Microsatellite Status and IκBα Expression Levels Predict Sensitivity to Pharmaceutical Curcumin in Colorectal Cancer Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Sensitivities to Curcumin and to Classical Chemotherapeutics Are Not Associated
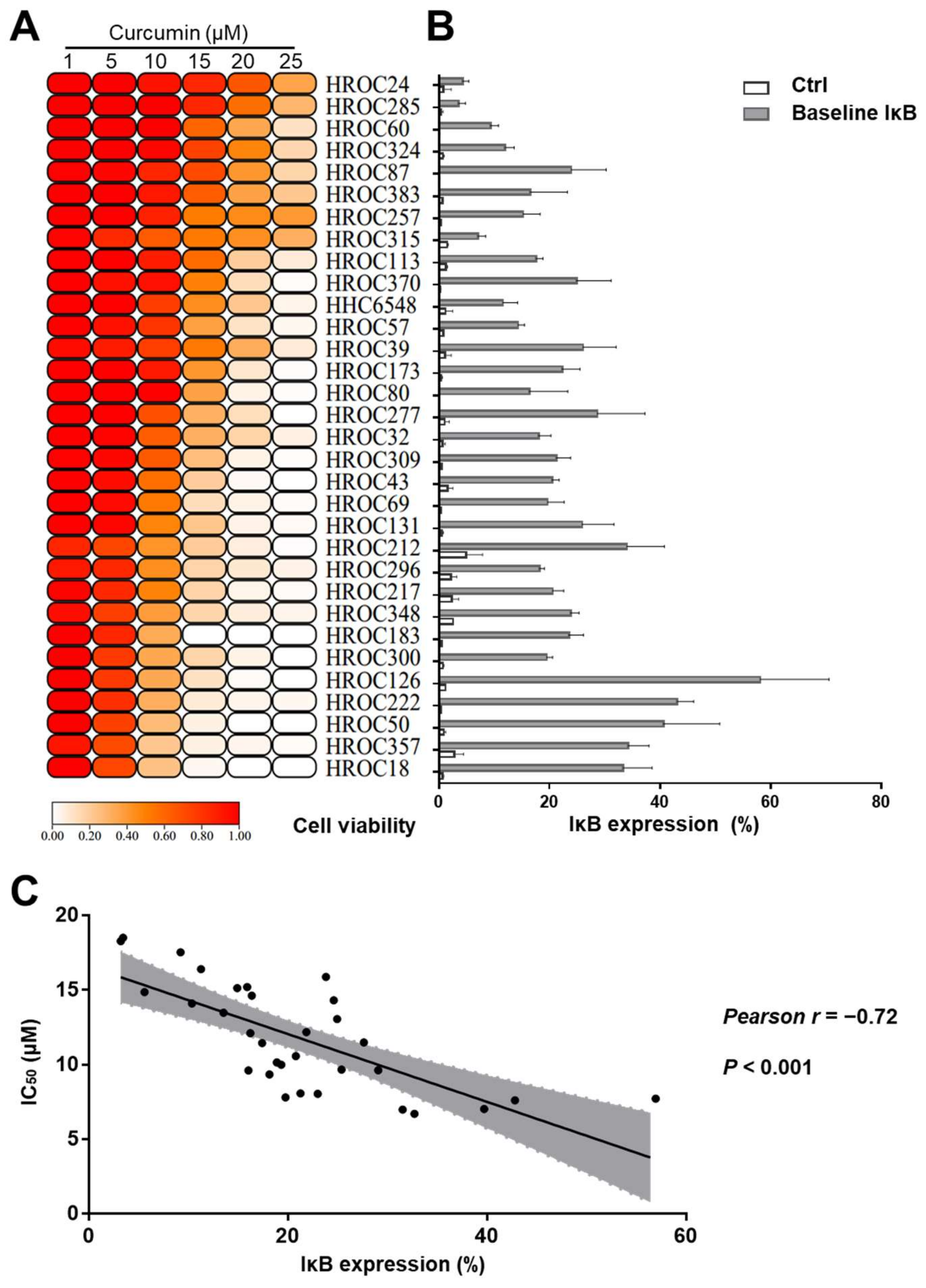
2.2. Curcumin-Sensitive CRC Cell Lines Expressed Higher IκBα Levels
2.3. Curcumin Induced Cell Death in Curcumin-Sensitive CRC Cell Lines
2.4. Curcumin Suppressed Migration in Curcumin-Sensitive CRC Cell Lines
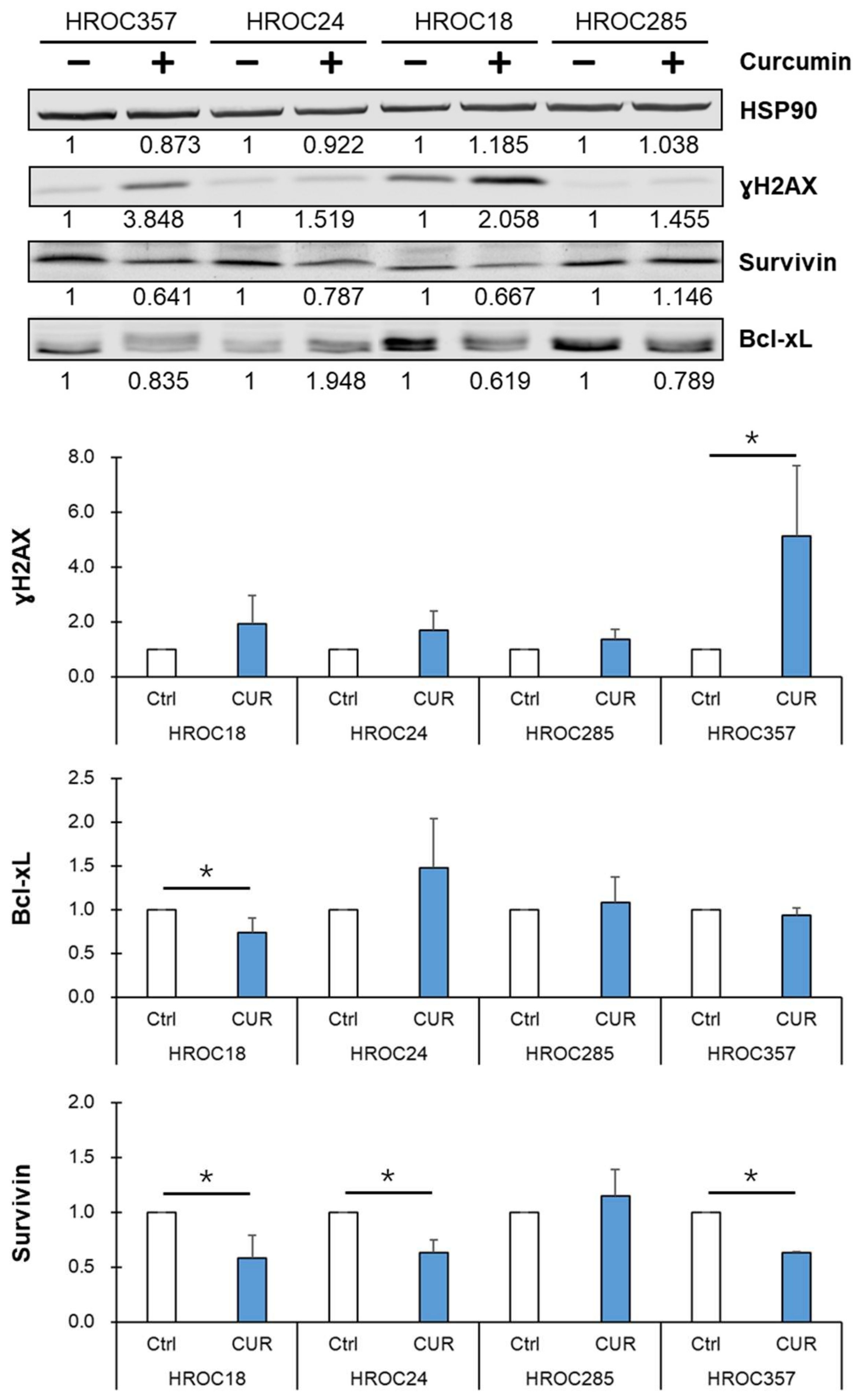
2.5. Curcumin Reduced NF-κB p65 Target Protein Survivin Expression and Led to an Accumulation of DNA Double-Strand Breaks in Curcumin-Sensitive CRC Cells
2.6. Curcumin Triggered Higher IκBα Levels in Curcumin-Sensitive CRC Cell Lines
2.7. Inverse Correlation of Cell Viability and IκBα Levels after Curcumin Treatment
2.8. Curcumin Downregulated NF-κB p65
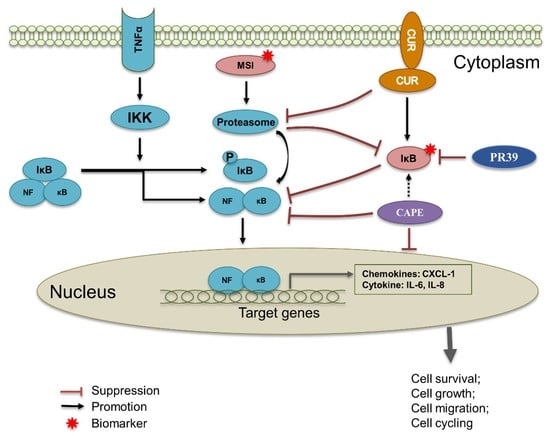
2.9. Curcumin Induced Cell Death by Inhibiting NF-κB p65 Activation via IκBα
2.10. Mutated SMAD4 Was Correlated with Curcumin Sensitivity in CRC Cell Lines
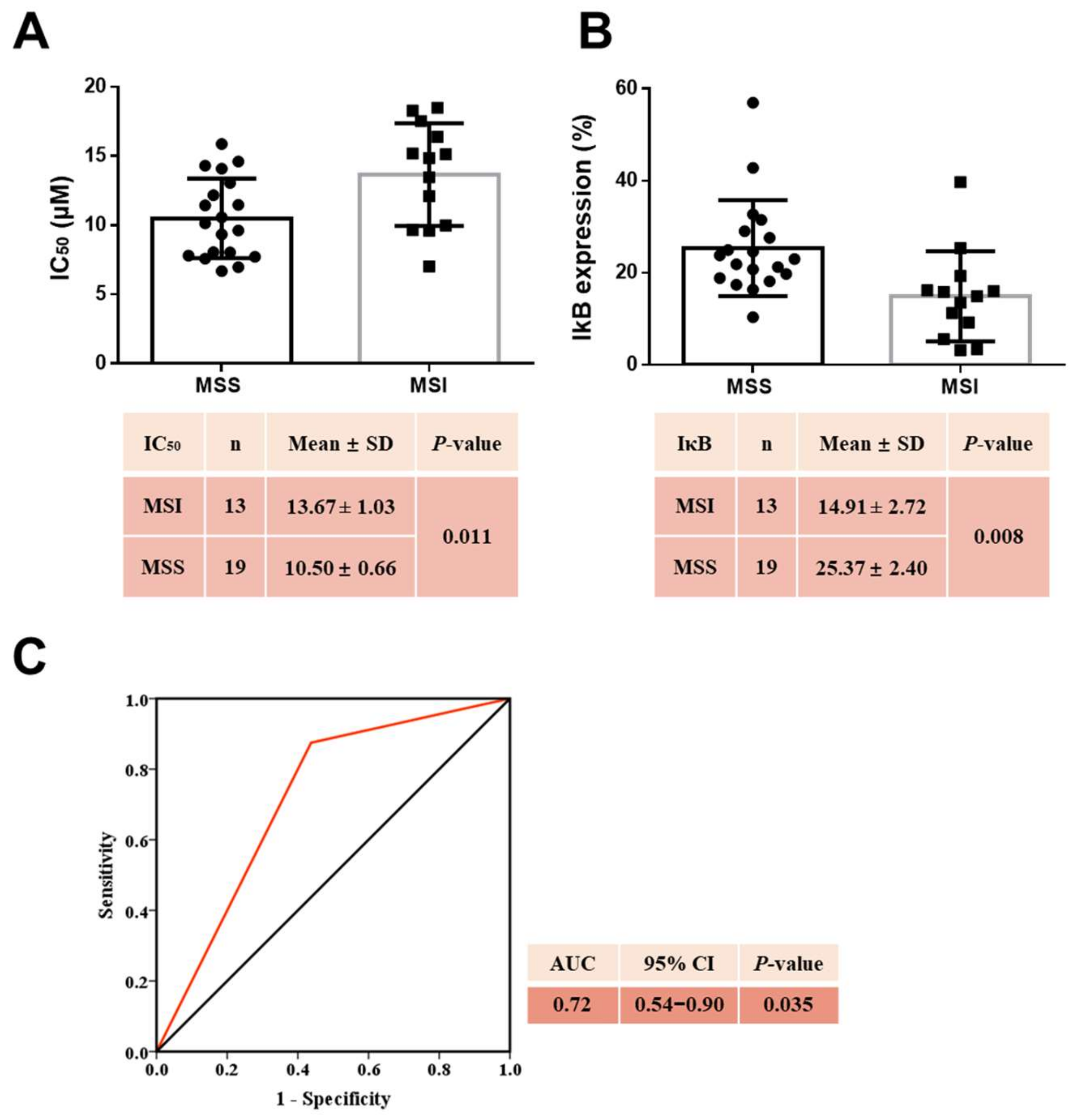
2.11. MSS CRC Cell Lines Were More Sensitive to Curcumin
2.12. MSS Combined with High Baseline IκBα Levels Estimated Curcumin Sensitivity
3. Discussion
4. Materials and Methods
4.1. CRC Cell Lines and Cell Culture
4.2. Chemo-Test and Crystal Violet Assay
4.3. Cell Death Measurement
4.4. Evaluation of IκBα in Cells
4.5. Wound-Healing Assay
4.6. Measurement of IL-6, IL-8, and CXCL-1 in Cells
4.7. Immunoblots
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer (2020). Available online: https://gco.iarc.fr/today (accessed on 1 July 2021).
- Lu, L.; Mullins, C.S.; Schafmayer, C.; Zeißig, S.; Linnebacger, M. A global assessment of recent trends in gastrointestinal cancer and lifestyle-associated risk factors. Cancer Commun. 2021, 41, 1137–1151, Epub ahead of print. [Google Scholar] [CrossRef]
- Kaiser, K.; Lyleroehr, M.; Shaunfield, S.; Lacson, L.; Corona, M.; Kircher, S.; Nittve, M.; Cella, D. Neuropathy experienced by colorectal cancer patients receiving oxaliplatin: A qualitative study to validate the Functional Assessment of Cancer Therapy/Gynecologic Oncology Group-Neurotoxicity scale. World J. Gastrointest Oncol. 2020, 12, 205–218. [Google Scholar] [CrossRef]
- Peng, J.; Dong, C.; Wang, C.; Li, W.; Yu, H.; Zhang, M.; Zhao, Q.; Zhu, B.; Zhang, J.; Li, W.; et al. Cardiotoxicity of 5-fluorouracil and capecitabine in Chinese patients: A prospective study. Cancer Commun. 2018, 38, 22. [Google Scholar] [CrossRef] [Green Version]
- Montagnani, F.; Chiriatti, A.; Turrisi, G.; Francini, G.; Fiorentini, G. A systematic review of FOLFOXIRI chemotherapy for the first-line treatment of metastatic colorectal cancer: Improved efficacy at the cost of increased toxicity. Colorectal Dis. 2011, 13, 846–852. [Google Scholar] [CrossRef]
- Shehzad, A.; Wahid, F.; Lee, Y.S. Curcumin in cancer chemoprevention: Molecular targets, pharmacokinetics, bioavailability, and clinical trials. Arch. Pharm. 2010, 343, 489–499. [Google Scholar] [CrossRef]
- Zheng, J.; Cheng, J.; Zheng, S.; Feng, Q.; Xiao, X. Curcumin, a Polyphenolic Curcuminoid With Its Protective Effects and Molecular Mechanisms in Diabetes and Diabetic Cardiomyopathy. Front. Pharmacol. 2018, 9, 472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pricci, M.; Girardi, B.; Giorgio, F.; Losurdo, G. Curcumin and Colorectal Cancer: From Basic to Clinical Evidences. Int. J. Mol. Sci. 2020, 21, 2364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismail, N.I.; Othman, I. Mechanism of Apoptosis Induced by Curcumin in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 2454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz de Porras, V.; Layos, L.; Martínez-Balibrea, E. Curcumin: A therapeutic strategy for colorectal cancer? Semin. Cancer Biol. 2021, 73, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Conteas, C.N. Anti-carcinogenic properties of curcumin on colorectal cancer. World J. Gastrointest Oncol. 2010, 2, 169–176. [Google Scholar] [CrossRef]
- Zhou, H.; Beevers, C.S.; Huang, S. The targets of curcumin. Curr. Drug Targets 2011, 12, 332–347. [Google Scholar] [CrossRef] [PubMed]
- Kunnumakkara, A.B.; Diagaradjane, P.; Anand, P.; Harikumar, K.B.; Deorukhkar, A.; Gelovani, J.; Guha, S.; Krishnan, S.; Aggarwal, B.B. Curcumin sensitizes human colorectal cancer to capecitabine by modulation of cyclin D1, COX-2, MMP-9, VEGF and CXCR4 expression in an orthotopic mouse model. Int. J. Cancer 2009, 125, 2187–2197. [Google Scholar] [CrossRef]
- Wong, K.E.; Ngai, S.C.; Chan, K.G.; Lee, L.H.; Goh, B.H.; Chuah, L.H. Curcumin Nanoformulations for Colorectal Cancer: A Review. Front. Pharmacol. 2019, 10, 152. [Google Scholar] [CrossRef]
- Howells, L.M.; Sale, S.; Sriramareddy, S.N.; Irving, G.R.; Jones, D.J.; Ottley, C.J.; Pearson, D.G.; Mann, C.D.; Manson, M.M.; Berry, D.P. Curcumin ameliorates oxaliplatin-induced chemoresistance in HCT116 colorectal cancer cells in vitro and in vivo. Int. J. Cancer 2011, 129, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ahmed, B.; Mehta, K.; Kurzrock, R. Liposomal curcumin with and without oxaliplatin: Effects on cell growth, apoptosis, and angiogenesis in colorectal cancer. Mol. Cancer Ther. 2007, 6, 1276–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.C.; Yang, B.L.; Chen, W.C.; Ho, A.S.; Sie, Z.L.; Lin, H.C.; Chang, C.C. STAT3 Mediated miR-30a-5p Inhibition Enhances Proliferation and Inhibits Apoptosis in Colorectal Cancer Cells. Int. J. Mol. Sci. 2020, 2, 7315. [Google Scholar] [CrossRef]
- Luo, Y.; Yang, Z.; Chen, Y.; Lu, X.; Quan, Y. Genomic and immunological features of microsatellite instability in colon cancer. Gene 2021, 781, 145534. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.C.; Xu, J.; Bian, X.; Wu, J.L.; Kang, W.; Qian, Y.; Li, W.; Chen, H.; Gou, H.; Liu, D.; et al. In Colorectal Cancer Cells With Mutant KRAS, SLC25A22-Mediated Glutaminolysis Reduces DNA Demethylation to Increase WNT Signaling, Stemness, and Drug Resistance. Gastroenterology 2020, 159, 2163–2180.e6. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Z.; Zhuang, J.; Wu, X.; Wang, Z.; Zhang, B.; Gao, G.; Zhang, Y.; Guo, C.; Xia, Q. Prognostic Value of Autophagy, Microsatellite Instability, and KRAS Mutations in Colorectal Cancer. J. Cancer 2021, 12, 3515–3528. [Google Scholar] [CrossRef]
- Kurasaka, C.; Ogino, Y. Molecular Mechanisms and Tumor Biological Aspects of 5-Fluorouracil Resistance in HCT116 Human Colorectal Cancer Cells. Int. J. Mol. Sci. 2021, 22, 2916. [Google Scholar] [CrossRef]
- Mullins, C.S.; Micheel, B.; Matschos, S.; Leuchter, M.; Bürtin, F.; Krohn, M.; Hühns, M.; Klar, E.; Prall, F.; Linnebacher, M. Integrated Biobanking and Tumor Model Establishment of Human Colorectal Carcinoma Provides Excellent Tools for Preclinical Research. Cancers 2019, 11, 1520. [Google Scholar] [CrossRef] [Green Version]
- Wagner, S.; Beger, N.T.; Matschos, S.; Szymanski, A.; Przybylla, R.; Bürtin, F.; Prall, F.; Linnebacher, M.; Mullins, C.S. Tumour-Derived Cell Lines and Their Potential for Therapy Prediction in Patients with Metastatic Colorectal Cancer. Cancers 2021, 13, 4717. [Google Scholar] [CrossRef] [PubMed]
- Gock, M.; Mullins, C.S.; Bergner, C.; Prall, F.; Ramer, R.; Göder, A.; Krämer, O.H.; Lange, F.; Krause, B.J.; Klar, E.; et al. Establishment, functional and genetic characterization of three novel patient-derived rectal cancer cell lines. World J. Gastroenterol. 2018, 24, 4880–4892. [Google Scholar] [CrossRef] [PubMed]
- Maletzki, C.; Gock, M.; Randow, M.; Klar, E.; Huehns, M.; Prall, F.; Linnebacher, M. Establishment and characterization of cell lines from chromosomal instable colorectal cancer. World J. Gastroenterol. 2015, 21, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Maletzki, C.; Stier, S.; Gruenert, U.; Gock, M.; Ostwald, C.; Prall, F.; Linnebacher, M. Establishment, characterization and chemosensitivity of three mismatch repair deficient cell lines from sporadic and inherited colorectal carcinomas. PLoS ONE 2012, 7, e52485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gock, M.; Mullins, C.S.; Harnack, C.; Prall, F.; Ramer, R.; Göder, A.; Krämer, O.H.; Klar, E.; Linnebacher, M. Establishment, functional and genetic characterization of a colon derived large cell neuroendocrine carcinoma cell line. World J. Gastroenterol. 2018, 24, 3749–3759. [Google Scholar] [CrossRef]
- Randon, G.; Fucà, G.; Rossini, D.; Raimondi, A.; Pagani, F.; Perrone, F.; Tamborini, E.; Busico, A.; Peverelli, G.; Morano, F. Prognostic impact of ATM mutations in patients with metastatic colorectal cancer. Sci. Rep. 2019, 9, 2858. [Google Scholar] [CrossRef] [Green Version]
- Perrone, F.; Lampis, A.; Orsenigo, M.; Di Bartolomeo, M.; Gevorgyan, A.; Losa, M.; Frattini, M.; Riva, C.; Andreola, S.; Bajetta, E.; et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann. Oncol. 2009, 20, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Taieb, J.; Zaanan, A.; Le Malicot, K.; Julié, C.; Blons, H.; Mineur, L.; Bennouna, J.; Tabernero, J.; Mini, E.; Folprecht, G.; et al. Prognostic Effect of BRAF and KRAS Mutations in Patients With Stage III Colon Cancer Treated With Leucovorin, Fluorouracil, and Oxaliplatin With or Without Cetuximab: A Post Hoc Analysis of the PETACC-8 Trial. JAMA Oncol. 2016, 2, 643–653. [Google Scholar] [CrossRef] [Green Version]
- Conlin, A.; Smith, G.; Carey, F.A.; Wolf, C.R.; Steele, R.J. The prognostic significance of K-ras, p53, and APC mutations in colorectal carcinoma. Gut 2005, 54, 1283–1286. [Google Scholar] [CrossRef] [Green Version]
- Gambardella, V.; Tarazona, N.; Cejalvo, J.M.; Lombardi, P.; Huerta, M.; Roselló, S.; Fleitas, T.; Roda, D.; Cervantes, A. Personalized Medicine: Recent Progress in Cancer Therapy. Cancers 2020, 12, 1009. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, A.; Kasinathan, A.; Ganesan, R.; Balasubramanian, A.; Bhaskaran, J.; Suresh, S.; Srinivasan, R.; Aravind, K.B.; Sivalingam, N. Curcumin induces apoptosis and cell cycle arrest via the activation of reactive oxygen species-independent mitochondrial apoptotic pathway in Smad4 and p53 mutated colon adenocarcinoma HT29 cells. Nutr. Res. 2018, 51, 67–81. [Google Scholar] [CrossRef]
- French, A.J.; Sargent, D.J.; Burgart, L.J.; Foster, N.R.; Kabat, B.F.; Goldberg, R.; Shepherd, L.; Windschitl, H.E.; Thibodeau, S.N. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin. Cancer Res. 2008, 14, 3408–3415. [Google Scholar] [CrossRef] [Green Version]
- Ribic, C.M.; Sargent, D.J.; Moore, M.J.; Thibodeau, S.N.; French, A.J.; Goldberg, R.M.; Hamilton, S.R.; Laurent-Puig, P.; Gryfe, R.; Shepherd, L.E.; et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N. Engl. J. Med. 2003, 349, 247–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsaleh, H.; Powell, B.; McCaul, K.; Grieu, F.; Grant, R.; Joseph, D.; Iacopetta, B. P53 alteration and microsatellite instability have predictive value for survival benefit from chemotherapy in stage III colorectal carcinoma. Clin. Cancer Res. 2001, 7, 1343–1349. [Google Scholar] [PubMed]
- Shakibaei, M.; Buhrmann, C.; Kraehe, P.; Shayan, P.; Lueders, C.; Goel, A. Curcumin chemosensitizes 5-fluorouracil resistant MMR-deficient human colon cancer cells in high density cultures. PLoS ONE 2014, 9, e85397. [Google Scholar] [CrossRef] [Green Version]
- Gavrilas, L.I.; Cruceriu, D.; Ionescu, C.; Miere, D.; Balacescu, O. Pro-apoptotic genes as new targets for single and combinatorial treatments with resveratrol and curcumin in colorectal cancer. Food Funct. 2019, 10, 3717–3726. [Google Scholar] [CrossRef]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087.e3. [Google Scholar] [CrossRef] [PubMed]
- Carethers, J.M.; Chauhan, D.P.; Fink, D.; Nebel, S.; Bresalier, R.S.; Howell, S.B.; Boland, C.R. Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology 1999, 117, 123–131. [Google Scholar] [CrossRef] [Green Version]
- Fishel, R. The selection for mismatch repair defects in hereditary nonpolyposis colorectal cancer: Revising the mutator hypothesis. Cancer Res. 2001, 61, 7369–7374. [Google Scholar] [PubMed]
- Slupphaug, G.; Kavli, B.; Krokan, H.E. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat. Res. 2003, 531, 231–251. [Google Scholar] [CrossRef] [PubMed]
- Bridge, G.; Rashid, S.; Martin, S.A. DNA mismatch repair and oxidative DNA damage: Implications for cancer biology and treatment. Cancers 2014, 6, 1597–1614. [Google Scholar] [CrossRef] [Green Version]
- Hsu, G.W.; Ober, M.; Carell, T.; Beese, L.S. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature 2004, 431, 217–221. [Google Scholar] [CrossRef]
- Aussem, A.; Ludwig, K. The Potential for Reducing Lynch Syndrome Cancer Risk with Nutritional Nrf2 Activators. Nutr. Cancer 2021, 73, 404–419. [Google Scholar] [CrossRef]
- Martín-López, J.V.; Fishel, R. The mechanism of mismatch repair and the functional analysis of mismatch repair defects in Lynch syndrome. Fam. Cancer 2013, 12, 159–168. [Google Scholar] [CrossRef] [Green Version]
- Majumdar, A.P.; Banerjee, S.; Nautiyal, J.; Patel, B.B.; Patel, V.; Du, J.; Yu, Y.; Elliott, A.A.; Levi, E.; Sarkar, F.H. Curcumin synergizes with resveratrol to inhibit colon cancer. Nutr. Cancer 2009, 61, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.C.; Lin, Y.S.; Tsao, P.N.; Wu-Tsai, J.J.; Wu, C.H.; Wong, J.M. Comparison of the anti-proliferation and apoptosis-induction activities of sulindac, celecoxib, curcumin, and nifedipine in mismatch repair-deficient cell lines. J. Formos. Med. Assoc. 2004, 103, 599–606. [Google Scholar]
- Jiang, Z.; Jin, S.; Yalowich, J.C.; Brown, K.D.; Rajasekaran, B. The mismatch repair system modulates curcumin sensitivity through induction of DNA strand breaks and activation of G2-M checkpoint. Mol. Cancer Ther. 2010, 9, 558–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandur, S.K.; Deorukhkar, A.; Pandey, M.K.; Pabón, A.M.; Shentu, S.; Guha, S.; Aggarwal, B.B.; Krishnan, S. Curcumin modulates the radiosensitivity of colorectal cancer cells by suppressing constitutive and inducible NF-kappaB activity. Int. J. Radiat. Oncol. Biol. Phys. 2009, 75, 534–542. [Google Scholar] [CrossRef] [Green Version]
- Naugler, W.E.; Karin, M. NF-kappaB and cancer-identifying targets and mechanisms. Curr. Opin. Genet. Dev. 2008, 18, 19–26. [Google Scholar] [CrossRef] [Green Version]
- Karin, M.; Ben-Neriah, Y. Phosphorylation meets ubiquitination: The control of NF-[kappa]B activity. Annu. Rev. Immunol. 2000, 18, 621–663. [Google Scholar] [CrossRef] [PubMed]
- Shakibaei, M.; Mobasheri, A.; Lueders, C.; Busch, F.; Shayan, P.; Goel, A. Curcumin enhances the effect of chemotherapy against colorectal cancer cells by inhibition of NF-κB and Src protein kinase signaling pathways. PLoS ONE 2013, 8, e57218. [Google Scholar] [CrossRef] [Green Version]
- Kunnumakkara, A.B.; Diagaradjane, P.; Guha, S.; Deorukhkar, A.; Shentu, S.; Aggarwal, B.B.; Krishnan, S. Curcumin sensitizes human colorectal cancer xenografts in nude mice to gamma-radiation by targeting nuclear factor-kappaB-regulated gene products. Clin. Cancer Res. 2008, 14, 2128–2136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jani, T.S.; DeVecchio, J.; Mazumdar, T.; Agyeman, A.; Houghton, J.A. Inhibition of NF-kappaB signaling by quinacrine is cytotoxic to human colon carcinoma cell lines and is synergistic in combination with tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) or oxaliplatin. J. Biol. Chem. 2010, 285, 19162–19172. [Google Scholar] [CrossRef] [Green Version]
- Ruiz de Porras, V.; Bystrup, S.; Martínez-Cardús, A.; Pluvinet, R.; Sumoy, L.; Howells, L.; James, M.I.; Iwuji, C.; Manzano, J.L.; Layos, L. Curcumin mediates oxaliplatin-acquired resistance reversion in colorectal cancer cell lines through modulation of CXC-Chemokine/NF-κB signalling pathway. Sci. Rep. 2016, 6, 24675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashem, S.; Nisar, S.; Sageena, G.; Macha, M.A.; Yadav, S.K.; Krishnankutty, R.; Uddin, S.; Haris, M.; Bhat, A.A. Therapeutic Effects of Curcumol in Several Diseases; An Overview. Nutr. Cancer 2021, 73, 181–195. [Google Scholar] [CrossRef]
- Chung, S.S.; Dutta, P.; Chard, N.; Wu, Y.; Chen, Q.H.; Chen, G.; Vadgama, J. A novel curcumin analog inhibits canonical and non-canonical functions of telomerase through STAT3 and NF-κB inactivation in colorectal cancer cells. Oncotarget 2019, 10, 4516–4531. [Google Scholar] [CrossRef] [Green Version]
- Amodio, V.; Mauri, G.; Reilly, N.M.; Sartore-Bianchi, A. Mechanisms of Immune Escape and Resistance to Checkpoint Inhibitor Therapies in Mismatch Repair Deficient Metastatic Colorectal Cancers. Cancers 2021, 13, 2638. [Google Scholar] [CrossRef]
- Momburg, F.; Tan, P. Tapasin-the keystone of the loading complex optimizing peptide binding by MHC class I molecules in the endoplasmic reticulum. Mol. Immunol. 2002, 39, 217–233. [Google Scholar] [CrossRef]
- Ji, Z.; He, L.; Regev, A.; Struhl, K. Inflammatory regulatory network mediated by the joint action of NF-kB, STAT3, and AP-1 factors is involved in many human cancers. Proc. Natl. Acad. Sci. USA 2019, 116, 9453–9462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamini, B.; Yu, X.; Dolan, M.E.; Wu, M.H.; Darga, T.E.; Kufe, D.W.; Weichselbaum, R.R. Inhibition of nuclear factor-kappaB activity by temozolomide involves O6-methylguanine induced inhibition of p65 DNA binding. Cancer Res. 2007, 67, 6889–6898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hühns, M.; Krohn, S.; Murua Escobar, H.; Prall, F. Genomic heterogeneity in primary colorectal carcinomas and their metastases: Born bad or brought up a villain? Hum. Pathol. 2018, 74, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Maletzki, C.; Huehns, M.; Bauer, I.; Ripperger, T.; Mork, M.M.; Vilar, E.; Klöcking, S.; Zettl, H.; Prall, F.; Linnebacher, M. Frameshift mutational target gene analysis identifies similarities and differences in constitutional mismatch repair-deficiency and Lynch syndrome. Mol. Carcinog. 2017, 56, 1753–1764. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Zhang, X.; Lu, L.; Jiang, K.; Krohn, M.; Matschos, S.; Mullins, C.S.; Vollmar, B.; Zechner, D.; Gong, P.; et al. Pharmaceutical immunoglobulin G impairs, anti-carcinoma activity of oxaliplatin in colon cancer cells. Br. J. Cancer 2021, 124, 1411–1420. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]






| Variables | n | % | |
|---|---|---|---|
| Age (years) 1 | 32 | 66.34 ± 17.85 | |
| Gender | Male | 15 | 46.88 |
| Female | 17 | 53.13 | |
| Sample location | Colon | 30 | 93.75 |
| Rectum | 2 | 6.25 | |
| T stage | T0-T2 | 4 | 12.50 |
| T3-T4 | 28 | 87.50 | |
| N stage | N0 | 14 | 43.75 |
| N1-2 | 18 | 56.25 | |
| M stage | M0 | 22 | 68.75 |
| M1 | 9 | 28.13 | |
| Mx | 1 | 3.13 | |
| R stage | R0 | 23 | 71.88 |
| R1-2 | 8 | 25.00 | |
| Unclear | 1 | 3.13 | |
| L stage | L0 | 21 | 65.63 |
| L1 | 9 | 28.13 | |
| Unclear | 2 | 6.25 | |
| V stage | V0 | 19 | 59.38 |
| V1-2 | 11 | 34.38 | |
| Unclear | 2 | 6.25 | |
| UICC 2 stage | I-II | 13 | 40.63 |
| III-IV | 18 | 56.25 | |
| Unclear | 1 | 3.13 | |
| Molecular type | spStd 3 | 14 | 43.75 |
| spMSI-H 4 | 8 | 25.00 | |
| CIMP-H 5 | 4 | 12.50 | |
| Lynch | 5 | 15.63 | |
| Neuroendocrine | 1 | 3.13 |
| Cell Line | IC50 (μM) | |||
|---|---|---|---|---|
| Curcumin | 5-FU | Irinotecan | Oxaliplatin | |
| HROC24 | 18.49 | 0.79 | 0.39 | 1.60 |
| HROC285 | 18.27 | 1.64 | 0.73 | 0.53 |
| HROC324 | 17.52 | 1.17 | 1.98 | 0.43 |
| HROC87 | 16.39 | 6.92 4 | 1.53 4 | 0.21 |
| HROC60 | 15.87 | 26.90 | 3.17 | 2.19 |
| HROC383 | 15.20 | 1.28 | 4.12 | 1.41 |
| HROC113 | 15.12 | 7.69 4 | 12.80 4 | 0.31 |
| HROC370 | 14.86 | 0.65 | 3.78 | <0.01 |
| HROC39 | 14.61 | 7.66 | 0.28 | 0.59 |
| HROC80 | 14.30 | 4.21 | 0.83 | 1.28 |
| HROC173 | 14.09 | 0.17 | 1.36 | 0.74 |
| HHC6548 | 13.47 | 0.23 | 1.40 | 0.31 |
| HROC57 | 13.05 | 80.95 5 | 1.27 5 | 1.76 5 |
| HROC277 | 12.17 | 9.20 1 | 0.10 1 | 2.90 1 |
| HROC257 | 12.11 | 9.35 | 1.16 | 4.09 |
| HROC309 | 11.48 | n.d. | n.d. | n.d. |
| HROC32 | 11.44 | 8.90 | 0.58 | 1.67 |
| HROC43 | 10.57 | 3.51 | 1.63 | 0.18 |
| HROC69 | 10.14 | 4.41 | 1.10 | 0.25 |
| HROC131 | 9.99 | 4.89 | 2.01 | 1.39 |
| HROC212 | 9.66 | 23.32 | 9.13 | 8.72 |
| HROC296 | 9.62 | n.d. | n.d. | n.d. |
| HROC315 | 9.60 | 1.08 | 0.04 | 0.53 |
| HROC217 | 9.34 | 2.09 | 1.13 | 0.85 |
| HROC348 | 8.06 | 4.40 1 | 2.10 1 | 2.20 1 |
| HROC183 | 8.04 | 1.05 | 1.48 | 0.93 |
| HROC300 | 7.80 | 51.19 | 2.15 | 1.83 |
| HROC126 | 7.72 | 2.30 | 0.72 2 | 0.55 |
| HROC222 | 7.60 | 18.47 | n.d. | 1.27 |
| HROC50 | 7.02 | 0.66 | 1.83 | 2.23 |
| HROC357 | 6.97 | 9.61 | 7.51 | 2.36 |
| HROC18 | 6.69 | 0.88 | 7.88 3 | 0.99 |
| Mean ± SD | 11.79 ± 3.55 | 9.83 ± 17.12 | 2.56 ± 3.03 | 1.48 ± 1.66 |
| p-value 6 | 0.543 | <0.001 | <0.001 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, L.; Przybylla, R.; Shang, Y.; Dai, M.; Krohn, M.; Krämer, O.H.; Mullins, C.S.; Linnebacher, M. Microsatellite Status and IκBα Expression Levels Predict Sensitivity to Pharmaceutical Curcumin in Colorectal Cancer Cells. Cancers 2022, 14, 1032. https://doi.org/10.3390/cancers14041032
Lu L, Przybylla R, Shang Y, Dai M, Krohn M, Krämer OH, Mullins CS, Linnebacher M. Microsatellite Status and IκBα Expression Levels Predict Sensitivity to Pharmaceutical Curcumin in Colorectal Cancer Cells. Cancers. 2022; 14(4):1032. https://doi.org/10.3390/cancers14041032
Chicago/Turabian StyleLu, Lili, Randy Przybylla, Yuru Shang, Meng Dai, Mathias Krohn, Oliver Holger Krämer, Christina Susanne Mullins, and Michael Linnebacher. 2022. "Microsatellite Status and IκBα Expression Levels Predict Sensitivity to Pharmaceutical Curcumin in Colorectal Cancer Cells" Cancers 14, no. 4: 1032. https://doi.org/10.3390/cancers14041032
APA StyleLu, L., Przybylla, R., Shang, Y., Dai, M., Krohn, M., Krämer, O. H., Mullins, C. S., & Linnebacher, M. (2022). Microsatellite Status and IκBα Expression Levels Predict Sensitivity to Pharmaceutical Curcumin in Colorectal Cancer Cells. Cancers, 14(4), 1032. https://doi.org/10.3390/cancers14041032