Simple Summary
The deubiquitinase-mediated cleavage of ubiquitin chains from substrate proteins plays a crucial role in regulating protein degradation, activities, interactions, and localization. The dysregulation of multiple deubiquitinases has been implicated in various human diseases, especially cancer. More importantly, many small molecules targeting oncogenic deubiquitinases have been discovered, some of which have exhibited promising anti-cancer effects and entered clinical trials. Gastric cancer remains one of the most common and fatal malignancies. In this review, we aim to summarize the multifaceted roles of deubiquitinases in gastric tumorigenesis. We also present the upstream regulation of specific deubiquitinases and the research progress of several deubiquitinase-associated small molecules for gastric cancer therapy. Together, this review will improve our understanding of the biological role of deubiquitinases as well as the therapeutic potential of targeting deubiquitinases in gastric cancer.
Abstract
Gastric cancers (GCs) are malignant tumors with a high incidence that threaten global public health. Despite advances in GC diagnosis and treatment, the prognosis remains poor. Therefore, the mechanisms underlying GC progression need to be identified to develop prognostic biomarkers and therapeutic targets. Ubiquitination, a post-translational modification that regulates the stability, activity, localization, and interactions of target proteins, can be reversed by deubiquitinases (DUBs), which can remove ubiquitin monomers or polymers from modified proteins. The dysfunction of DUBs has been closely linked to tumorigenesis in various cancer types, and targeting certain DUBs may provide a potential option for cancer therapy. Multiple DUBs have been demonstrated to function as oncogenes or tumor suppressors in GC. In this review, we summarize the DUBs involved in GC and their associated upstream regulation and downstream mechanisms and present the benefits of targeting DUBs for GC treatment, which could provide new insights for GC diagnosis and therapy.
1. Introduction
Gastric cancer (GC) is the fifth most common cancer and the fourth leading cause of cancer-related mortality worldwide. Based on estimates from the GLOBOCAN database, more than 1 million new GC cases and an estimated 769,000 GC deaths occurred worldwide in 2020 []. As with other cancer types, GC development is a multistage process involving genetic and epigenetic alterations [,]. In addition to host factors, other etiological factors, including a high-salt diet, tobacco use, and infectious agents, such as Helicobacter pylori (H. pylori) and Epstein–Barr virus (EBV), also play a significant role in the initiation and progression of GC []. Despite advances in understanding the mechanisms underlying GC and improved therapies, almost one-third of patients with GC are diagnosed at a late stage, with a 5-year survival rate below 20% [,,]. Therefore, there is an urgent need to develop novel biomarkers and therapeutic targets for treating GC.
Deubiquitinases (DUBs) are isopeptidases that can cleave a single ubiquitin or entire ubiquitin chains from target proteins, thereby counteracting protein ubiquitylation, a post-translational modification fundamental for the regulation of protein stability, activity, subcellular localization, and interactions [,]. DUBs are involved in many physiological processes, such as apoptosis, autophagy, and the cell cycle, and pathological processes, such as neurodegenerative diseases and cancers [,,,,,]. Hence, DUBs have attracted attention as therapeutic targets, and DUB inhibitors have been developed, with some now in preclinical development or clinical trials []. To date, approximately 100 DUBs have been reported and are classified into seven subfamilies based on their sequence and structural similarities as follows: ubiquitin-specific proteases (USPs), ubiquitin carboxy-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Jab1/MPN domain-associated metalloenzymes (JAMMs), Machado–Joseph disease proteases (MJDs), the monocyte chemotactic protein-induced protease family (MINDYs), and Zn-finger and UFSP domain proteins (ZUFSPs) (Figure 1) [].
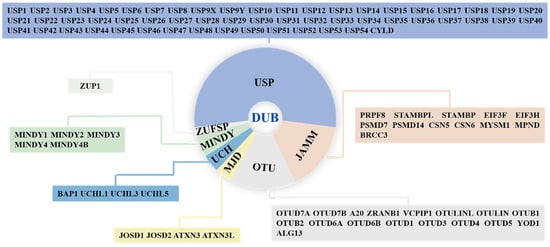
Figure 1.
The classification and members of DUB family. DUBs are divided into seven subfamilies: ubiquitin-specific proteases (USPs), ubiquitin carboxy-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Jab1/MPN domain-associated metalloenzymes (JAMMs), Machado–Joseph disease proteases (MJDs), monocyte chemotactic protein-induced protease family (MINDYs), and Zn-finger and UFSP domain proteins (ZUFSPs).
During the past decades, many studies have demonstrated that multiple DUBs are implicated in the development of GC. In this review, we first summarize and discuss the DUBs related to GC, with a summary of their mechanisms of action and regulation (Table 1). In addition, we document the potential of pharmacological interventions that target DUBs (Table 2). Taken together, the present review aims to provide a better understanding of the molecular mechanisms underlying GC development associated with DUBs and how they can be targeted for GC treatment.

Table 1.
The list of DUBs involved in GC.

Table 2.
Compounds that suppress GC by modulating DUBs.
2. USPs and GC
USPs constitute the largest family of human DUBs, with 56 members. As shown in Figure 2, at least 25 USPs are linked to GC.
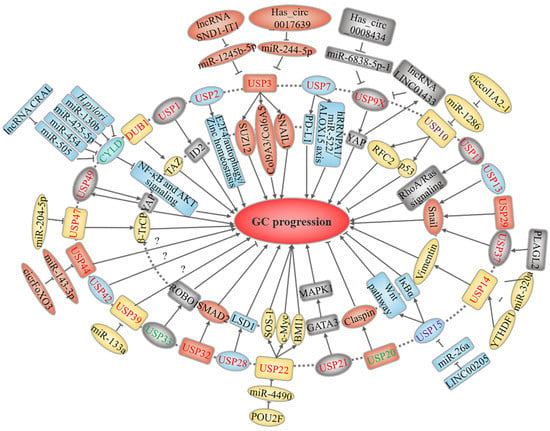
Figure 2.
USP-related upstream regulation and downstream mechanisms in GC. USPs marked in red and green represent oncoproteins and tumor suppressors, respectively, while purple-labeled USPs play a dual role. The text in the outer and inner circles of the USPs describes the upstream regulatory events and downstream substrates or signaling pathways, respectively.
2.1. USP3
Studies have highlighted the role of USP3 in GC progression. USP3 is overexpressed in GC cells and tissues and can serve as a prognostic marker for patients [,]. USP3 facilitates GC cell growth and metastasis in vitro and in vivo by modulating cell cycle control and epithelial–mesenchymal transition (EMT)-related molecules []. Wu et al. identified differentially expressed proteins in BGC-823 cells stably expressing USP3 []. Among them, SUZ12, a scaffolding component of the PRC2 complex, and COL9A3/COL6A5, collagen family members, were deubiquitinated and stabilized by USP3, which accounts for its role in promoting invasion and migration [,]. In addition to SUZ12, the core PRC2 complex also comprises the histone methyltransferase EZH2 and the scaffolding component EED. As an epigenetic regulator complex, PRC2 trimethylates lysine 27 on histone H3 tails to modulate its several thousand target genes, including various genes involved in cancer proliferation, migration, and invasion, such as E-cadherin and p14ARF []. However, it is unknown whether these three proteins are involved in the USP3-directed proliferation of GC cells, and future studies should examine their oncogenic roles in GC.
Additionally, non-coding RNAs, including circular RNAs, long non-coding RNAs (lncRNAs), and microRNAs (miRNAs), play critical roles in modulating GC [,,,]. Hsa_circ_0017639, a circular RNA, is increased in GC cell lines and promotes proliferation and migration by increasing USP3 expression by sponging miR-224-5p []. In addition, Jin et al. demonstrated that exosomal lncRNA SND1-IT1 secreted from GC cells not only recruited DDX54, a DEAD-box RNA helicase that binds to specific RNAs, to enhance USP3 mRNA stability, but also bound to miR-1245b-5p to upregulate USP3 expression, thus leading to SNAIL1 stabilization and inducing the malignant transformation of gastric mucosa cells []. Taken together, these studies suggest that targeting USP3 may be a potential treatment for GC.
2.2. USP7 and USP11
USP7, also called herpes-virus-associated ubiquitin-specific protease, deubiquitinates many substrate proteins involved in the cell cycle, DNA damage responses, and immune responses [,]. Accordingly, the aberrant expression and activity of USP7 have been found in GC. Wang et al. showed that USP7 is highly expressed in GC and increases the expression of programmed cell death protein 1 (PD-L1), a pivotal immune checkpoint molecule that decreases T-cell immune responses []. Furthermore, USP7 knockdown or inhibition with its inhibitor Almac4, developed by Gavory et al., conferred GC cell sensitivity to T-cell cytotoxicity, reduced proliferation, and induced cell cycle arrest by stabilizing p53, which interacts with USP7 [,,]. Similarly, the USP7 inhibitor C9, a quinazolin-4(3H)-one derivative, also suppressed GC cell proliferation by upregulating p53 and its downstream target p21 [].
To better understand the USP7 binding network in tumor cells, Anna et al. performed affinity purification coupled with mass spectrometry to identify interactions in GC cells overexpressing USP7. In addition to several reported binding proteins (such as USP11 and TRIP12), this study also identified DHX40 and DDX24, two DEAD/DEAH-box RNA helicases, as novel targets of USP7, providing preliminary evidence that USP7 may regulate RNA metabolism []. Furthermore, these interactions were confirmed with nasopharyngeal carcinoma cells. However, the role of these USP7 substrates in GC carcinogenesis were not investigated []. Interestingly, a recent study suggested that USP11 might play an oncogenic role in GC by affecting RhoA and Ras-mediated signaling pathways []. Moreover, a previous study indicated that H. pylori decreases the expression and activity of USP7 in infected GC cells. However, the exact function of H. pylori in regulating USP7 expression remains unclear [].
In addition, Zhang et al. found that USP7 stabilizes hnRNPA1 in cancer-associated fibroblasts (CAFs) to contribute to the packaging and release of exosomes containing miR-522. Paclitaxel and cisplatin also promoted miR-522 secretion by activating the USP7/hnRNPA1 axis. The lipoxygenase ALOX15 is important in mediating the lipid peroxidation that drives ferroptosis. Further, exosomal miR-522 repressed ALOX15 expression and lipid peroxide accumulation, inhibited ferroptosis, and ultimately lead to acquired chemoresistance in GC cells [].
2.3. USP9X, USP36, and USP49
USP9X has been shown to function as either an oncogene or tumor suppressor, depending on the type of cancer []. Increased USP9X expression is associated with a poor prognosis in patients with GC, suggesting an oncogenic role []. Consistent with this evidence, another study showed that silencing USP9X represses the migration, invasion, and colony formation ability of GC cells. Moreover, hsa_circ_0008434, a miRNA sponge for miR-6838-5p, increased USP9X expression and promoted GC progression [].
Hippo signaling has been implicated in regulating cell growth, metastasis, and chemoresistance in GC []. As the core downstream effectors, YAP and TAZ activity is controlled by a conserved kinase cassette. In mammals, once Hippo signaling is activated, MST1/2 kinases phosphorylate and activate LATS1/2 kinases, which further phosphorylate YAP/TAZ for cytoplasmic sequestration or degradation. Inhibiting the Hippo pathway leads to the nuclear translocation of YAP/TAZ, which regulate target gene expression by binding to TEAD coactivators [,]. Additionally, several DUBs activate or repress Hippo signaling through different mechanisms []. For instance, USP9X promotes breast cancer cell survival and attenuates cell sensitivity to chemotherapy by deubiquitinating and stabilizing YAP []. Zhang et al. demonstrated that LINC01433, a lncRNA positively related to GC progression, increased YAP stability by enhancing its interaction with USP9X and decreased YAP phosphorylation by weakening its association with LATS1. Because YAP binds to the LINC01433 promoter and activates its transcription, this positive feedback loop could be a therapeutic target for GC treatment [].
Two studies have suggested that DUB1 and USP49 also contribute to GC development by regulating the Hippo signaling pathway [,]. DUB1 is a short form of USP36, but its role in tumorigenesis remains largely unknown [,]. However, it has a role in GC progression. Wang et al. found that DUB1 is capable of interacting with, deubiquitinating, and stabilizing the Hippo signaling effector TAZ []. USP49 silencing decreases cell proliferation, migration, and invasion and enhances GC cell sensitivity to chemotherapy; however, this effect can be reversed by YAP1 overexpression. Because USP49 is a target gene modulated by YAP1/TEAD4, this result indicates that USP49 and YAP1 form a positive feedback loop to support the malignant progression of GC [].
2.4. USP10
USP10 plays a dual role in different human cancer types []. Zeng et al. showed that USP10 expression was lower in GC cell lines and clinical samples than in their noncancerous counterparts. More importantly, decreased USP10 expression indicates several highly malignant clinicopathological features and poor survival in patients with GC, suggesting that USP10 may be a prognostic biomarker []. Additionally, the calcium-binding protein S100A12 was found to be a GC prognostic marker, and its levels correlated with USP10 [,]. Given that USP10 and S100A12 are both located in the cytoplasm, USP10 may regulate the stability of S100A12 via deubiquitylation []. Therefore, future studies should investigate the role of USP10 in GC proliferation and metastasis. Moreover, the USP10-mediated stabilization of p53 likely contributes to the sensitivity of GC cells to 3-deazaneplanocin A, a histone methylation inhibitor that depletes EZH2, the enzymatic component of the PRC2 complex that catalyzes H3K27me3 []. As p53 turnover is regulated by a variety of DUBs [], whether these DUBs influence chemotherapy resistance in GC cells warrants further study. While these results indicate that USP10 functions as a tumor suppressor, one study showed that USP10 promoted GC cell migration and invasion by stabilizing replication factor C subunit 2 []. Collectively, the function of USP10 in GC remains unclear and requires further studies.
2.5. USP13, USP29, and USP37
As a master transcriptional factor that induces EMT, Snail is frequently overexpressed in tumors and drives tumor progression, cell survival, metastasis, and stem cell properties []. E3 ligases and DUBs are involved in the phosphorylation-dependent ubiquitination and proteasomal degradation of Snail []. For instance, GSK-3β phosphorylates Snail at six Ser sites. Snail phosphorylation at four Ser sites (Ser-107, 111, 115, and 119) causes cytoplasmic localization, whereas phosphorylation at two Ser sites (Ser-96 and 100) promotes ubiquitination and degradation by β-TrCP []. USP13, USP29, and USP37 were reported to promote the metastasis of GC by deubiquitinating and stabilizing Snail [,,,]. In addition, the upstream events of USP29 and USP37 are relevant for Snail regulation. Specifically, USP29, induced by TGFβ, TNFα, and hypoxia, increases the interaction between Snail and phosphatase SCP1, resulting in the dephosphorylation and deubiquitination of Snail, preventing its degradation []. USP37, which is transcriptionally activated by PLAG1-like zinc finger 2, stabilizes Snail in a GSK-3β phosphorylation-dependent manner []. Overall, these studies demonstrate the complexity of Snail regulation and provide potential therapeutic targets for the treatment of GC.
2.6. USP14
USP14 is a proteasome-associated DUB that plays a dual role in regulating protein degradation. Although it cleaves ubiquitin, it also promotes protein degradation by activating the proteasome [,]. USP14 has been reported to be an oncogene in GC []. USP14 levels are elevated in GC and may be an independent disease-free survival marker in patients [,]. In one study, USP14 was found to promote GC cell proliferation, invasion, and migration by stabilizing the EMT protein vimentin. Furthermore, the authors showed that miR-320a acts as a tumor suppressor by reducing vimentin expression and preventing USP14 from stabilizing this protein []. However, Fu et al. showed that USP14 depletion did not lead to cell death but sensitized GC cells to cisplatin-induced apoptosis []. As one chemical modification commonly found in eukaryotic mRNAs, N6-methyladenosine (m6A) plays a crucial role in modulating mRNA processing; studies have indicated that dysregulated m6A regulators are involved in cancer progression []. The m6A reader YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) promotes gastric carcinogenesis by modulating the translation of FDZ7, a key Wnt receptor []. YTHDF1 also increased USP14 levels in an m6A-dependent manner, and USP14 overexpression reversed the tumor-suppressive effects elicited by YTHDF1 silencing []. Additionally, the USP14 inhibitor IU1 restricted GC cell viability and metastasis triggered by YTHDF1 overexpression [,]. Together, these findings provide novel insights into the USP14-related mechanisms underlying malignancy in GC.
2.7. USP15
The role of USP15 in GC is less defined and contradictory. Zheng et al. suggested that the ectopic expression of USP15 could suppress GC cell growth, migration, and invasion through the deubiquitination of IκBα by the COP9 signalosome (CSN) complex, thereby hindering NF-κB activity [,]. In contrast, USP15 knockdown attenuated the activity of Wnt/β-catenin signaling and inhibited GC progression both in vitro and in vivo, but the mechanism through which USP15 regulates this pathway remains unclear []. As reviewed by Das et al., USP15 can either activate or suppress NF-κB and Wnt/β-catenin signaling depending on its effects on different proteins in various cellular contexts []. Furthermore, Huangfu et al. suggested that USP15 may assist in GC development by acting as a target that is regulated by the LINC00205/miR-26a axis []. In short, the effects of USP15 on specific signaling pathways, such as NF-κB and Wnt/β-catenin, and on tumorigenesis and metastasis in GC require further study.
2.8. USP22
USP22 has been identified as producing cancer-stem-cell-like qualities, including aggressive growth, metastasis, and therapy resistance []. Unlike other DUBs, the USP22 zinc-finger ubiquitin-binding domain does not directly bind to ubiquitin and instead is recruited and activated by the SAGA complex, forming a subcomplex (DUB module) with other proteins [,]. By regulating a broad range of histone and non-histone protein substrates, USP22 is associated with oncogenesis in GC []. Several studies have suggested that USP22 expression is upregulated in GC tissues and cells [,,,,,,]. In gastric tumors, USP22 expression was found to be positively correlated with the expression of the three well-known oncoproteins BMI1, c-Myc, and HSP90, which better predicted GC progression and prognosis than other methods [,,]. Notably, USP22 stabilizes BMI1 and c-Myc, but not HSP90 [,,]. USP22 maintained GC cell stemness by stabilizing BMI1 and promoted proliferation and metastasis by activating the FOXO1 and YAP signaling pathway [,]. However, these studies did not provide evidence that USP22 can stabilize BMI1 or c-Myc through deubiquitination, but this role was later identified in glioma and breast cancer cells [,]. As a guanine nucleotide exchange factor, son of sevenless 1 (SOS1) catalyzes GTP-bound RAS formation, whose activation elicits oncogenic signaling pathways []. Lim et al. proposed that USP22 upregulates SOS1 expression in GC, thus leading to the activation of RAS/ERK and RAS/PI3K/AKT pathways []. USP22 can also be modulated by the POU2F1/miR-4490 axis to further increase GC proliferation and metastasis [].
To target these USP22-regulated stemness properties, Yang et al. constructed USP22 siRNA-loaded nanoliposomes modified with anti-CD44 (USP22-NLs-CD44), which targets a stem-cell-associated marker. USP22-NLs-CD44 impaired tumorsphere formation and reduced the percentage of CD44+ cells in two human GC cell lines, MKN45 and NCI-N87, presenting an approach to eliminating GC stem cells [].
Although the aforementioned studies indicate that USP22 is a GC oncogene, a meta-analysis noted contradictions between USP22 expression, tumor size, differentiation state, tumor stage, and clinical outcomes in patients with gastric tumors. However, these inconsistencies may be due to the small sample size, suggesting that the role of USP22 should be analyzed in larger randomized controlled clinical trials []. Moreover, there are discrepancies in USP22′s effects on malignant behaviors. Ma et al. reported that USP22 only affects cell proliferation, whereas other authors have shown that USP22 affects GC cell proliferation, cell migration, and apoptosis [,,,]. Such controversial conclusions may be explained by differences in the cell context, including variations in USP22 levels among cell lines. Taken together, the role and downstream effectors of USP22 in GC warrant further investigation.
2.9. USP28
USP28 has been validated as an oncoprotein and therapeutic target in various cancer types, and a list of oncogenic substrates of USP28 have been reported, such as JUN, NOTCH1, CCNE, and LSD1 [,]. For instance, USP28 was identified as a DUB of LSD1 and conferred stem-cell-like characteristics in breast cancer []. Likewise, Zhao et al. found that USP28 was highly expressed in GC and was conducive to proliferation and metastasis, mainly due to its ability to increase LSD1 levels []. The same group subsequently designed and synthesized new [,,] triazolo [4,5-d] pyrimidine derivatives as potent USP28 inhibitors. Among them, compound 19 potently inhibited USP28 activity with an IC50 of 1.10 ± 0.02 μM and induced its degradation at higher doses, thereby exhibiting cytotoxic effects against GC cells []. In addition to LSD1, compound 19 also promotes c-Myc degradation, another substrate of USP28 []. Hu et al. showed that two GC cell lines, MKN-45 and SGC-7901, were sensitive to lanatoside C, an FDA-approved cardiac glycoside derived from Digitalis lanata. They also showed that lanatoside C partially promoted c-Myc degradation by reducing its interactions with USP28 []. These studies suggest that c-Myc is a potential downstream effector responsible for the oncogenic role of USP28 in GC.
2.10. USP32
USP32 was reported to be one protein that contributed to the chimeric Tre2 (USP6) oncogene []. The pro-cancer effects of USP32 have been observed in breast cancer and glioblastoma [,]. Additionally, USP32 promotes GC cell growth, metastasis, and chemoresistance by upregulating SMAD2, an important protein in the TGF-β signaling pathway []. However, it is unclear whether USP32 regulates the ubiquitination level of SMAD2.
2.11. USP33
USP33 expression is reduced in GC tissues and cell lines, which correlates with poor patient survival [,]. Additionally, USP33 overexpression suppresses the proliferation, migration, and invasion of gastric adenocarcinoma cells []. In addition, Xia et al. demonstrated that USP33 deubiquitinates and stabilizes ROBO1, which is required for the SLIT2-mediated inhibition of EMT and GC cell metastasis [].
2.12. USP39
Although USP39 was initially identified as a DUB without ubiquitin hydrolysis activity, other studies have suggested that it plays a role in RNA splicing and oncogenesis in multiple malignant tumors [,]. Intriguingly, USP39 stabilizes SP1, ZEB1, and FOXM1 through its deubiquitination activity, thus supporting the progression of hepatocellular carcinoma and breast cancer [,,]. Silencing USP39 expression hinders MGC-803 GC cell proliferation and induces G2/M arrest and PARP cleavage []. In line with these results, USP39 knockdown also decreased the proliferation of MGC-803 and HGC-27 cells. Moreover, miR-133a can suppress USP39 expression by directly targeting its 3′ UTR, thereby acting as a tumor suppressor in GC []. However, the mechanisms by which USP39 depletion inhibits GC proliferation require further investigation.
2.13. USP44
Aneuploidy is a common characteristic of tumor cells []. Several studies have demonstrated that USP44 dysregulation results in aneuploidy, which has also been observed in GC tissues [,]. Furthermore, the combination of elevated USP44 expression and DNA aneuploidy provides useful prognostic information []. In addition, USP44 is regulated by the circFOXO3/miR-143-3p axis in GC, in which circFOXO3 functions as a miR-143-3p sponge to promote USP44-directed malignancy in gastric carcinoma [].
2.14. USP47
USP47 is inhibited by miR-204-5p, which represses GC cell proliferation []. Interaction between USP47 and β-TrCP, a subunit of the SCF β-TrCP E3 ubiquitin ligase complex, is involved in a number of signaling pathways [,,]. Naghavi et al. found that USP47 stabilizes β-TrCP-modulated NF-κB activity by promoting RelA phosphorylation. Their findings also showed that USP47 knockdown sensitized NCI-N87 cells to camptothecin- or etoposide-induced apoptosis []. These data indicate that USP47 may be involved in NF-κB-dependent chemoresistance in some cellular contexts.
2.15. CYLD
CYLD was initially described in cylindromatosis and is currently considered a tumor suppressor that negatively regulates multiple signaling pathways, such as NF-κB, AKT, and Wnt [,,]. Ghadami et al. showed that CYLD expression was decreased in GC tissues owing to the hypermethylation of its promoter. They also revealed a direct correlation between H. pylori, EBV, and CMV infections and CYLD hypermethylation and downregulated protein expression, suggesting that infectious-agent-induced CYLD hypermethylation may be a significant mechanism of GC development []. Several studies have shown that CYLD can also be regulated by non-coding RNAs and serve as a downstream effector by modulating the progression and chemoresistance of GC. For example, miR-130b, miR-425-5p, miR-454, and exosomal miR-588 from M2-polarized macrophages negatively regulated CYLD expression by targeting its 3′-UTR, thus promoting GC cell proliferation, migration, invasion, and treatment resistance [,,,]. In addition, the activation of NF-κB by miR-362, miR-500, and miR-20a plays a significant role in the survival and cisplatin resistance of GC cells [,,]. Notably, miR-500 also activates NF-κB signaling by directly repressing two other negative regulators of NF-κB, OTUD7B, and TAX1BP1 []. miR-505 also targets the 3′-UTR of CYLD mRNA. However, miR-505 can be sponged by CRAL and increase CYLD levels, which decreases AKT signaling and increases treatment susceptibility in GC cells []. Furthermore, the ALKBH5/ZNF333/CYLD axis participates in the development of gastric intestinal metaplasia, a precursor of GC. In bile-acid-induced gastric intestinal metaplasia, ZNF333 activates NF-κB signaling by repressing CYLD expression []. Taken together, these studies suggest that targeting proteins and miRNA that reduce CYLD levels may be a useful therapeutic strategy for GC.
2.16. Other USPs Involved in GC
In addition to the USPs mentioned previously herein, there are several other GC-associated USPs, including USP1, USP2, USP20, USP21, and USP42. USP1 levels were increased in GC cell lines and clinical samples, and its overexpression confers poor patient survival rates. USP1 knockdown inhibits their proliferation, migration, and invasion [,]. Mechanistically, USP1 promotes metastasis by stabilizing the inhibitor of DNA binding-2 protein []. USP2 was recently reported to harbor oncogenic properties by promoting E2F4-mediated cytoprotective autophagy and zinc homeostasis. By blocking the USP2-E2F4 interaction, emetine inhibited autophagy and GC aggressiveness, suggesting the therapeutic potential of targeting the USP2-E2F4 axis [].
USP20 promotes the tumorigenesis of several cancer types, such as breast and cervical cancer; however, it exhibits inhibitory effects on GC []. Past reports have indicated that USP20 overexpression inhibited cell proliferation and delayed the cell cycle transition from the G1 to S phase by stabilizing Claspin [,]. Additionally, USP21 elevates MAPK1 expression through the transcription factor GATA3, thereby contributing to tumor growth and stemness in GC []. Hou et al. showed that higher USP42 expression was associated with poor prognosis in patients with GC. In vitro and in vivo studies showed that USP42 knockdown inhibited GC cell survival and invasion, suggesting that USP42 may be a therapeutic target [].
3. UCHs and GC
All members of this family have been reported to be associated with GC (Figure 3). UCHL1, also called PGP9.5, controls intracellular ubiquitin levels and is related to tumorigenesis in various cancer types []. However, UCHL1 has a paradoxical role in GC. UCHL1 promoter hypermethylation is a common event in several types of primary digestive tumors, including GC [,,,,]. Moreover, UCHL1 hypermethylation is more frequent in advanced-stage GC [,]. In addition, this hypermethylation is associated with poor prognosis []. Furthermore, galangin cytotoxicity in SNU-484 cells may occur through increasing UCHL1 levels []. These studies indicate that UCHL1 may serve as a tumor suppressor and a diagnostic marker for GC. However, two other studies reported higher UCHL1 expression in liver metastases from GC and gastric cardiac adenocarcinoma, likely because UCHL1 overexpression increases the proliferation, migration, and invasion capabilities of GC cells [,]. Lastly, the pro-cancer effect of UCHL1 is mechanistically correlated with activated Akt and ERK1/2 pathways []. Thus, the role of UCHL1 in GC requires further investigation.
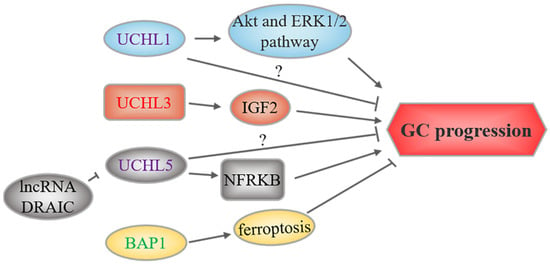
Figure 3.
UCH-related upstream regulation and downstream mechanisms in GC. UCHs marked in red and green represent oncoproteins and tumor suppressors, respectively, while purple-labeled UCHs play a dual role. The text to the left and right of the UCHs corresponds to the upstream regulatory events and downstream substrates or signaling pathways, respectively.
Similar to USP14 and PSMD14, UCHL5 is also a proteasome-associated DUB. Higher UCHL5 expression is a predictor of increased survival in a subgroup of patients with early-stage GC []. However, another study suggested that UCHL5 stabilizing NFRKB, a chromatin-remodeling protein, may promote GC cell proliferation and metastasis []. Therefore, the biological function of UCHL5 requires more investigation.
One recent study reported that UCHL3 promotes GC cell migration and invasion by upregulating IGF2 []. In addition, Yan et al. found that BAP1 levels are decreased in GC, which was linked to advanced tumor features and unfavorable survival, suggesting its role as a tumor suppressor []. Moreover, another study suggested that BAP1 upregulation is essential for ferroptosis induced by 3,3′-diindolylmethane in BGC-823 cells, suggesting that BAP1-induced ferroptosis could be one of the potential mechanisms by which it suppresses GC progression [].
4. OTUs and GC
Three members of this family, including OTUB1, OTUB2, and A20, have been linked to GC (Figure 4). Weng et al. found that patients with GC with high OTUB1 expression were associated with several advanced clinical features, such as invasion depth, lymph node status, and nerve invasion, and these patients had lower disease-specific survival rates. OTUB1 was found to be active in GC cell invasion and migration [], but the underlying mechanism is unknown. Similar to OTUB1, OTUB2 was also found to be overexpressed in GC tissues and cell lines and predicted a poor prognosis [,]. Silencing OTUB2 inhibited GC cell growth, metastasis, and sphere formation. Mechanistically, OTUB2 acts as a potential driver oncogene in GC by deubiquitinating and stabilizing the demethylase KDM1A, and epithelial keratin KRT80 [,]. As previously reported, KDM1A and KRT80 contributed to GC progression by regulating KLF2 expression and the PI3K/Akt pathway, respectively [,].
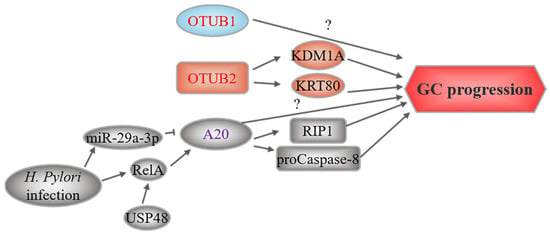
Figure 4.
OTU-related upstream regulation and downstream mechanisms in GC. OTUs marked in red represent oncoproteins, while purple-labeled OTUs play a dual role. The text to the left and right of the OTUs corresponds to the upstream regulatory events and downstream substrates, respectively.
A20, also known as TNFAIP3, is a ubiquitin-editing enzyme with both DUB and E3 ligase activities []. A20 expression is increased in GC tissues and cell lines, which is associated with poor clinical outcomes [,]. In vitro studies have suggested that A20 downregulation suppresses the proliferation, migration, and invasion of MGC-803 GC cells []. In addition, A20 was found to be the target of miR-200a in GC cells (MGC-803 and SGC7901) resistant to TRAIL-induced apoptosis. Overexpressing miR-200a or depleting A20 enhanced apoptosis by reducing RIP1 polyubiquitination and promoting caspase-8 cleavage []. These results were consistent with a previous study showing that A20 mediates the polyubiquitination of RIP1, which in turn binds to and inhibits caspase-8 activation []. Later studies suggested that RIP1 is involved in multiple cellular signaling pathways and processes, such as NF-κB activation and apoptosis, and promotes GC growth and invasion []. Moreover, the DUB activity of A20 counteracted the ubiquitination of procaspase-8, thereby restricting caspase-8 activity and apoptosis []. Consistently, NF-κB activation induced A20 upregulation during H. pylori infection, mainly because USP48 stabilizes nuclear RelA and promotes its transcriptional activity [,,,]. Together, USP48 and A20 promoted GC cell survival during H. pylori infection and suggested an oncogenic role for A20. Conversely, one study reported that H. pylori infection increased miR-29a-3p, which promoted the migration of gastric epithelial cells by reducing A20 expression, indicating that A20 silencing may induce EMT to promote GC progression []. Thus, the role of A20 in GC might be context-dependent and requires further investigation.
5. JAMMs and GC
Three members of this family, PSMD14, CSN5, and BRCC3, have been found to be associated with GC (Figure 5). PSMD14, also known as Rpn11 and POH1, is a subunit of the proteasomal 19S regulatory particle, which functions as a DUB []. PSMD14 overexpression has been reported to be tumorigenic and promote cancer progression through multiple mechanisms [,,,,], such as stabilizing the alternative splicing factor PTBP1 to promote GC tumorigenesis [].
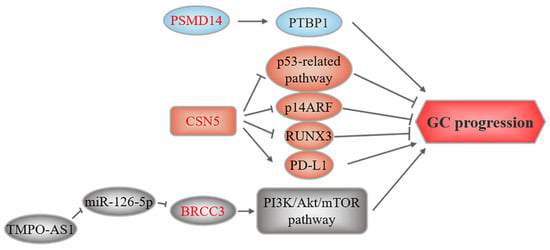
Figure 5.
JAMM-related upstream regulation and downstream mechanisms in GC. These three members of JAMMs all play an oncogenic role. The text to the left and right of the JAMMs corresponds to the upstream regulatory events and downstream substrates, respectively.
CSN5, the catalytic subunit of the COP9 signalosome, also called COPS5 or JAB1, may play a role in GC []. CSN5 overexpression contributes to GC by modulating the stability or expression of several tumorigenic proteins. Silencing CSN5 suppresses GC cell growth and induces apoptosis by regulating P53 and BAX expression []. Additionally, CSN5 induces non-ubiquitin proteasomal degradation of the tumor suppressor p14ARF []. CSN5 also facilitates the nuclear export and degradation of the tumor suppressor RUNX3 []. Previous studies suggested that TNFα and CCL5 increase CSN5, stabilize PD-L1, and facilitate the immune escape of breast cancer cells and colorectal cancer cells [,]. CSN5 also stabilizes PD-L1 in GC cells. Furthermore, CSN5 activity in GC cells is regulated by the DAPK1/IKKβ axis []. Collectively, these studies indicate that CSN5 could be a novel therapeutic target in GC.
Lastly, Hu et al. reported that BRCC3 is upregulated in GC and is regulated by the lncRNA TMPO-AS1/miR-126-5p axis. TMPO-AS1 sponges miR-126-5p to upregulate BRCC3 expression, thereby activating the PI3K/Akt/mTOR pathway, which leads to malignancy [].
6. MJDs and GC
In the MJD family, only Ataxin-3 has been reported to be associated with GC tumorigenesis. Ataxin-3 levels were found to be decreased in GC tissues and cells, which correlated with clinicopathological characteristics, including tumor size, Lauren classification, histologic differentiation, and p53 mutation status []. However, the molecular mechanisms underlying this process have not yet been elucidated.
7. Conclusions and Perspectives
In the past few years, DUBs and tumorigenesis have been linked. Here, we summarize the regulatory roles of DUBs in the occurrence and development of GC. As shown in Figure 2, Figure 3, Figure 4 and Figure 5, most DUBs promote GC progression, whereas several DUBs play an inhibitory role or exert context-dependent effects. Moreover, the regulatory mechanisms of DUBs are complicated and involve multiple targets and signaling pathways, yet some regulatory mechanisms have not been discovered (Table 1). Although the USP subfamily has received the most attention in GC research, other subfamilies, namely, OTUs, UCHs, JAMMs, and MJDs, have received attention as well. However, there have been no reports on MINDYs and ZUFSPs to date. Therefore, we speculate that USPs may be the most promising biomarkers for GC diagnosis and treatment, and we believe that the function of other DUB subfamilies requires further in-depth exploration.
In addition, DUB regulation is complex. Although the regulatory mechanisms of some GC-related DUBs remain to be discovered, non-coding RNAs have been reported to modulate the expression of DUBs, such as USP3 and CYLD, in GC (Table 1). Intriguingly, three studies discovered that non-coding RNAs are also key mediators of interactions between GC cells and other cell types, including gastric mucosa cells, CAFs, and M2-polarized macrophages, and that USP3, USP7, and CYLD are linked to this process [,,]. Furthermore, several infectious agents, cytokines, and antitumor agents also affect the expression of several DUBs, such as USP3, USP7, USP10, USP29, CYLD, and A20 [,,,,,,,] (Table 1), which implicates unknown pro-cancer effects under certain circumstances.
Finally, research on the therapeutic potential of DUB inhibitors for GC therapy is limited despite the efforts made in their development. Some inhibitors show promising therapeutic effects in various cancer types [,]. For instance, pharmacological inhibition of USP28, USP1, and USP7 with their inhibitors efficiently hindered in vitro and in vivo tumor growth or metastasis in squamous cell carcinoma, breast cancer, and colon cancer, respectively [,,]. However, only a few studies have demonstrated the antitumor effects of USP7, USP14, and USP28 inhibitors on GC cells. Moreover, compounds that upregulate DUB tumor suppressors also exhibited activity against GC cells, providing another direction for drug development (Table 2). In conclusion, the investigation of DUBs in the pathogenesis and treatment of GC requires more work, which may provide clues for GC treatment in the future.
Author Contributions
Conceptualization, T.A. and J.H.; writing—original draft preparation, T.A. and Y.L.; writing—review and editing, G.T. and J.H.; visualization, Z.G., Y.W. and C.S.; funding acquisition, T.A., Y.L. and J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the Start-up Fund for Tao An from Qilu University of Technology (Shandong Academy of Sciences) (No. 81110573), the Science, Education and Industry Integration Innovation Pilot Project from Qilu University of Technology (Shandong Academy of Sciences) (2022JBZ02-04), the Shandong Provincial Natural Science Foundation (ZR2020QH329), and the Foundation of Xiamen Municipal Bureau of Science and Technology (No. 3502Z20214ZD1034).
Conflicts of Interest
No conflict of interest needs to be declared.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Garcia, E.; Garcia-Puga, M.; Arevalo, S.; Matheu, A. Towards precision medicine: Linking genetic and cellular heterogeneity in gastric cancer. Ther. Adv. Med. Oncol. 2018, 10, 1758835918794628. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.N.; Teng, Q.X.; Tian, Q.; Chen, W.; Xie, Y.; Wu, K.; Zeng, Q.; Zeng, L.; Pan, Y.; Chen, Z.S.; et al. Signaling pathways and therapeutic interventions in gastric cancer. Signal Transduct. Target. Ther. 2022, 7, 358. [Google Scholar] [CrossRef] [PubMed]
- Correa, P. Gastric cancer: Overview. Gastroenterol. Clin. N. Am. 2013, 42, 211–217. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Z.; Soutto, M.; Wang, W.; Piazuelo, M.B.; Zhu, S.; Guo, Y.; Maturana, M.J.; Corvalan, A.H.; Chen, X.; et al. Integrated Analysis of Mouse and Human Gastric Neoplasms Identifies Conserved microRNA Networks in Gastric Carcinogenesis. Gastroenterology 2019, 156, 1127–1139.e8. [Google Scholar] [CrossRef]
- Zhang, D.; Chu, Y.; Qian, H.; Qian, L.; Shao, J.; Xu, Q.; Yu, L.; Li, R.; Zhang, Q.; Wu, F.; et al. Antitumor Activity of Thermosensitive Hydrogels Packaging Gambogic Acid Nanoparticles and Tumor-Penetrating Peptide iRGD Against Gastric Cancer. Int. J. Nanomed. 2020, 15, 735–747. [Google Scholar] [CrossRef]
- Komander, D.; Clague, M.J.; Urbe, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef]
- Kwon, Y.T.; Ciechanover, A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 2017, 42, 873–886. [Google Scholar] [CrossRef]
- Csizmadia, T.; Low, P. The Role of Deubiquitinating Enzymes in the Various Forms of Autophagy. Int. J. Mol. Sci. 2020, 21, 4196. [Google Scholar] [CrossRef]
- Sun, T.; Liu, Z.; Yang, Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol. Cancer 2020, 19, 146. [Google Scholar] [CrossRef]
- Kaushal, K.; Ramakrishna, S. Deubiquitinating Enzyme-Mediated Signaling Networks in Cancer Stem Cells. Cancers 2020, 12, 3253. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Wang, R.; Zhang, Y.; Li, X.; Gan, Y.; Gao, F.; Rong, P.; Wang, W.; Li, W. The role of ubiquitination and deubiquitination in tumor invasion and metastasis. Int. J. Biol. Sci. 2022, 18, 2292–2303. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Kim, S.; Hwang, G.; Song, J. Deubiquitinases: Modulators of Different Types of Regulated Cell Death. Int. J. Mol. Sci. 2021, 22, 4352. [Google Scholar] [CrossRef] [PubMed]
- Jolly, L.A.; Kumar, R.; Penzes, P.; Piper, M.; Gecz, J. The DUB Club: Deubiquitinating Enzymes and Neurodevelopmental Disorders. Biol. Psychiatry 2022, 92, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Harrigan, J.A.; Jacq, X.; Martin, N.M.; Jackson, S.P. Deubiquitylating enzymes and drug discovery: Emerging opportunities. Nat. Rev. Drug Discov. 2018, 17, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ye, C.; Tian, T.; Jiang, Q.; Zhao, P.; Wang, X.; Liu, F.; Shan, J.; Ruan, J. The emerging role of ubiquitin-specific protease 20 in tumorigenesis and cancer therapeutics. Cell Death Dis. 2022, 13, 434. [Google Scholar] [CrossRef]
- Meng, D.; Li, D. Ubiquitin-specific protease 1 overexpression indicates poor prognosis and promotes proliferation, migration, and invasion of gastric cancer cells. Tissue Cell 2022, 74, 101723. [Google Scholar] [CrossRef]
- Li, N.; Wu, L.; Zuo, X.; Luo, H.; Sheng, Y.; Yan, J. USP1 Promotes GC Metastasis via Stabilizing ID2. Dis. Markers 2021, 2021, 3771990. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, J.; Wang, X.; Cai, S.; Guo, Y.; Ye, L.; Li, D.; Hu, A.; Jin, S.; Yuan, B.; et al. Therapeutic targeting of the USP2-E2F4 axis inhibits autophagic machinery essential for zinc homeostasis in cancer progression. Autophagy 2022, 18, 2615–2635. [Google Scholar] [CrossRef]
- Fang, C.L.; Lin, C.C.; Chen, H.K.; Hseu, Y.C.; Hung, S.T.; Sun, D.P.; Uen, Y.H.; Lin, K.Y. Ubiquitin-specific protease 3 overexpression promotes gastric carcinogenesis and is predictive of poor patient prognosis. Cancer Sci. 2018, 109, 3438–3449. [Google Scholar] [CrossRef]
- Wu, X.; Liu, M.; Zhu, H.; Wang, J.; Dai, W.; Li, J.; Zhu, D.; Tang, W.; Xiao, Y.; Lin, J.; et al. Ubiquitin-specific protease 3 promotes cell migration and invasion by interacting with and deubiquitinating SUZ12 in gastric cancer. J. Exp. Clin. Cancer Res. 2019, 38, 277. [Google Scholar] [CrossRef]
- Wu, X.; Wang, H.; Zhu, D.; Chai, Y.; Wang, J.; Dai, W.; Xiao, Y.; Tang, W.; Li, J.; Hong, L.; et al. USP3 promotes gastric cancer progression and metastasis by deubiquitination-dependent COL9A3/COL6A5 stabilisation. Cell Death Dis. 2021, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Jin, M.; Cao, F.; Li, J.; Wu, J.; Xu, L.; Liu, X.; Shi, Y.; Chen, W. Hsa_circ_0017639 expression promotes gastric cancer proliferation and metastasis by sponging miR-224-5p and upregulating USP3. Gene 2020, 750, 144753. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Zhang, J.; Cao, T.; Chen, B.; Tian, Y.; Shi, Y. Exosome-mediated lncRNA SND1-IT1 from gastric cancer cells enhances malignant transformation of gastric mucosa cells via up-regulating SNAIL1. J. Transl. Med. 2022, 20, 284. [Google Scholar] [CrossRef] [PubMed]
- Georges, A.; Marcon, E.; Greenblatt, J.; Frappier, L. Identification and Characterization of USP7 Targets in Cancer Cells. Sci. Rep. 2018, 8, 15833. [Google Scholar] [CrossRef]
- Wang, Z.; Kang, W.; Li, O.; Qi, F.; Wang, J.; You, Y.; He, P.; Suo, Z.; Zheng, Y.; Liu, H.M. Abrogation of USP7 is an alternative strategy to downregulate PD-L1 and sensitize gastric cancer cells to T cells killing. Acta Pharm. Sin. B 2021, 11, 694–707. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, T.; Liu, R.; Ning, T.; Yang, H.; Liu, D.; Zhang, Q.; Lin, D.; Ge, S.; Bai, M.; et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol. Cancer 2020, 19, 43. [Google Scholar] [CrossRef]
- Fu, X.; Xie, W.; Song, X.; Wu, K.; Xiao, L.; Liu, Y.; Zhang, L. Aberrant expression of deubiquitylating enzyme USP9X predicts poor prognosis in gastric cancer. Clin. Res. Hepatol. Gastroenterol. 2017, 41, 687–692. [Google Scholar] [CrossRef]
- Xu, X.; Wang, S.; Wang, H.; Pan, C.; Yang, W.; Yu, J. Hsa_circ_0008434 regulates USP9X expression by sponging miR-6838-5p to promote gastric cancer growth, migration and invasion. BMC Cancer 2021, 21, 1289. [Google Scholar] [CrossRef]
- Zhang, C.; Qian, H.; Liu, K.; Zhao, W.; Wang, L. A Feedback Loop Regulation of LINC01433 And YAP Promotes Malignant Behavior In Gastric Cancer Cells. OncoTargets Ther. 2019, 12, 7949–7962. [Google Scholar] [CrossRef]
- Zeng, Z.; Wu, H.X.; Zhan, N.; Huang, Y.B.; Wang, Z.S.; Yang, G.F.; Wang, P.; Fu, G.H. Prognostic significance of USP10 as a tumor-associated marker in gastric carcinoma. Tumor Biol. 2014, 35, 3845–3853. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.L.; Itahana, Y.; Lei, Z.D.; Chia, N.Y.; Wu, Y.; Yu, Y.; Zhang, S.L.; Thike, A.A.; Pandey, A.; Rozen, S.; et al. TP53 genomic status regulates sensitivity of gastric cancer cells to the histone methylation inhibitor 3-deazaneplanocin A (DZNep). Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012, 18, 4201–4212. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chai, L.; Ding, Z.; He, H. CircCOL1A2 Sponges MiR-1286 to Promote Cell Invasion and Migration of Gastric Cancer by Elevating Expression of USP10 to Downregulate RFC2 Ubiquitination Level. J. Microbiol. Biotechnol. 2022, 32, 938–948. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, M.; He, B.; Li, Q. Inhibition of USP11 sensitizes gastric cancer to chemotherapy via suppressing RhoA and Ras-mediated signaling pathways. Clin. Res. Hepatol. Gastroenterol. 2022, 46, 101779. [Google Scholar] [CrossRef] [PubMed]
- Biterge Sut, B. Molecular profiling of immune cell-enriched Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) interacting protein USP13. Life Sci. 2020, 258, 118170. [Google Scholar] [CrossRef]
- Zhang, T.; Zheng, J.; Qiao, L.; Zhao, W. Deubiquitinase USP13 promotes the epithelial-mesenchymal transition and metastasis in gastric cancer by maintaining Snail protein. Pathol. Res. Pract. 2022, 229, 153705. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, Y.; Sui, Z.; Zhang, Y.; Liu, M.; Tang, H. USP14 de-ubiquitinates vimentin and miR-320a modulates USP14 and vimentin to contribute to malignancy in gastric cancer cells. Oncotarget 2017, 8, 48725–48736. [Google Scholar] [CrossRef]
- Fu, Y.; Ma, G.; Liu, G.; Li, B.; Li, H.; Hao, X.; Liu, L. USP14 as a novel prognostic marker promotes cisplatin resistance via Akt/ERK signaling pathways in gastric cancer. Cancer Med. 2018, 7, 5577–5588. [Google Scholar] [CrossRef]
- Chen, X.Y.; Liang, R.; Yi, Y.C.; Fan, H.N.; Chen, M.; Zhang, J.; Zhu, J.S. The m(6)A Reader YTHDF1 Facilitates the Tumorigenesis and Metastasis of Gastric Cancer via USP14 Translation in an m(6)A-Dependent Manner. Front. Cell Dev. Biol. 2021, 9, 647702. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, H.; Ma, R.; Liu, H.; Gao, P. Long non-coding RNA KRT19P3 suppresses proliferation and metastasis through COPS7A-mediated NF-kappaB pathway in gastric cancer. Oncogene 2019, 38, 7073–7088. [Google Scholar] [CrossRef]
- Zhong, M.; Zhou, L.; Fang, Z.; Yao, Y.Y.; Zou, J.P.; Xiong, J.P.; Xiang, X.J.; Deng, J. Ubiquitin-specific protease 15 contributes to gastric cancer progression by regulating the Wnt/beta-catenin signaling pathway. World J. Gastroenterol. 2021, 27, 4221–4235. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, L.; Fan, B.; Wang, G.; Gan, X.; Tian, S.; He, Q.; Yao, Q.; Shi, J.; Li, X.; Du, H.; et al. Novel prognostic marker LINC00205 promotes tumorigenesis and metastasis by competitively suppressing miRNA-26a in gastric cancer. Cell Death Discov. 2022, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, C.; Ji, J.; Jiang, J.; Shi, M.; Cai, Q.; Yu, Y.; Zhu, Z.; Zhang, J. Deubiquitinating enzyme USP20 is a positive regulator of Claspin and suppresses the malignant characteristics of gastric cancer cells. Int. J. Oncol. 2017, 50, 1136–1146. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Shi, D.; Lin, L.; Li, H.; Wei, Y.; Li, B.; Wu, D. De-Ubiquitinating Enzymes USP21 Regulate MAPK1 Expression by Binding to Transcription Factor GATA3 to Regulate Tumor Growth and Cell Stemness of Gastric Cancer. Front. Cell Dev. Biol. 2021, 9, 641981. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.D.; Cui, B.B.; Sun, L.Y.; Zheng, H.Q.; Huang, Q.; Tong, J.X.; Zhang, Q.F. The co-expression of USP22 and BMI-1 may promote cancer progression and predict therapy failure in gastric carcinoma. Cell Biochem. Biophys. 2011, 61, 703–710. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Jin, Y.J.; Zhang, Y.H.; Meng, H.X.; Zhao, B.S.; Jiang, Y.; Zhu, J.W.; Liang, G.Y.; Kong, D.; Jin, X.M. Ubiquitin-specific peptidase 22 overexpression may promote cancer progression and poor prognosis in human gastric carcinoma. Transl. Res. 2015, 165, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Fu, H.L.; Wang, Z.; Huang, H.; Ni, J.; Song, J.; Xia, Y.; Jin, W.L.; Cui, D.X. USP22 maintains gastric cancer stem cell stemness and promotes gastric cancer progression by stabilizing BMI1 protein. Oncotarget 2017, 8, 33329–33342. [Google Scholar] [CrossRef]
- Yang, F.; Zheng, Z.; Xue, X.; Zheng, L.; Qin, J.; Li, H.; Zhou, Y.; Fang, G. Targeted eradication of gastric cancer stem cells by CD44 targeting USP22 small interfering RNA-loaded nanoliposomes. Future Oncol. 2019, 15, 281–295. [Google Scholar] [CrossRef]
- Zheng, H.; Yu, J.; Li, W.; Yang, D.; Gao, C.; Zhang, Q.; Xu, L. Is co-expression of USP22 and HSP90 more effective in predicting prognosis of gastric cancer? Pathol. Res. Pract. 2019, 215, 653–659. [Google Scholar] [CrossRef]
- Liu, H.; Liu, N.; Zhao, Y.; Zhu, X.; Wang, C.; Liu, Q.; Gao, C.; Zhao, X.; Li, J. Oncogenic USP22 supports gastric cancer growth and metastasis by activating c-Myc/NAMPT/SIRT1-dependent FOXO1 and YAP signaling. Aging 2019, 11, 9643–9660. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, S.; Li, J.; Dai, W.; Tang, W.; Xiang, L.; Zhang, W.; Lin, J.; Wang, J.; Wu, X.; et al. The POU2F1/miR-4490/USP22 axis regulates cell proliferation and metastasis in gastric cancer. Cell. Oncol. (Dordr) 2020, 43, 1017–1033. [Google Scholar] [CrossRef]
- Lim, C.; Xu, J.C.; Chen, T.Y.; Xu, J.X.; Chen, W.F.; Hu, J.W.; Li, Q.L.; Zhang, Y.Q. Ubiquitin-specific peptide 22 acts as an oncogene in gastric cancer in a son of sevenless 1-dependent manner. Cancer Cell Int. 2020, 20, 45. [Google Scholar] [CrossRef]
- Zhao, L.J.; Zhang, T.; Feng, X.J.; Chang, J.; Suo, F.Z.; Ma, J.L.; Liu, Y.J.; Liu, Y.; Zheng, Y.C.; Liu, H.M. USP28 contributes to the proliferation and metastasis of gastric cancer. J. Cell. Biochem. 2018, 120, 7657–7666. [Google Scholar] [CrossRef]
- Qian, W.; Li, Q.; Wu, X.; Li, W.; Li, Q.; Zhang, J.; Li, M.; Zhang, D.; Zhao, H.; Zou, X.; et al. Deubiquitinase USP29 promotes gastric cancer cell migration by cooperating with phosphatase SCP1 to stabilize Snail protein. Oncogene 2020, 39, 6802–6815. [Google Scholar] [CrossRef]
- Dou, N.; Hu, Q.; Li, L.; Wu, Q.; Li, Y.; Gao, Y. USP32 promotes tumorigenesis and chemoresistance in gastric carcinoma via upregulation of SMAD2. Int. J. Biol. Sci. 2020, 16, 1648–1657. [Google Scholar] [CrossRef]
- Chen, Y.; Pang, X.; Ji, L.; Sun, Y.; Ji, Y. Reduced Expression of Deubiquitinase USP33 Is Associated with Tumor Progression and Poor Prognosis of Gastric Adenocarcinoma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 3496–3505. [Google Scholar] [CrossRef]
- Xia, Y.; Wang, L.; Xu, Z.; Kong, R.; Wang, F.; Yin, K.; Xu, J.; Li, B.; He, Z.; Wang, L.; et al. Reduced USP33 expression in gastric cancer decreases inhibitory effects of Slit2-Robo1 signalling on cell migration and EMT. Cell Prolif. 2019, 52, e12606. [Google Scholar] [CrossRef]
- Wang, D.; Li, Z.; Li, X.; Yan, C.; Yang, H.; Zhuang, T.; Wang, X.; Zang, Y.; Liu, Z.; Wang, T.; et al. DUB1 suppresses Hippo signaling by modulating TAZ protein expression in gastric cancer. J. Exp. Clin. Cancer Res. 2022, 41, 219. [Google Scholar] [CrossRef]
- Wu, L.; Zhao, N.; Zhou, Z.; Chen, J.; Han, S.; Zhang, X.; Bao, H.; Yuan, W.; Shu, X. PLAGL2 promotes the proliferation and migration of gastric cancer cells via USP37-mediated deubiquitination of Snail1. Theranostics 2021, 11, 700–714. [Google Scholar] [CrossRef]
- Wang, X.; Yu, Q.; Huang, L.; Yu, P. Lentivirus-mediated inhibition of USP39 suppresses the growth of gastric cancer cells via PARP activation. Mol. Med. Rep. 2016, 14, 301–306. [Google Scholar] [CrossRef]
- Dong, X.; Su, H.; Jiang, F.; Li, H.; Shi, G.; Fan, L. miR-133a, directly targeted USP39, suppresses cell proliferation and predicts prognosis of gastric cancer. Oncol. Lett. 2018, 15, 8311–8318. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Zhu, Z.; Wang, Y.; Zhang, C.; Yu, S.; Zhu, Q.; Yan, B. Overexpression and Biological Function of Ubiquitin-Specific Protease 42 in Gastric Cancer. PLoS ONE 2016, 11, e0152997. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, S.; Oki, E.; Ando, K.; Iimori, M.; Nakaji, Y.; Nakashima, Y.; Saeki, H.; Oda, Y.; Maehara, Y. High ubiquitin-specific protease 44 expression induces DNA aneuploidy and provides independent prognostic information in gastric cancer. Cancer Med. 2017, 6, 1453–1464. [Google Scholar] [CrossRef] [PubMed]
- Xiang, T.; Jiang, H.S.; Zhang, B.T.; Liu, G. CircFOXO3 functions as a molecular sponge for miR-143-3p to promote the progression of gastric carcinoma via upregulating USP44. Gene 2020, 753, 144798. [Google Scholar] [CrossRef]
- Zhang, B.; Yin, Y.; Hu, Y.; Zhang, J.; Bian, Z.; Song, M.; Hua, D.; Huang, Z. MicroRNA-204-5p inhibits gastric cancer cell proliferation by downregulating USP47 and RAB22A. Med. Oncol. 2015, 32, 331. [Google Scholar] [CrossRef]
- Naghavi, L.; Schwalbe, M.; Ghanem, A.; Naumann, M. Deubiquitinylase USP47 Promotes RelA Phosphorylation and Survival in Gastric Cancer Cells. Biomedicines 2018, 6, 62. [Google Scholar] [CrossRef]
- Liu, Z.; Li, J.; Ding, Y.; Ma, M.; Chen, J.; Lei, W.; Li, L.; Yao, Y.; Yu, X.; Zhong, M.; et al. USP49 mediates tumor progression and poor prognosis through a YAP1-dependent feedback loop in gastric cancer. Oncogene 2022, 41, 2555–2570. [Google Scholar] [CrossRef]
- Xia, J.T.; Chen, L.Z.; Jian, W.H.; Wang, K.B.; Yang, Y.Z.; He, W.L.; He, Y.L.; Chen, D.; Li, W. MicroRNA-362 induces cell proliferation and apoptosis resistance in gastric cancer by activation of NF-kappaB signaling. J. Transl. Med. 2014, 12, 33. [Google Scholar] [CrossRef]
- Zhang, L.; Ding, Y.; Yuan, Z.; Liu, J.; Sun, J.; Lei, F.; Wu, S.; Li, S.; Zhang, D. MicroRNA-500 sustains nuclear factor-kappaB activation and induces gastric cancer cell proliferation and resistance to apoptosis. Oncotarget 2015, 6, 2483–2495. [Google Scholar] [CrossRef]
- Sun, B.; Li, L.; Ma, W.; Wang, S.; Huang, C. MiR-130b inhibits proliferation and induces apoptosis of gastric cancer cells via CYLD. Tumor Biol. 2016, 37, 7981–7987. [Google Scholar] [CrossRef]
- Zhu, M.; Zhou, X.; Du, Y.; Huang, Z.; Zhu, J.; Xu, J.; Cheng, G.; Shu, Y.; Liu, P.; Zhu, W.; et al. miR-20a induces cisplatin resistance of a human gastric cancer cell line via targeting CYLD. Mol. Med. Rep. 2016, 14, 1742–1750. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.F.; Gong, F.M.; Wang, B.S.; Zheng, W. MiR-425-5p promotes tumor progression via modulation of CYLD in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2130–2136. [Google Scholar] [PubMed]
- Ghadami, E.; Nikbakhsh, N.; Fattahi, S.; Kosari-Monfared, M.; Ranaee, M.; Taheri, H.; Amjadi-Moheb, F.; Godazandeh, G.; Shafaei, S.; Nosrati, A.; et al. Epigenetic alterations of CYLD promoter modulate its expression in gastric adenocarcinoma: A footprint of infections. J. Cell. Physiol. 2019, 234, 4115–4124. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Q.; Xu, G.; Meng, N.; Huang, X.; Jiang, Z.; Chen, C.; Zhang, Y.; Chen, J.; Li, A.; et al. The long noncoding RNA CRAL reverses cisplatin resistance via the miR-505/CYLD/AKT axis in human gastric cancer cells. RNA Biol. 2020, 17, 1576–1589. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Liu, J.; Pan, X.; Peng, C.; Xiong, B.; Feng, M.; Yang, X. miR-454 promotes survival and induces oxaliplatin resistance in gastric carcinoma cells by targeting CYLD. Exp. Ther. Med. 2020, 19, 3604–3610. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.Y.; Rong, J.S.; Chen, J.; Guo, J.; Zhu, J.Q.; Ruan, M.; Zuo, R.R.; Zhang, S.S.; Qi, J.M.; Zhang, B.H. Exosomal microRNA-588 from M2 polarized macrophages contributes to cisplatin resistance of gastric cancer cells. World J. Gastroenterol. 2021, 27, 6079–6092. [Google Scholar] [CrossRef]
- Yue, B.; Cui, R.; Zheng, R.; Jin, W.; Song, C.; Bao, T.; Wang, M.; Yu, F.; Zhao, E. Essential role of ALKBH5-mediated RNA demethylation modification in bile acid-induced gastric intestinal metaplasia. Mol. Ther. Nucleic Acids 2021, 26, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, K.; Park, H.L.; Kim, M.S.; Osada, M.; Tokumaru, Y.; Inoue, H.; Mori, M.; Sidransky, D. PGP9.5 methylation in diffuse-type gastric cancer. Cancer Res. 2006, 66, 3921–3927. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Yamashita, K.; Kim, M.S.; Park, H.L.; Osada, M.; Mori, M.; Sidransky, D. The role of PGP9.5 as a tumor suppressor gene in human cancer. Int. J. Cancer 2008, 123, 753–759. [Google Scholar] [CrossRef]
- Yu, J.; Tao, Q.; Cheung, K.F.; Jin, H.; Poon, F.F.; Wang, X.; Li, H.; Cheng, Y.Y.; Rocken, C.; Ebert, M.P.; et al. Epigenetic identification of ubiquitin carboxyl-terminal hydrolase L1 as a functional tumor suppressor and biomarker for hepatocellular carcinoma and other digestive tumors. Hepatology 2008, 48, 508–518. [Google Scholar] [CrossRef]
- Kim, J.H.; Jung, E.J.; Lee, H.S.; Kim, M.A.; Kim, W.H. Comparative analysis of DNA methylation between primary and metastatic gastric carcinoma. Oncol. Rep. 2009, 21, 1251–1259. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, W.; Zhou, B.; Jin, C.; Wang, Z.; Yang, Y.; Wang, Z.; Chen, Y.; Feng, X. The diagnosis value of promoter methylation of UCHL1 in the serum for progression of gastric cancer. Biomed. Res. Int. 2015, 2015, 741030. [Google Scholar] [CrossRef][Green Version]
- Gu, Y.Y.; Yang, M.; Zhao, M.; Luo, Q.; Yang, L.; Peng, H.; Wang, J.; Huang, S.K.; Zheng, Z.X.; Yuan, X.H.; et al. The de-ubiquitinase UCHL1 promotes gastric cancer metastasis via the Akt and Erk1/2 pathways. Tumor Biol. 2015, 36, 8379–8387. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, C.; Fang, S.; Ou, R.; Li, W.; Xu, Y. UCH-LI acts as a novel prognostic biomarker in gastric cardiac adenocarcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 13957–13967. [Google Scholar]
- Peng, J.; Meng, Q.; Shu, Y.; Qu, Z. UCHL3 stimulates metastasis of gastric cancer through upregulating IGF2. Minerva Gastroenterol. (Torino) 2022, 68, 343–344. [Google Scholar] [CrossRef]
- Arpalahti, L.; Laitinen, A.; Hagstrom, J.; Mustonen, H.; Kokkola, A.; Bockelman, C.; Haglund, C.; Holmberg, C.I. Positive cytoplasmic UCHL5 tumor expression in gastric cancer is linked to improved prognosis. PLoS ONE 2018, 13, e0193125. [Google Scholar] [CrossRef]
- Zhang, Z.; Hu, X.; Kuang, J.; Liao, J.; Yuan, Q. LncRNA DRAIC inhibits proliferation and metastasis of gastric cancer cells through interfering with NFRKB deubiquitination mediated by UCHL5. Cell. Mol. Biol. Lett. 2020, 25, 29. [Google Scholar] [CrossRef]
- Yan, S.; He, F.; Luo, R.; Wu, H.; Huang, M.; Huang, C.; Li, Y.; Zhou, Z. Decreased expression of BRCA1-associated protein 1 predicts unfavorable survival in gastric adenocarcinoma. Tumor Biol. 2016, 37, 6125–6133. [Google Scholar] [CrossRef]
- Weng, W.; Zhang, Q.; Xu, M.; Wu, Y.; Zhang, M.; Shen, C.; Chen, X.; Wang, Y.; Sheng, W. OTUB1 promotes tumor invasion and predicts a poor prognosis in gastric adenocarcinoma. Am. J. Transl. Res. 2016, 8, 2234–2244. [Google Scholar]
- Ouyang, S.; Zeng, Z.; Liu, Z.; Zhang, Z.; Sun, J.; Wang, X.; Ma, M.; Ye, X.; Yu, J.; Kang, W. OTUB2 regulates KRT80 stability via deubiquitination and promotes tumour proliferation in gastric cancer. Cell Death Discov. 2022, 8, 45. [Google Scholar] [CrossRef]
- Liu, G.; Guo, W.; Qin, J.; Lin, Z. OTUB2 Facilitates Tumorigenesis of Gastric Cancer Through Promoting KDM1A-Mediated Stem Cell-Like Properties. Front. Oncol. 2021, 11, 711735. [Google Scholar] [CrossRef]
- Wisnieski, F.; Santos, L.C.; Calcagno, D.Q.; Geraldis, J.C.; Gigek, C.O.; Anauate, A.C.; Chen, E.S.; Rasmussen, L.T.; Payao, S.L.M.; Artigiani, R.; et al. The impact of DNA demethylation on the upregulation of the NRN1 and TNFAIP3 genes associated with advanced gastric cancer. J. Mol. Med. 2020, 98, 707–717. [Google Scholar] [CrossRef]
- Du, B.; Liu, M.; Li, C.; Geng, X.; Zhang, X.; Ning, D.; Liu, M. The potential role of TNFAIP3 in malignant transformation of gastric carcinoma. Pathol. Res. Pract. 2019, 215, 152471. [Google Scholar] [CrossRef]
- Guo, T.; Zhang, Y.; Qu, X.; Che, X.; Li, C.; Fan, Y.; Wan, X.; Ma, R.; Hou, K.; Zhou, H.; et al. miR-200a enhances TRAIL-induced apoptosis in gastric cancer cells by targeting A20. Cell Biol. Int. 2018, 42, 506–514. [Google Scholar] [CrossRef]
- Maeda, S.; Otsuka, M.; Hirata, Y.; Mitsuno, Y.; Yoshida, H.; Shiratori, Y.; Masuho, Y.; Muramatsu, M.; Seki, N.; Omata, M. cDNA microarray analysis of Helicobacter pylori-mediated alteration of gene expression in gastric cancer cells. Biochem. Biophy.s Res. Commun. 2001, 284, 443–449. [Google Scholar] [CrossRef]
- Lim, M.C.C.; Maubach, G.; Sokolova, O.; Feige, M.H.; Diezko, R.; Buchbinder, J.; Backert, S.; Schluter, D.; Lavrik, I.N.; Naumann, M. Pathogen-induced ubiquitin-editing enzyme A20 bifunctionally shuts off NF-kappaB and caspase-8-dependent apoptotic cell death. Cell Death Differ. 2017, 24, 1621–1631. [Google Scholar] [CrossRef]
- Jantaree, P.; Chaithongyot, S.; Sokolova, O.; Naumann, M. USP48 and A20 synergistically promote cell survival in Helicobacter pylori infection. Cell. Mol. Life Sci. 2022, 79, 461. [Google Scholar] [CrossRef]
- Jantaree, P.; Yu, Y.; Chaithongyot, S.; Tager, C.; Sarabi, M.A.; Meyer, T.F.; Boccellato, F.; Maubach, G.; Naumann, M. Human gastric fibroblasts ameliorate A20-dependent cell survival in co-cultured gastric epithelial cells infected by Helicobacter pylori. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119364. [Google Scholar] [CrossRef]
- Sun, F.; Ni, Y.; Zhu, H.; Fang, J.; Wang, H.; Xia, J.; Ding, F.; Shen, H.; Shao, S. microRNA-29a-3p, Up-Regulated in Human Gastric Cells and Tissues with H.Pylori Infection, Promotes the Migration of GES-1 Cells via A20-Mediated EMT Pathway. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 51, 1250–1263. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Xu, F.; Sun, B.; Yang, L.; Wang, H. Deubiquitinating enzyme PSMD14 facilitates gastric carcinogenesis through stabilizing PTBP1. Exp. Cell Res. 2022, 415, 113148. [Google Scholar] [CrossRef]
- Kim, J.H.; Choi, J.K.; Cinghu, S.; Jang, J.W.; Lee, Y.S.; Li, Y.H.; Goh, Y.M.; Chi, X.Z.; Lee, K.S.; Wee, H.; et al. Jab1/CSN5 induces the cytoplasmic localization and degradation of RUNX3. J. Cell. Biochem. 2009, 107, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Sang, M.M.; Du, W.Q.; Zhang, R.Y.; Zheng, J.N.; Pei, D.S. Suppression of CSN5 promotes the apoptosis of gastric cancer cells through regulating p53-related apoptotic pathways. Bioorg. Med. Chem. Lett. 2015, 25, 2897–2901. [Google Scholar] [CrossRef]
- Wang, L.; Du, W.Q.; Xie, M.; Liu, M.R.; Huo, F.C.; Yang, J.; Pei, D.S. Jab1 promotes gastric cancer tumorigenesis via non-ubiquitin proteasomal degradation of p14ARF. Gastric Cancer 2020, 23, 1003–1017. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zhou, C.; Zhou, L.; Wang, Z.; Zhu, X.; Mu, X. Overexpression of DAPK1-mediated inhibition of IKKbeta/CSN5/PD-L1 axis enhances natural killer cell killing ability and inhibits tumor immune evasion in gastric cancer. Cell. Immunol. 2022, 372, 104469. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, Y.; Ding, M.; Xu, R. Long noncoding RNA TMPO-AS1/miR-126-5p/BRCC3 axis accelerates gastric cancer progression and angiogenesis via activating PI3K/Akt/mTOR pathway. J. Gastroenterol. Hepatol. 2021, 36, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.X.; Tang, Y.; Ma, Y. Ataxin-3 expression correlates with the clinicopathologic features of gastric cancer. Int. J. Clin. Exp. Med. 2014, 7, 973–981. [Google Scholar] [PubMed]
- Gavory, G.; O’Dowd, C.R.; Helm, M.D.; Flasz, J.; Arkoudis, E.; Dossang, A.; Hughes, C.; Cassidy, E.; McClelland, K.; Odrzywol, E.; et al. Discovery and characterization of highly potent and selective allosteric USP7 inhibitors. Nat. Chem. Biol. 2018, 14, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, Y.; Yang, H.; Liu, H.M. Design, synthesis, biological evaluation and structure-activity relationship study of quinazolin-4(3H)-one derivatives as novel USP7 inhibitors. Eur. J. Med. Chem. 2021, 216, 113291. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Lee, M.J.; Park, S.; Oh, D.C.; Elsasser, S.; Chen, P.C.; Gartner, C.; Dimova, N.; Hanna, J.; Gygi, S.P.; et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 2010, 467, 179–184. [Google Scholar] [CrossRef]
- Liu, Z.; Zhao, T.; Li, Z.; Sun, K.; Fu, Y.; Cheng, T.; Guo, J.; Yu, B.; Shi, X.; Liu, H. Discovery of [1,2,3]triazolo[4,5-d]pyrimidine derivatives as highly potent, selective, and cellularly active USP28 inhibitors. Acta Pharm. Sin. B 2020, 10, 1476–1491. [Google Scholar] [CrossRef]
- Hu, Y.; Yu, K.; Wang, G.; Zhang, D.; Shi, C.; Ding, Y.; Hong, D.; Zhang, D.; He, H.; Sun, L.; et al. Lanatoside C inhibits cell proliferation and induces apoptosis through attenuating Wnt/beta-catenin/c-Myc signaling pathway in human gastric cancer cell. Biochem. Pharmacol. 2018, 150, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.A.; Jeon, Y.K.; Nam, M.J. Galangin induces apoptosis in gastric cancer cells via regulation of ubiquitin carboxy-terminal hydrolase isozyme L1 and glutathione S-transferase P. Food Chem. Toxicol. 2012, 50, 684–688. [Google Scholar] [CrossRef]
- Ye, Y.; Li, X.; Feng, G.; Ma, Y.; Ye, F.; Shen, H.; Sun, K.; Lu, R.; Miao, S. 3,3′-Diindolylmethane induces ferroptosis by BAP1-IP3R axis in BGC-823 gastric cancer cells. Anticancer Drugs 2022, 33, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Comet, I.; Riising, E.M.; Leblanc, B.; Helin, K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer 2016, 16, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Ping, S.; Xu, Y.; Wang, M.; Jiang, X.; Xiong, L.; Zhang, L.; Yu, H.; Xiong, Z. Non-Coding RNAs in Gastric Cancer: From Malignant Hallmarks to Clinical Applications. Front. Cell Dev. Biol. 2021, 9, 732036. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, J.; Cao, G.; Hua, J.; Shan, G.; Lin, W. Emerging roles of circular RNAs in gastric cancer metastasis and drug resistance. J. Exp. Clin. Cancer Res. 2022, 41, 218. [Google Scholar] [CrossRef]
- Lu, Y.; Li, K.; Gao, Y.; Liang, W.; Wang, X.; Chen, L. CircRNAs in gastric cancer: Current research and potential clinical implications. FEBS Lett. 2021, 595, 2644–2654. [Google Scholar] [CrossRef]
- Gong, Y.Q.; Lu, T.L.; Hou, F.T.; Chen, C.W. Antisense long non-coding RNAs in gastric cancer. Clin. Chim. Acta Int. J. Clin. Chem. 2022, 534, 128–137. [Google Scholar] [CrossRef]
- Al-Eidan, A.; Wang, Y.; Skipp, P.; Ewing, R.M. The USP7 protein interaction network and its roles in tumorigenesis. Genes Dis. 2022, 9, 41–50. [Google Scholar] [CrossRef]
- Nininahazwe, L.; Liu, B.; He, C.; Zhang, H.; Chen, Z.S. The emerging nature of Ubiquitin-specific protease 7 (USP7): A new target in cancer therapy. Drug Discov. Today 2021, 26, 490–502. [Google Scholar] [CrossRef]
- Harakandi, C.; Nininahazwe, L.; Xu, H.; Liu, B.; He, C.; Zheng, Y.C.; Zhang, H. Recent advances on the intervention sites targeting USP7-MDM2-p53 in cancer therapy. Bioorg. Chem. 2021, 116, 105273. [Google Scholar] [CrossRef] [PubMed]
- Coombs, N.; Sompallae, R.; Olbermann, P.; Gastaldello, S.; Goppel, D.; Masucci, M.G.; Josenhans, C. Helicobacter pylori affects the cellular deubiquitinase USP7 and ubiquitin-regulated components TRAF6 and the tumour suppressor p53. Int. J. Med. Microbiol. 2011, 301, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, M.; Jolly, L.A.; Gecz, J.; Wood, S.A. La FAM fatale: USP9X in development and disease. Cell. Mol. Life Sci. 2015, 72, 2075–2089. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.; Cheng, A.S.; Yu, J.; To, K.F. Emerging role of Hippo pathway in gastric and other gastrointestinal cancers. World J. Gastroenterol. 2016, 22, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Hergovich, A. Mammalian Hippo signalling: A kinase network regulated by protein-protein interactions. Biochem. Soc. Trans. 2012, 40, 124–128. [Google Scholar] [CrossRef]
- Zhao, B.; Li, L.; Tumaneng, K.; Wang, C.Y.; Guan, K.L. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 2010, 24, 72–85. [Google Scholar] [CrossRef]
- Mussell, A.; Frangou, C.; Zhang, J. Regulation of the Hippo signaling pathway by deubiquitinating enzymes in cancer. Genes Dis. 2019, 6, 335–341. [Google Scholar] [CrossRef]
- Li, L.; Liu, T.; Li, Y.; Wu, C.; Luo, K.; Yin, Y.; Chen, Y.; Nowsheen, S.; Wu, J.; Lou, Z.; et al. The deubiquitinase USP9X promotes tumor cell survival and confers chemoresistance through YAP1 stabilization. Oncogene 2018, 37, 2422–2431. [Google Scholar] [CrossRef]
- Kim, M.S.; Yoo, K.J.; Kang, I.; Chung, H.M.; Baek, K.H. A novel cysteine protease HeLa DUB-1 responsible for cleaving the ubiquitin in human ovarian cancer cells. Int. J. Oncol. 2004, 25, 373–379. [Google Scholar] [CrossRef]
- Kim, M.S.; Kim, Y.K.; Kim, Y.S.; Seong, M.; Choi, J.K.; Baek, K.H. Deubiquitinating enzyme USP36 contains the PEST motif and is polyubiquitinated. Biochem. Biophys. Res. Commun. 2005, 330, 797–804. [Google Scholar] [CrossRef]
- Tao, L.; Liu, X.; Jiang, X.; Zhang, K.; Wang, Y.; Li, X.; Jiang, S.; Han, T. USP10 as a Potential Therapeutic Target in Human Cancers. Genes 2022, 13, 831. [Google Scholar] [CrossRef]
- Li, D.; Zeng, Z.; Yu, T.; Qin, J.; Wu, J.; Song, J.C.; Zhou, Z.Y.; Yuan, J.P. Expression and clinical implication of S100A12 in gastric carcinoma. Tumor Biol. 2016, 37, 6551–6559. [Google Scholar] [CrossRef]
- Peng, X.C.; Zeng, Z.; Huang, Y.N.; Deng, Y.C.; Fu, G.H. Clinical significance of TM4SF1 as a tumor suppressor gene in gastric cancer. Cancer Med. 2018, 7, 2592–2600. [Google Scholar] [CrossRef]
- Kwon, S.K.; Saindane, M.; Baek, K.H. p53 stability is regulated by diverse deubiquitinating enzymes. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 404–411. [Google Scholar] [CrossRef]
- Dong, B.; Wu, Y. Epigenetic Regulation and Post-Translational Modifications of SNAI1 in Cancer Metastasis. Int. J. Mol. Sci. 2021, 22, 11062. [Google Scholar] [CrossRef]
- Zhou, B.P.; Deng, J.; Xia, W.; Xu, J.; Li, Y.M.; Gunduz, M.; Hung, M.C. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat. Cell Biol. 2004, 6, 931–940. [Google Scholar] [CrossRef]
- de Poot, S.A.H.; Tian, G.; Finley, D. Meddling with Fate: The Proteasomal Deubiquitinating Enzymes. J. Mol. Biol. 2017, 429, 3525–3545. [Google Scholar] [CrossRef]
- Wang, F.; Ning, S.; Yu, B.; Wang, Y. USP14: Structure, Function, and Target Inhibition. Front. Pharmacol. 2021, 12, 801328. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, C.; Gu, C.; Li, Q.; Wu, N. Function of Deubiquitinating Enzyme USP14 as Oncogene in Different Types of Cancer. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2016, 38, 993–1002. [Google Scholar] [CrossRef]
- Anita, R.; Paramasivam, A.; Priyadharsini, J.V.; Chitra, S. The m6A readers YTHDF1 and YTHDF3 aberrations associated with metastasis and predict poor prognosis in breast cancer patients. Am. J. Cancer Res. 2020, 10, 2546–2554. [Google Scholar]
- Pi, J.; Wang, W.; Ji, M.; Wang, X.; Wei, X.; Jin, J.; Liu, T.; Qiang, J.; Qi, Z.; Li, F.; et al. YTHDF1 Promotes Gastric Carcinogenesis by Controlling Translation of FZD7. Cancer Res. 2021, 81, 2651–2665. [Google Scholar] [CrossRef]
- Schweitzer, K.; Bozko, P.M.; Dubiel, W.; Naumann, M. CSN controls NF-kappaB by deubiquitinylation of IkappaBalpha. EMBO J. 2007, 26, 1532–1541. [Google Scholar] [CrossRef]
- Das, T.; Song, E.J.; Kim, E.E. The Multifaceted Roles of USP15 in Signal Transduction. Int. J. Mol. Sci. 2021, 22, 4728. [Google Scholar] [CrossRef]
- Glinsky, G.V. Genomic models of metastatic cancer: Functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle 2006, 5, 1208–1216. [Google Scholar] [CrossRef]
- Feng, T.; Ling, S.; Xu, C.; Ying, L.; Su, D.; Xu, X. Ubiquitin-specific peptidase 22 in cancer. Cancer Lett. 2021, 514, 30–37. [Google Scholar] [CrossRef]
- Atanassov, B.S.; Mohan, R.D.; Lan, X.; Kuang, X.; Lu, Y.; Lin, K.; McIvor, E.; Li, W.; Zhang, Y.; Florens, L.; et al. ATXN7L3 and ENY2 Coordinate Activity of Multiple H2B Deubiquitinases Important for Cellular Proliferation and Tumor Growth. Mol. Cell 2016, 62, 558–571. [Google Scholar] [CrossRef]
- Qiu, G.Z.; Mao, X.Y.; Ma, Y.; Gao, X.C.; Wang, Z.; Jin, M.Z.; Sun, W.; Zou, Y.X.; Lin, J.; Fu, H.L.; et al. Ubiquitin-specific protease 22 acts as an oncoprotein to maintain glioma malignancy through deubiquitinating B cell-specific Moloney murine leukemia virus integration site 1 for stabilization. Cancer Sci. 2018, 109, 2199–2210. [Google Scholar] [CrossRef]
- Kim, D.; Hong, A.; Park, H.I.; Shin, W.H.; Yoo, L.; Jeon, S.J.; Chung, K.C. Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. J. Cell. Physiol. 2017, 232, 3664–3676. [Google Scholar] [CrossRef]
- Vigil, D.; Cherfils, J.; Rossman, K.L.; Der, C.J. Ras superfamily GEFs and GAPs: Validated and tractable targets for cancer therapy? Nat. Rev. Cancer 2010, 10, 842–857. [Google Scholar] [CrossRef]
- Wang, Y.; Jia, Z.; Gao, J.; Zhou, T.; Zhang, X.; Zu, G. Clinicopathological and Prognostic Value of USP22 Expression in Gastric Cancer: A Systematic Review and Meta-Analysis and Database Validation. Front. Surg. 2022, 9, 920595. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Z.; Zhang, L.; Yang, Z.; Chen, X.; Luo, J.; Zhou, Z.; Mei, X.; Yu, X.; Shao, Z.; et al. Targeting deubiquitinase USP28 for cancer therapy. Cell Death Dis. 2018, 9, 186. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Garcia, C.; Tomaskovic, I.; Shah, V.J.; Dikic, I.; Diefenbacher, M. USP28: Oncogene or Tumor Suppressor? A Unifying Paradigm for Squamous Cell Carcinoma. Cells 2021, 10, 2652. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, Y.; Yang, X.H.; Kang, T.; Zhao, Y.; Wang, C.; Evers, B.M.; Zhou, B.P. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep. 2013, 5, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Paulding, C.A.; Ruvolo, M.; Haber, D.A. The Tre2 (USP6) oncogene is a hominoid-specific gene. Proc. Natl. Acad. Sci. USA 2003, 100, 2507–2511. [Google Scholar] [CrossRef]
- Akhavantabasi, S.; Akman, H.B.; Sapmaz, A.; Keller, J.; Petty, E.M.; Erson, A.E. USP32 is an active, membrane-bound ubiquitin protease overexpressed in breast cancers. Mamm. Genome 2010, 21, 388–397. [Google Scholar] [CrossRef]
- Chen, S.; Chen, X.; Li, Z.; Mao, J.; Jiang, W.; Zhu, Z.; Li, Y.; Jiang, Z.; Zhao, W.; Tan, G.; et al. Identification of ubiquitin-specific protease 32 as an oncogene in glioblastoma and the underlying mechanisms. Sci. Rep. 2022, 12, 6445. [Google Scholar] [CrossRef]
- Van Leuken, R.J.; Luna-Vargas, M.P.; Sixma, T.K.; Wolthuis, R.M.; Medema, R.H. Usp39 is essential for mitotic spindle checkpoint integrity and controls mRNA-levels of aurora B. Cell Cycle 2008, 7, 2710–2719. [Google Scholar] [CrossRef]
- Ding, K.; Ji, J.; Zhang, X.; Huang, B.; Chen, A.; Zhang, D.; Li, X.; Wang, X.; Wang, J. RNA splicing factor USP39 promotes glioma progression by inducing TAZ mRNA maturation. Oncogene 2019, 38, 6414–6428. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Liu, Z.; Zhang, E.; Zhang, P.; Wang, Y.; Hang, J.; Li, Q. USP39 promotes tumorigenesis by stabilizing and deubiquitinating SP1 protein in hepatocellular carcinoma. Cell Signal. 2021, 85, 110068. [Google Scholar] [CrossRef]
- Li, X.; Yuan, J.; Song, C.; Lei, Y.; Xu, J.; Zhang, G.; Wang, W.; Song, G. Deubiquitinase USP39 and E3 ligase TRIM26 balance the level of ZEB1 ubiquitination and thereby determine the progression of hepatocellular carcinoma. Cell Death Differ. 2021, 28, 2315–2332. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, W.; Bao, X.; Sun, T.; Wang, J.; Li, M.; Liu, C. USP39 facilitates breast cancer cell proliferation through stabilization of FOXM1. Am. J. Cancer Res. 2022, 12, 3644–3661. [Google Scholar]
- Giam, M.; Rancati, G. Aneuploidy and chromosomal instability in cancer: A jackpot to chaos. Cell Div. 2015, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Foreman, O.; Wigle, D.A.; Kosari, F.; Vasmatzis, G.; Salisbury, J.L.; van Deursen, J.; Galardy, P.J. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J. Clin. Investig. 2012, 122, 4362–4374. [Google Scholar] [CrossRef] [PubMed]
- Peschiaroli, A.; Skaar, J.R.; Pagano, M.; Melino, G. The ubiquitin-specific protease USP47 is a novel beta-TRCP interactor regulating cell survival. Oncogene 2010, 29, 1384–1393. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Liu, Y.; Xu, X.; Zhang, W.; Yu, T.; Jia, J.; Liu, C. Deubiquitinase USP47/UBP64E Regulates beta-Catenin Ubiquitination and Degradation and Plays a Positive Role in Wnt Signaling. Mol. Cell. Biol. 2015, 35, 3301–3311. [Google Scholar] [CrossRef]
- Bufalieri, F.; Infante, P.; Bernardi, F.; Caimano, M.; Romania, P.; Moretti, M.; Lospinoso Severini, L.; Talbot, J.; Melaiu, O.; Tanori, M.; et al. ERAP1 promotes Hedgehog-dependent tumorigenesis by controlling USP47-mediated degradation of betaTrCP. Nat. Commun. 2019, 10, 3304. [Google Scholar] [CrossRef]
- Bignell, G.R.; Warren, W.; Seal, S.; Takahashi, M.; Rapley, E.; Barfoot, R.; Green, H.; Brown, C.; Biggs, P.J.; Lakhani, S.R.; et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 2000, 25, 160–165. [Google Scholar] [CrossRef]
- Mathis, B.J.; Lai, Y.; Qu, C.; Janicki, J.S.; Cui, T. CYLD-mediated signaling and diseases. Curr. Drug Targets 2015, 16, 284–294. [Google Scholar] [CrossRef]
- Yang, W.L.; Jin, G.; Li, C.F.; Jeong, Y.S.; Moten, A.; Xu, D.; Feng, Z.; Chen, W.; Cai, Z.; Darnay, B.; et al. Cycles of ubiquitination and deubiquitination critically regulate growth factor-mediated activation of Akt signaling. Sci. Signal 2013, 6, ra3. [Google Scholar] [CrossRef]
- Zhu, M.; Zhao, H.; Liao, J.; Xu, X. HERC2/USP20 coordinates CHK1 activation by modulating CLASPIN stability. Nucleic Acids Res. 2014, 42, 13074–13081. [Google Scholar] [CrossRef]
- Pfoh, R.; Lacdao, I.K.; Saridakis, V. Deubiquitinases and the new therapeutic opportunities offered to cancer. Endocr. Relat. Cancer 2015, 22, T35–T54. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Xu, J.; Lin, H.; Xu, X.; Tian, F. The histone demethylase lysine-specific demethylase-1-mediated epigenetic silence of KLF2 contributes to gastric cancer cell proliferation, migration, and invasion. Tumor Biol. 2017, 39, 1010428317698356. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Xu, Y.; Xu, T.; Fan, R.; Jiang, T.; Cao, M.; Shi, L.; Song, J. CircPIP5K1A activates KRT80 and PI3K/AKT pathway to promote gastric cancer development through sponging miR-671-5p. Biomed. Pharmacother. 2020, 126, 109941. [Google Scholar] [CrossRef]
- Malynn, B.A.; Ma, A. A20: A multifunctional tool for regulating immunity and preventing disease. Cell. Immunol. 2019, 340, 103914. [Google Scholar] [CrossRef] [PubMed]
- Bellail, A.C.; Olson, J.J.; Yang, X.; Chen, Z.J.; Hao, C. A20 ubiquitin ligase-mediated polyubiquitination of RIP1 inhibits caspase-8 cleavage and TRAIL-induced apoptosis in glioblastoma. Cancer Discov. 2012, 2, 140–155. [Google Scholar] [CrossRef]
- Zhu, G.; Ye, J.; Huang, Y.; Zheng, W.; Hua, J.; Yang, S.; Zhuang, J.; Wang, J. Receptor-interacting protein-1 promotes the growth and invasion in gastric cancer. Int. J. Oncol. 2016, 48, 2387–2398. [Google Scholar] [CrossRef][Green Version]
- Spataro, V.; Buetti-Dinh, A. POH1/Rpn11/PSMD14: A journey from basic research in fission yeast to a prognostic marker and a druggable target in cancer cells. Br. J. Cancer 2022, 127, 788–799. [Google Scholar] [CrossRef]
- Sun, T.; Liu, Z.; Bi, F.; Yang, Q. Deubiquitinase PSMD14 promotes ovarian cancer progression by decreasing enzymatic activity of PKM2. Mol. Oncol. 2021, 15, 3639–3658. [Google Scholar] [CrossRef]
- Lv, J.; Zhang, S.; Wu, H.; Lu, J.; Lu, Y.; Wang, F.; Zhao, W.; Zhan, P.; Lu, J.; Fang, Q.; et al. Deubiquitinase PSMD14 enhances hepatocellular carcinoma growth and metastasis by stabilizing GRB2. Cancer Lett. 2020, 469, 22–34. [Google Scholar] [CrossRef]
- Zhi, T.; Jiang, K.; Xu, X.; Yu, T.; Zhou, F.; Wang, Y.; Liu, N.; Zhang, J. ECT2/PSMD14/PTTG1 axis promotes the proliferation of glioma through stabilizing E2F1. Neuro-oncology 2019, 21, 462–473. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, D.M.; Seo, M.J.; Kang, H.C.; Kwon, S.K.; Choi, K.S. PSMD14 Targeting Triggers Paraptosis in Breast Cancer Cells by Inducing Proteasome Inhibition and Ca(2+) Imbalance. Int. J. Mol. Sci. 2022, 23, 2648. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Liu, X.; Zhou, N.; Wu, Q.; Zhou, L.; Li, Q. Gene expression profiling and bioinformatics analysis of gastric carcinoma. Exp. Mol. Pathol. 2014, 96, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.O.; Li, C.W.; Xia, W.; Cha, J.H.; Chan, L.C.; Wu, Y.; Chang, S.S.; Lin, W.C.; Hsu, J.M.; Hsu, Y.H.; et al. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell 2016, 30, 925–939. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yao, Z.; Wang, J.; Zhang, W.; Yang, Y.; Zhang, Y.; Qu, X.; Zhu, Y.; Zou, J.; Peng, S.; et al. Macrophage-derived CCL5 facilitates immune escape of colorectal cancer cells via the p65/STAT3-CSN5-PD-L1 pathway. Cell Death Differ. 2020, 27, 1765–1781. [Google Scholar] [CrossRef]
- Chen, S.; Liu, Y.; Zhou, H. Advances in the Development Ubiquitin-Specific Peptidase (USP) Inhibitors. Int. J. Mol. Sci. 2021, 22, 4546. [Google Scholar] [CrossRef]
- Prieto-Garcia, C.; Hartmann, O.; Reissland, M.; Braun, F.; Fischer, T.; Walz, S.; Schulein-Volk, C.; Eilers, U.; Ade, C.P.; Calzado, M.A.; et al. Maintaining protein stability of Np63 via USP28 is required by squamous cancer cells. EMBO Mol. Med. 2020, 12, e11101. [Google Scholar] [CrossRef]
- Ma, A.; Tang, M.; Zhang, L.; Wang, B.; Yang, Z.; Liu, Y.; Xu, G.; Wu, L.; Jing, T.; Xu, X.; et al. USP1 inhibition destabilizes KPNA2 and suppresses breast cancer metastasis. Oncogene 2019, 38, 2405–2419. [Google Scholar] [CrossRef]
- Fu, C.; Zhu, X.; Xu, P.; Li, Y. Pharmacological inhibition of USP7 promotes antitumor immunity and contributes to colon cancer therapy. Onco. Targets Ther. 2019, 12, 609–617. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).