid Xplorer: A software for consensual cross-platform lipidomics. PLoS ONE 2012, 7, e29851. [Google Scholar] [CrossRef]
- Sullivan, G.M.; Feinn, R. Using Effect Size-or Why the P Value Is Not Enough. J. Grad. Med Educ. 2012, 4, 279–282. [Google Scholar] [CrossRef]
- Cureton, E.E. Rank-biserial correlation. Psychometrika 1956, 21, 287–290. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 7, 1–7. [Google Scholar] [CrossRef]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2019, 1, D498–D503. [Google Scholar] [CrossRef]
- Yin, T.; Xin, H.; Yu, J.; Teng, F. The role of exosomes in tumour immunity under radiotherapy: Eliciting abscopal effects? Biomark. Res. 2021, 9, 1–12. [Google Scholar] [CrossRef]
- Fang, J.; Zhou, S.-H.; Fan, J.; Yan, S.-X. Roles of glucose transporter-1 and the phosphatidylinositol 3kinase/protein kinase B pathway in cancer radioresistance (Review). Mol. Med. Rep. 2014, 11, 1573–1581. [Google Scholar] [CrossRef]
- Tang, L.; Wei, F.; Wu, Y.; He, Y.; Shi, L.; Xiong, F.; Gong, Z.; Guo, C.; Li, X.; Deng, H.; et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J. Exp. Clin. Cancer Res. 2018, 37, 87. [Google Scholar] [CrossRef]
- Multhoff, G.; Radons, J. Radiation, inflammation, and immune responses in cancer. Front. Oncol. 2012, 2, 58. [Google Scholar] [CrossRef]
- Cho, S.H.; Shim, H.J.; Park, M.R.; Choi, J.-N.; Akanda, M.D.; Hwang, J.-E.; Bae, W.-K.; Lee, K.-H.; Sun, E.-G.; Chung, I.-J. Lgals3bp suppresses colon inflammation and tumorigenesis through the downregulation of TAK1-NF-κB signaling. Cell Death Discov. 2021, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Sanjurjo, L.; Aran, G.; Téllez, É.; Amézaga, N.; Armengol, C.; López, D.; Prats, C.; Sarrias, M.-R. CD5L Promotes M2 Macrophage Polarization through Autophagy-Mediated Upregulation of ID3. Front. Immunol. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Zebrowska, A.; Skowronek, A.; Wojakowska, A.; Widlak, P.; Pietrowska, M. Metabolome of Exosomes: Focus on Vesicles Released by Cancer Cells and Present in Human Body Fluids. Int. J. Mol. Sci. 2019, 20, 3461. [Google Scholar] [CrossRef] [PubMed]
- Heeran, A.B.; Berrigan, H.P.; Buckley, C.E.; Bottu, H.M.; Prendiville, O.; Buckley, A.M.; Clarke, N.; Donlon, N.E.; Nugent, T.S.; Durand, M.; et al. Radiation-induced Bystander Effect (RIBE) alters mitochondrial metabolism using a human rectal cancer ex vivo explant model. Transl. Oncol. 2021, 14, 100882. [Google Scholar] [CrossRef]
- Jelonek, K.; Krzywon, A.; Jablonska, P.; Slominska, E.M.; Smolenski, R.T.; Polanska, J.; Rutkowski, T.; Mrochem-Kwarciak, J.; Skladowski, K.; Widlak, P. Systemic Effects of Radiotherapy and Concurrent Chemo-Radiotherapy in Head and Neck Cancer Patients—Comparison of Serum Metabolome Profiles. Metabolites 2020, 10, 60. [Google Scholar] [CrossRef]
- Ytting, H.; Christensen, I.J.; Thiel, S.; Jensenius, J.C.; Nielsen, H.J. Serum mannan-binding lectin-associated serine protease 2 levels in colorectal cancer: Relation to recurrence and mortality. Clin. Cancer Res. 2005, 11, 1441–1446. [Google Scholar] [CrossRef][Green Version]
- Żmigrodzka, M.; Witkowska-Piłaszewicz, O.; Winnicka, A. Platelets Extracellular Vesicles as Regulators of Cancer Progression-An Updated Perspective. Int. J. Mol. Sci. 2020, 21, 5195. [Google Scholar] [CrossRef]
- Gay, L.J.; Felding-Habermann, B. Contribution of platelets to tumour metastasis. Nat. Rev. Cancer 2011, 11, 123–134. [Google Scholar] [CrossRef]
- Lazar, I.; Clement, E.; Attane, C.; Muller, C.; Nieto, L. Thematic review series: Exosomes and microvesicles: Lipids as key components of their biogenesis and functions: A new role for extracellular vesicles: How small vesicles can feed tumors’ big appetite. J. Lipid Res. 2018, 59, 1793–1804. [Google Scholar] [CrossRef]
- Laiakis, E.C.; Mak, T.D.; Anizan, S.; Amundson, S.A.; Barker, C.A.; Wolden, S.L.; Brenner, D.J.; Fornace, A.J. Development of a metabolomic radiation signature in urine from patients undergoing total body irradiation. Radiat. Res. 2014, 181, 350–361. [Google Scholar] [CrossRef]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell–cell communication and various pathophysiologies. Biochim. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2014, 1841, 108–120. [Google Scholar] [CrossRef]
- Butler, L.M.; Perone, Y.; Dehairs, J.; Lupien, L.E.; de Laat, V.; Talebi, A.; Loda, M.; Kinlaw, W.B.; Swinnen, J.V. Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv. Drug Deliv. Rev. 2020, 159, 245–293. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; Hewapathirana, S.; García-Seisdedos, D.; Kamatchinathan, S.; Kundu, D.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucl. Acids Res. 2021, 50, D543–D552. [Google Scholar] [CrossRef]
| Total n (%) | Good Responders n (%) | Poor Responders n (%) | Difference p-Value (Test) | ||
|---|---|---|---|---|---|
| Sex | Females | 15 (37.5) | 5 (29.4) | 10 (43.5) | 0.57 (Chi2) |
| Males | 25 (62.5) | 12 (70.6) | 13 (56.5) | ||
| Age (years) | mean (S.D.) median | 65.9 (9.8) 65.5 | 64.9 (12.2) 67 | 66.5 (7.8) 65 | 0.98 (M-W U) |
| BMI | mean (SD) | 26.2 (3.5) | 25.0 (3.5) | 27.0 (3.3) | 0.047 (M-W U) |
| Clinical Stage | II | 13 (32.5) | 5 (29.4) | 8 (34.8) | 1.0 (Fisher) |
| III | 25 (62.5) | 11 (64.7) | 14 (60.9) | ||
| IV | 2 (5.0) | 1 (5.9) | 1 (4.3) | ||
| RT scheme | 39 Gy | 17 (42.5) | 8 (47.1) | 9 (39.1) | 0.1 (Fisher) |
| 42 Gy | 16 (40.0) | 4 (23.5) | 12 (52.2) | ||
| 54 Gy | 7 (17.5) | 5 (29.4) | 2 (8.7) | ||
| RT/CT | 20 | 10 | 10 | ||
| Time RT/S (days) | mean (SD) median | 52.7 (20.3) 49 | 54.6 (20.2) 52 | 51.3 (20.7) 49 | 0.53 (M-W U) |
| Surgery mode | AR | 26 (65.0) | 10 (58.8) | 16 (69.6) | 0.7 (Chi2) |
| APR | 14 (35.0) | 7 (41.2) | 7 (30.4) | ||
| ypT | 0–2 | 13 (32.5) | 7 (41.2) | 6 (26.1) | 0.5 (Chi2) |
| 3 | 27 (67.5) | 10 (58.8) | 17 (73.9) | ||
| ypN | negative | 24 (60.0) | 13 (76.5) | 11 (47.8) | 0.1 (Fisher) |
| positive | 16 (40.0) | 4 (23.5) | 12 (52.2) | ||
| LNY | mean (SD) | 12.3 (5.8) | 12.5 (5.1) | 12.1 (6.4) | 0.59 (M-W U) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strybel, U.; Marczak, L.; Zeman, M.; Polanski, K.; Mielańczyk, Ł.; Klymenko, O.; Samelak-Czajka, A.; Jackowiak, P.; Smolarz, M.; Chekan, M.; et al. Molecular Composition of Serum Exosomes Could Discriminate Rectal Cancer Patients with Different Responses to Neoadjuvant Radiotherapy. Cancers 2022, 14, 993. https://doi.org/10.3390/cancers14040993
Strybel U, Marczak L, Zeman M, Polanski K, Mielańczyk Ł, Klymenko O, Samelak-Czajka A, Jackowiak P, Smolarz M, Chekan M, et al. Molecular Composition of Serum Exosomes Could Discriminate Rectal Cancer Patients with Different Responses to Neoadjuvant Radiotherapy. Cancers. 2022; 14(4):993. https://doi.org/10.3390/cancers14040993
Chicago/Turabian StyleStrybel, Urszula, Lukasz Marczak, Marcin Zeman, Krzysztof Polanski, Łukasz Mielańczyk, Olesya Klymenko, Anna Samelak-Czajka, Paulina Jackowiak, Mateusz Smolarz, Mykola Chekan, and et al. 2022. "Molecular Composition of Serum Exosomes Could Discriminate Rectal Cancer Patients with Different Responses to Neoadjuvant Radiotherapy" Cancers 14, no. 4: 993. https://doi.org/10.3390/cancers14040993
APA StyleStrybel, U., Marczak, L., Zeman, M., Polanski, K., Mielańczyk, Ł., Klymenko, O., Samelak-Czajka, A., Jackowiak, P., Smolarz, M., Chekan, M., Zembala-Nożyńska, E., Widlak, P., Pietrowska, M., & Wojakowska, A. (2022). Molecular Composition of Serum Exosomes Could Discriminate Rectal Cancer Patients with Different Responses to Neoadjuvant Radiotherapy. Cancers, 14(4), 993. https://doi.org/10.3390/cancers14040993








