External Validation of a Radiomics Model for the Prediction of Complete Response to Neoadjuvant Chemoradiotherapy in Rectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
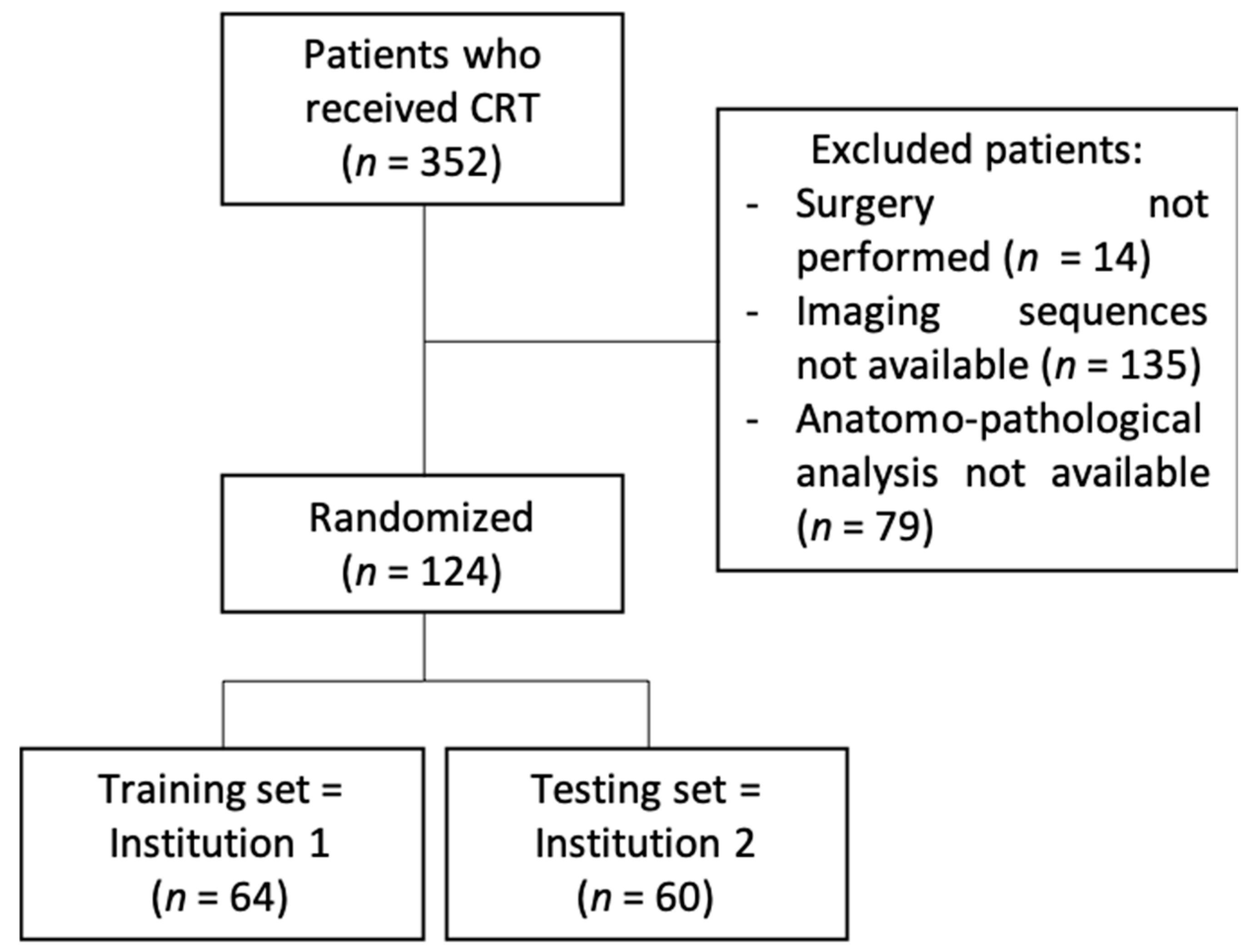
2.1. Patient Population
2.2. Outcome
2.3. MRI
2.4. Contrast-Enhanced CT Scan
2.5. Clinical Features
2.6. Tumor Delineation
2.7. Radiomic Features
2.8. Harmonization Method
2.9. Statistical Analysis
2.10. Inter-Individual Variability
2.11. Radiomics Quality Score
2.12. Ethical Considerations
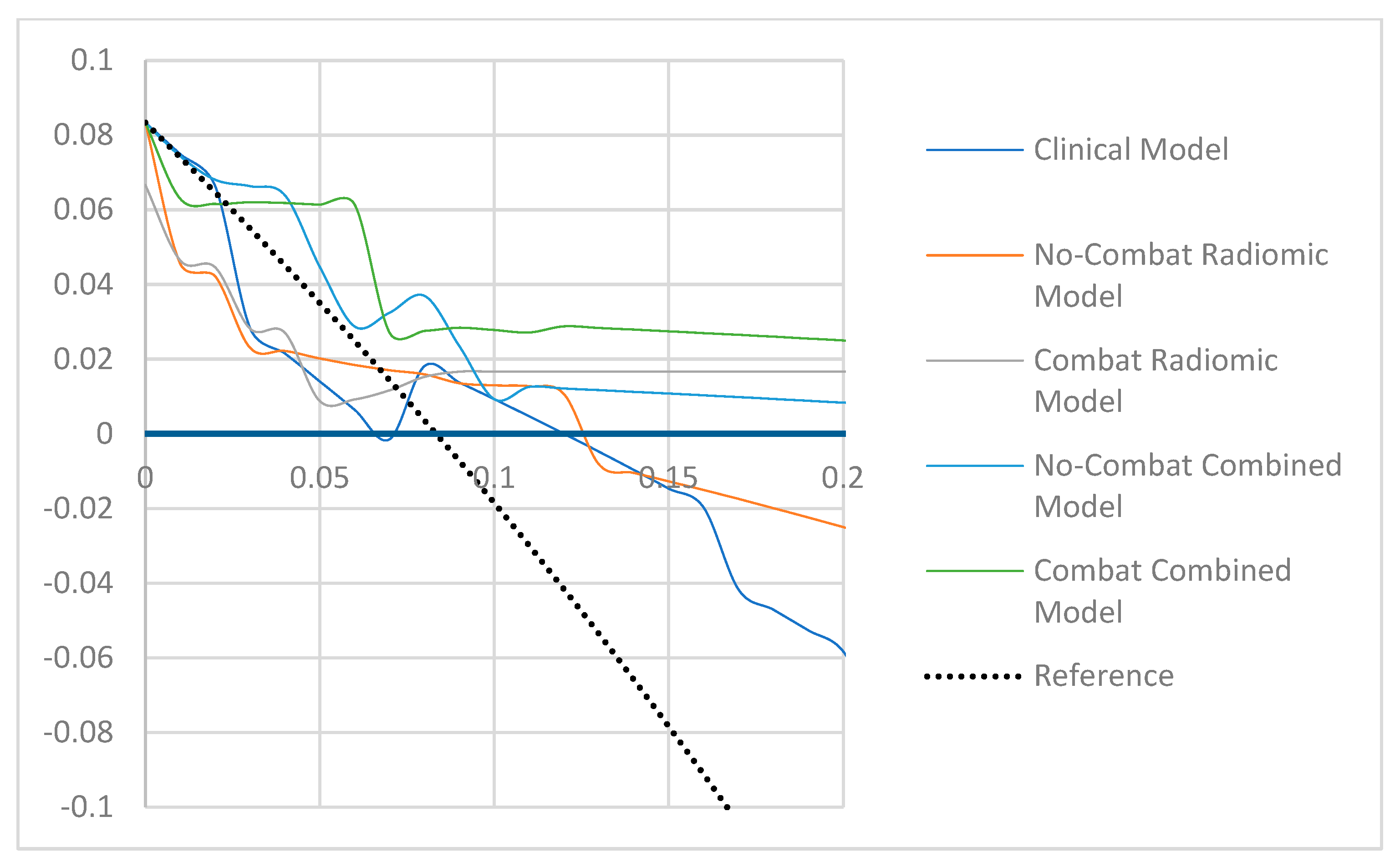
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- ECIS—European Cancer Information System. Available online: https://ecis.jrc.ec.europa.eu (accessed on 9 January 2021).
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA A Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Bahadoer, R.R.; Dijkstra, E.A.; van Etten, B.; Marijnen, C.A.M.; Putter, H.; Kranenbarg, E.M.-K.; Roodvoets, A.G.H.; Nagtegaal, I.D.; Beets-Tan, R.G.H.; Blomqvist, L.K.; et al. Short-course radiotherapy followed by chemotherapy before total mesorectal excision (TME) versus preoperative chemoradiotherapy, TME, and optional adjuvant chemotherapy in locally advanced rectal cancer (RAPIDO): A randomised, open-label, phase 3 trial. Lancet Oncol. 2021, 22, 29–42. [Google Scholar] [CrossRef]
- Conroy, T.; Bosset, J.-F.; Etienne, P.-L.; Rio, E.; François, E.; Mesgouez-Nebout, N.; Vendrely, V.; Artignan, X.; Bouché, O.; Gargot, D.; et al. Neoadjuvant chemotherapy with FOLFIRINOX and preoperative chemoradiotherapy for patients with locally advanced rectal cancer (UNICANCER-PRODIGE 23): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2021, 22, 702–715. [Google Scholar] [CrossRef]
- Gérard, J.-P.; André, T.; Bibeau, F.; Conroy, T.; Legoux, J.-L.; Portier, G.; Bosset, J.-F.; Cadiot, G.; Bouché, O.; Bedenne, L. Rectal cancer: French Intergroup clinical practice guidelines for diagnosis, treatments and follow-up (SNFGE, FFCD, GERCOR, UNICANCER, SFCD, SFED, SFRO). Dig. Liver Dis. 2017, 49, 359–367. [Google Scholar] [CrossRef]
- Ma, B.; Gao, P.; Wang, H.; Xu, Q.; Song, Y.; Huang, X.; Sun, J.; Zhao, J.; Luo, J.; Sun, Y.; et al. What has preoperative radio(chemo)therapy brought to localized rectal cancer patients in terms of perioperative and long-term outcomes over the past decades? A systematic review and meta-analysis based on 41,121 patients. Int. J. Cancer 2017, 141, 1052–1065. [Google Scholar] [CrossRef]
- Maas, M.; Nelemans, P.J.; Valentini, V.; Das, P.; Rödel, C.; Kuo, L.-J.; Calvo, F.A.; García-Aguilar, J.; Glynne-Jones, R.; Haustermans, K.; et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: A pooled analysis of individual patient data. Lancet Oncol. 2010, 11, 835–844. [Google Scholar] [CrossRef]
- Dossa, F.; Chesney, T.R.; Acuna, S.; Baxter, N.N. A watch-and-wait approach for locally advanced rectal cancer after a clinical complete response following neoadjuvant chemoradiation: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2017, 2, 501–513. [Google Scholar] [CrossRef]
- Maas, M.; Beets-Tan, R.G.; Lambregts, D.M.; Lammering, G.; Nelemans, P.J.; Engelen, S.M.; van Dam, R.M.; Jansen, R.L.; Sosef, M.; Leijtens, J.W.; et al. Wait-and-See Policy for Clinical Complete Responders After Chemoradiation for Rectal Cancer. J. Clin. Oncol. 2011, 29, 4633–4640. [Google Scholar] [CrossRef]
- Renehan, A.G.; Malcomson, L.; Emsley, R.; Scott, N.; O’Dwyer, S.T. OnCoRe project i: Watch-and-wait versus surgical resection for patients with rectal cancer—Authors’ reply. Lancet Oncol. 2016, 17, e134–e135. [Google Scholar] [CrossRef][Green Version]
- Davnall, F.; Yip, C.S.P.; Ljungqvist, G.; Selmi, M.; Ng, F.; Sanghera, B.; Ganeshan, B.; Miles, K.A.; Cook, G.; Goh, V. Assessment of tumor heterogeneity: An emerging imaging tool for clinical practice? Insights Imaging 2012, 3, 573–589. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Bourbonne, V.; Fournier, G.; Vallières, M.; Lucia, F.; Doucet, L.; Tissot, V.; Cuvelier, G.; Hue, S.; Du, H.L.P.; Perdriel, L.; et al. External Validation of an MRI-Derived Radiomics Model to Predict Biochemical Recurrence after Surgery for High-Risk Prostate Cancer. Cancers 2020, 12, 814. [Google Scholar] [CrossRef] [PubMed]
- Lucia, F.; Visvikis, D.; Desseroit, M.-C.; Miranda, O.; Malhaire, J.-P.; Robin, P.; Pradier, O.; Hatt, M.; Schick, U. Prediction of outcome using pretreatment 18F-FDG PET/CT and MRI radiomics in locally advanced cervical cancer treated with chemoradiotherapy. Eur. J. Pediatr. 2018, 45, 768–786. [Google Scholar] [CrossRef] [PubMed]
- Vandendorpe, B.; Durot, C.; Lebellec, L.; Le Deley, M.-C.; Sylla, D.; Bimbai, A.-M.; Amroun, K.; Ramiandrisoa, F.; Cordoba, A.; Mirabel, X.; et al. Prognostic value of the texture analysis parameters of the initial computed tomographic scan for response to neoadjuvant chemoradiation therapy in patients with locally advanced rectal cancer. Radiother. Oncol. 2019, 135, 153–160. [Google Scholar] [CrossRef]
- Giannini, V.; Mazzetti, S.; Bertotto, I.; Chiarenza, C.; Cauda, S.; Delmastro, E.; Bracco, C.; Di Dia, A.; Leone, F.; Medico, E.; et al. Predicting locally advanced rectal cancer response to neoadjuvant therapy with 18F-FDG PET and MRI radiomics features. Eur. J. Pediatr. 2019, 46, 878–888. [Google Scholar] [CrossRef]
- Glynne-Jones, R.; Wyrwicz, L.; Tiret, E.; Brown, G.; Rödel, C.; Cervantes, A.; Arnold, D. Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017, 28 (Suppl. 4), iv22–iv40. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
- Fortin, J.-P.; Parker, D.; Tunç, B.; Watanabe, T.; Elliott, M.A.; Ruparel, K.; Roalf, D.R.; Satterthwaite, T.D.; Gur, R.C.; Gur, R.E.; et al. Harmonization of multi-site diffusion tensor imaging data. NeuroImage 2017, 161, 149–170. [Google Scholar] [CrossRef]
- Johnson, W.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2006, 8, 118–127. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Bibault, J.E.; Giraud, P.; Housset, M.; Durdux, C.; Taieb, J.; Berger, A.; Coriat, R.; Chaussade, S.; Dousset, B.; Nordlinger, B.; et al. Deep Learning and Radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci. Rep. 2018, 8, 12611. [Google Scholar] [CrossRef] [PubMed]
- Shaish, H.; Aukerman, A.; Vanguri, R.; Spinelli, A.; Armenta, P.; Jambawalikar, S.; Makkar, J.; Bentley-Hibbert, S.; Del Portillo, A.; Kiran, R.; et al. Radiomics of MRI for pretreatment prediction of pathologic complete response, tumor regression grade, and neoadjuvant rectal score in patients with locally advanced rectal cancer undergoing neoadjuvant chemoradiation: An international multicenter study. Eur. Radiol. 2020, 30, 6263–6273. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Yang, X.; Shi, Z.; Yang, Z.; Du, X.; Zhao, Z.; Cheng, X. Radiomics analysis of multiparametric MRI for prediction of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Eur. Radiol. 2019, 29, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yi, Y.; Liu, Z.; Cao, W.; Lai, B.; Sun, K.; Li, L.; Zhou, Z.; Feng, Y.; Tian, J. Radiomics-Based Pretherapeutic Prediction of Non-response to Neoadjuvant Therapy in Locally Advanced Rectal Cancer. Ann. Surg. Oncol. 2019, 26, 1676–1684. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, X.-Y.; Shi, Y.-J.; Wang, L.; Zhu, H.-T.; Tang, Z.; Wang, S.; Li, X.-T.; Tian, J.; Sun, Y.-S. Radiomics Analysis for Evaluation of Pathological Complete Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Clin. Cancer Res. 2017, 23, 7253–7262. [Google Scholar] [CrossRef]
- Horvat, N.; Veeraraghavan, H.; Khan, M.; Blazic, I.; Zheng, J.; Capanu, M.; Sala, E.; Garcia-Aguilar, J.; Gollub, M.J.; Petkovska, I. MR Imaging of Rectal Cancer: Radiomics Analysis to Assess Treatment Response after Neoadjuvant Therapy. Radiology 2018, 287, 833–843. [Google Scholar] [CrossRef]
- Machackova, T.; Trachtova, K.; Prochazka, V.; Grolich, T.; Farkasova, M.; Fiala, L.; Sefr, R.; Kiss, I.; Skrovina, M.; Dosoudil, M.; et al. Tumor microRNAs Identified by Small RNA Sequencing as Potential Response Predictors in Locally Advanced Rectal Cancer Patients Treated With Neoadjuvant Chemoradiotherapy. Cancer Genom. Proteom. 2020, 17, 249–257. [Google Scholar] [CrossRef]
- Pazdirek, F.; Minarik, M.; Benesova, L.; Halkova, T.; Belsanova, B.; Macek, M.; Stepanek, L.; Hoch, J. Monitoring of Early Changes of Circulating Tumor DNA in the Plasma of Rectal Cancer Patients Receiving Neoadjuvant Concomitant Chemoradiotherapy: Evaluation for Prognosis and Prediction of Therapeutic Response. Front. Oncol. 2020, 10, 1028. [Google Scholar] [CrossRef]
- Badic, B.; Hatt, M.; Durand, S.; Le Jossic-Corcos, C.; Simon, B.; Visvikis, D.; Corcos, L. Radiogenomics-based cancer prognosis in colorectal cancer. Sci. Rep. 2019, 9, 9743. [Google Scholar] [CrossRef]
- Park, I.J.; Yu, Y.S.; Mustafa, B.; Park, J.Y.; Seo, Y.B.; Kim, G.-D.; Kim, J.; Kim, C.M.; Noh, H.D.; Hong, S.-M.; et al. A Nine-Gene Signature for Predicting the Response to Preoperative Chemoradiotherapy in Patients with Locally Advanced Rectal Cancer. Cancers 2020, 12, 800. [Google Scholar] [CrossRef] [PubMed]
- Issa, N.; Murninkas, A.; Schmilovitz-Weiss, H.; Agbarya, A.; Powsner, E. Transanal Endoscopic Microsurgery After Neoadjuvant Chemoradiotherapy for Rectal Cancer. J. Laparoendosc. Adv. Surg. Tech. 2015, 25, 617–624. [Google Scholar] [CrossRef] [PubMed]

| Variable | Total Cohort n = 124 | Training Set n = 64 | Testing Set n = 60 | p-Value |
|---|---|---|---|---|
| Mean age at diagnosis (years) | 65 (SD: 10.75) | 62 (SD: 11.8) | 68 (SD: 8.4) | 0.65 |
| Gender (male/female) | 76/47 | 37/27 | 40/20 | 0.91 |
| Degree of differentiation | ||||
| Well differentiated (%) | 43 (35%) | 26 (40.6%) | 19 (31.7%) | 0.82 |
| Moderately differentiated (%) | 58 (47%) | 32 (50%) | 23 (38.3%) | 0.43 |
| Undifferentiated (%) | 15 (12%) | 1 (1.5%) | 14 (23.3%) | 0.59 |
| High-grade dysplasia (%) | 8 (6%) | 4 (6.3%) | 6 (10%) | 0.59 |
| Mean ACE rate (ng/mL) | 8 (SD: 12.27) | 6.8 (SD: 7.2) | 9.7 (SD: 16.8) | 0.80 |
| cT stage | ||||
| cT 1 (%) | 1 (0.8%) | 0 (0%) | 1 (1.6%) | 0.99 |
| cT 2 (%) | 16 (13%) | 7 (10.9%) | 9 (15%) | 0.96 |
| cT 3 (%) | 97 (78.2%) | 52 (81.3%) | 28 (46.7%) | 0.23 |
| cT 4 (%) | 10 (8%) | 5 (7.8%) | 4 (6.6%) | 0.82 |
| N+ (%) | 95 (76%) | 50 (78%) | 44 (73%) | 0.99 |
| pCR (%) | 14 (11%) | 9 (14%) | 5 (8%) | 0.75 |
| Radiotherapy | 124 (100%) | |||
| 3D-CRT | 70 (56.5%) | 53 (82.8%) | 17 (28.3%) | <0.0001 |
| IMRT | 54 (43.5%) | 11 (17.2%) | 43 (71.7%) | |
| 45 Gy to the pelvis only | 70 (56%) | 59 (92%) | 10 (16.7%) | 0.04 |
| 45 Gy to the pelvis + boost up to 50.4 Gy to the rectal tumor | 54 (44%) | 5 (8%) | 50 (83.3%) | 0.03 |
| Concomitant chemotherapy | 118 (95%) | 64 (100%) | 54 (90%) | 0.39 |
| Capecitabine | 97 (78%) | 56 (88%) | 41 (68%) | 0.37 |
| Folfox | 21 (17%) | 8 (12%) | 13 (21.7%) | 0.42 |
| Duration of neoadjuvant therapy (mean, days) | 39 (SD: 4.71) | 38 (SD: 4.67) | 39 (SD: 6.11) | 0.93 |
| Delay between the end of treatment and surgery (mean, days) | 58 (SD: 13.19) | 59 (SD: 12.08) | 56 (SD: 15.05) | 0.82 |
| Model | AUC | p | Cut-Off (%) | Se (%) | Sp (%) | Bacc (%) | Below the Cut-Off | Above the Cut-Off | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total (n, %) | TN (n, %) | FN (n, %) | Total (n, %) | FP (n, %) | TP (n, %) | |||||||
| Clinical | 0.77 | 0.001 | 8.0 | 71.2 | 77.8 | 65.5 | 38 (59.4) | 36 (94.7) | 2 (5.3) | 26 (40.6) | 19 (73.1) | 7 (26.9) |
| Radiomic | 1.00 | <0.0001 | 23.0 | 100.0 | 96.4 | 98.2 | 53 (82.8) | 53 (100.0) | 0 (0.0) | 11 (17.2) | 2 (18.2) | 9 (81.8) |
| Combined | 0.97 | <0.0001 | 5.0 | 100.0 | 87.3 | 93.6 | 48 (75.0) | 48 (100.0) | 0 (0.0) | 16 (25.0) | 7 (43.7) | 9 (56.2) |
| ComBat_Radiomic | 1.00 | <0.0001 | 17 | 100.0 | 100.0 | 100.0 | 55 (85.9) | 55 (100.0) | 0 (0.0) | 9 (14.1) | 0 (0.0) | 9 (100.0) |
| ComBat_Combined | 0.95 | <0.0001 | 6.0 | 100.0 | 80.0 | 90.0 | 44 (68.7) | 44 (100.0) | 0 (0.0) | 20 (31.2) | 11 (55.0) | 9 (45.0) |
| Model | AUC | p | Cut-Off (%) | Se (%) | Sp (%) | Bacc (%) | Below the Cut-Off | Above the Cut-Off | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total (n, %) | TN (n, %) | FN (n, %) | Total (n, %) | FP (n, %) | TP (n, %) | |||||||
| Clinical | 0.50 | 1.00 | 8.0 | 60.0 | 60.0 | 60.0 | 35 (58.3) | 33 (94.3) | 2 (5.7) | 25 (41.7) | 22 (88.0) | 3 (12.0) |
| Radiomic | 0.69 | 0.07 | 23.0 | 20.0 | 81.8 | 50.9 | 49 (81.7) | 45 (91.8) | 4 (8.2) | 11 (18.3) | 10 (90.9) | 1 (9.1) |
| Combined | 0.77 | 0.004 | 5.0 | 80.0 | 60.0 | 70.0 | 34 (56.7) | 33 (91.1) | 1 (2.9) | 26 (43.3) | 22 (84.6) | 4 (15.4) |
| ComBat_Radiomic | 0.62 | 0.49 | 17 | 20.0 | 100.0 | 60.0 | 59 (98.3) | 55 (93.2) | 4 (6.8) | 1 (1.7) | 0 (0.0) | 1 (100.0) |
| ComBat_Combined | 0.81 | 0.03 | 6.0 | 80.0 | 90.9 | 85.5 | 51 (85.0) | 50 (98.0) | 1 (2.0) | 9 (15.0) | 5 (55.6) | 4 (44.4) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bordron, A.; Rio, E.; Badic, B.; Miranda, O.; Pradier, O.; Hatt, M.; Visvikis, D.; Lucia, F.; Schick, U.; Bourbonne, V. External Validation of a Radiomics Model for the Prediction of Complete Response to Neoadjuvant Chemoradiotherapy in Rectal Cancer. Cancers 2022, 14, 1079. https://doi.org/10.3390/cancers14041079
Bordron A, Rio E, Badic B, Miranda O, Pradier O, Hatt M, Visvikis D, Lucia F, Schick U, Bourbonne V. External Validation of a Radiomics Model for the Prediction of Complete Response to Neoadjuvant Chemoradiotherapy in Rectal Cancer. Cancers. 2022; 14(4):1079. https://doi.org/10.3390/cancers14041079
Chicago/Turabian StyleBordron, Anaïs, Emmanuel Rio, Bogdan Badic, Omar Miranda, Olivier Pradier, Mathieu Hatt, Dimitris Visvikis, François Lucia, Ulrike Schick, and Vincent Bourbonne. 2022. "External Validation of a Radiomics Model for the Prediction of Complete Response to Neoadjuvant Chemoradiotherapy in Rectal Cancer" Cancers 14, no. 4: 1079. https://doi.org/10.3390/cancers14041079
APA StyleBordron, A., Rio, E., Badic, B., Miranda, O., Pradier, O., Hatt, M., Visvikis, D., Lucia, F., Schick, U., & Bourbonne, V. (2022). External Validation of a Radiomics Model for the Prediction of Complete Response to Neoadjuvant Chemoradiotherapy in Rectal Cancer. Cancers, 14(4), 1079. https://doi.org/10.3390/cancers14041079






