Choice of High-Throughput Proteomics Method Affects Data Integration with Transcriptomics and the Potential Use in Biomarker Discovery
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Estimation of Protein Concentration
2.3. Preparation of Samples for Tryptic Digestion
2.4. Automated HILIC Digestion
2.5. Liquid Chromatography-Mass Spectrometry
2.6. DDA Data Processing
2.7. DIA Data Processing
2.8. Data Analysis
2.9. Gene Set Enrichment Analysis
2.10. Decision Tree
3. Results
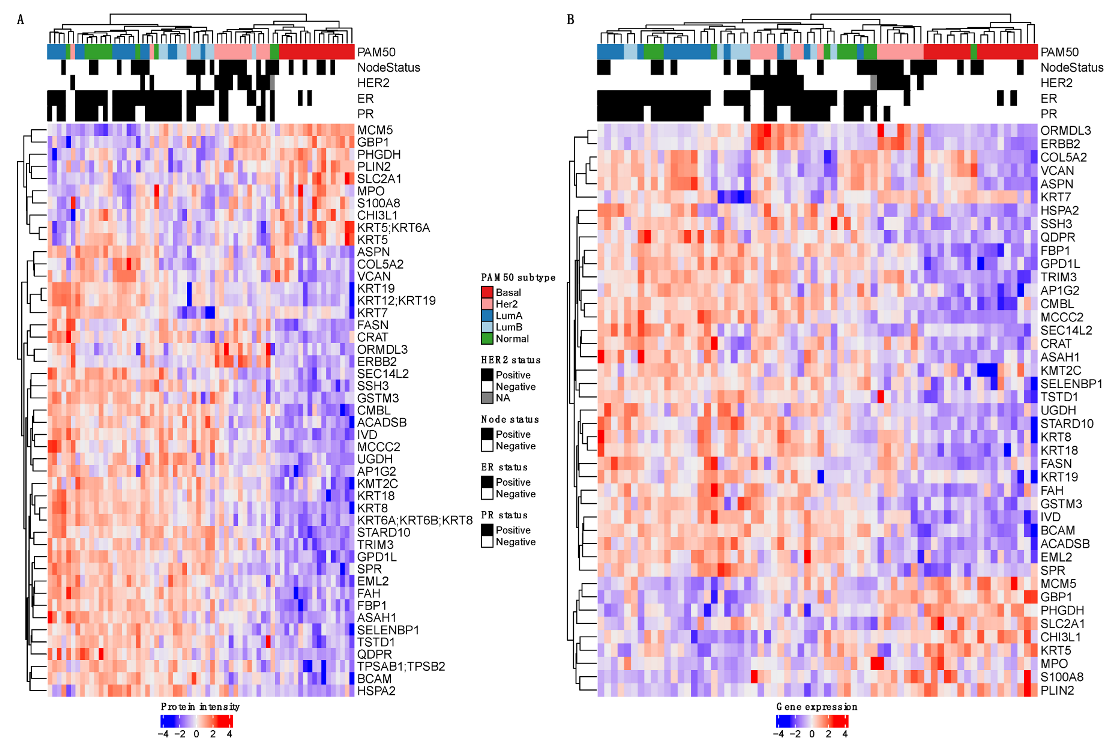
3.1. Protocol Development and Generation of Protein Datasets
3.2. Performance Evaluation of Different LC-MS/MS Methods
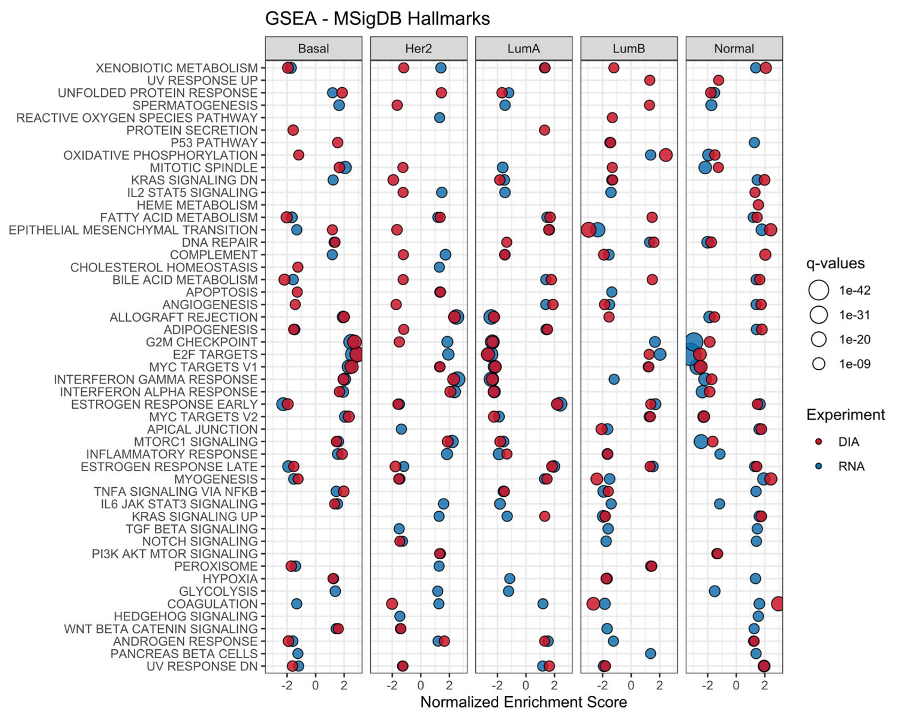
3.3. Gene Set Enrichment Analysis of DIA-NN and RNA-Seq Data
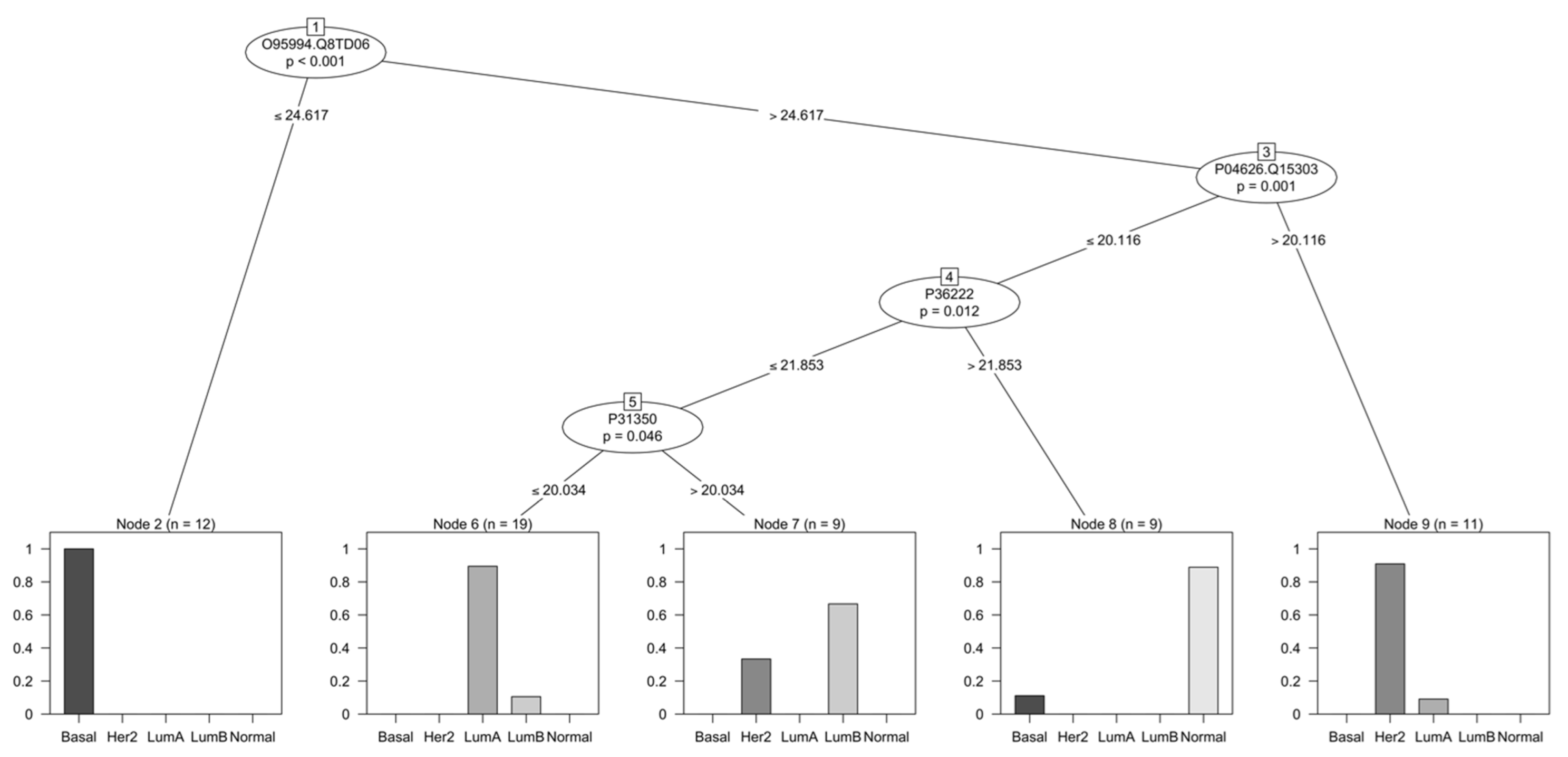
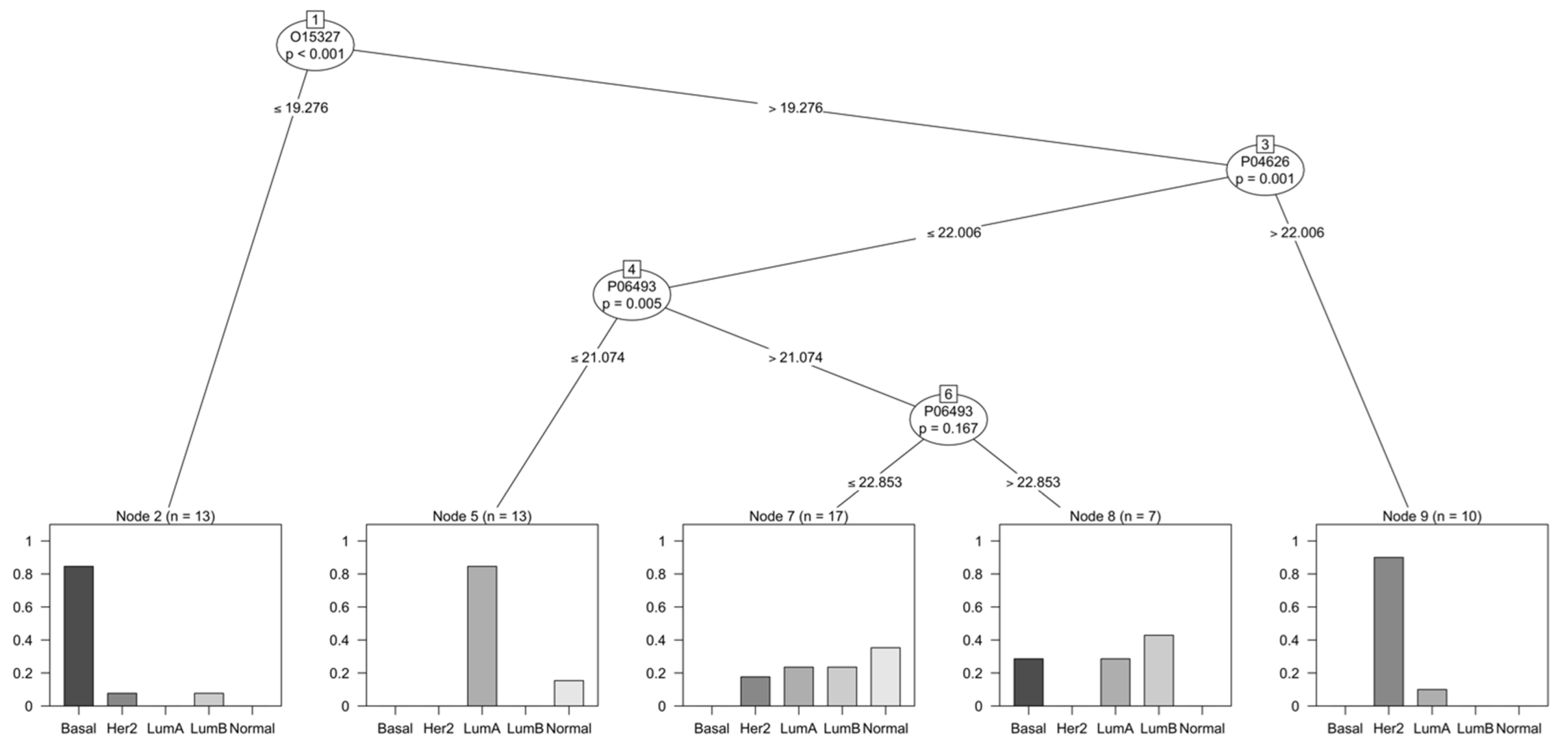
3.4. Selection of Discriminant Proteins through a Decision Tree Classifier
4. Discussion
4.1. Label-Free LC-MS/MS for Cancer Classification
4.2. Functional Differences between Proteomics and Transcriptomics
4.3. Relevance of Proteins Found by Decision Tree
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Lam, S.W.; Jimenez, C.R.; Boven, E. Breast cancer classification by proteomic technologies: Current state of knowledge. Cancer Treat. Rev. 2014, 40, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Parise, C.A.; Caggiano, V. Breast Cancer Survival Defined by the ER/PR/HER2 Subtypes and a Surrogate Classification according to Tumor Grade and Immunohistochemical Biomarkers. J. Cancer Epidemiol. 2014, 2014, 469251. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Tibshirani, R.; Parker, J.; Hastie, T.; Marron, J.S.; Nobel, A.; Deng, S.; Johnsen, H.; Pesich, R.; Geisler, S.; et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA 2003, 100, 8418–8423. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Brouckaert, O.; Schoneveld, A.; Truyers, C.; Kellen, E.; Van Ongeval, C.; Vergote, I.; Moerman, P.; Floris, G.; Wildiers, H.; Christiaens, M.R.; et al. Breast cancer phenotype, nodal status and palpability may be useful in the detection of overdiagnosed screening-detected breast cancers. Ann. Oncol. 2013, 24, 1847–1852. [Google Scholar] [CrossRef]
- Saal, L.H.; Vallon-Christersson, J.; Hakkinen, J.; Hegardt, C.; Grabau, D.; Winter, C.; Brueffer, C.; Tang, M.H.; Reutersward, C.; Schulz, R.; et al. The Sweden Cancerome Analysis Network - Breast (SCAN-B) Initiative: A large-scale multicenter infrastructure towards implementation of breast cancer genomic analyses in the clinical routine. Genome Med. 2015, 7, 20. [Google Scholar] [CrossRef]
- Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Vallon-Christersson, J.; Hakkinen, J.; Hegardt, C.; Saal, L.H.; Larsson, C.; Ehinger, A.; Lindman, H.; Olofsson, H.; Sjoblom, T.; Warnberg, F.; et al. Cross comparison and prognostic assessment of breast cancer multigene signatures in a large population-based contemporary clinical series. Sci. Rep. 2019, 9, 12184. [Google Scholar] [CrossRef] [PubMed]
- Staaf, J.; Hakkinen, J.; Hegardt, C.; Saal, L.H.; Kimbung, S.; Hedenfalk, I.; Lien, T.; Sorlie, T.; Naume, B.; Russnes, H.; et al. RNA sequencing-based single sample predictors of molecular subtype and risk of recurrence for clinical assessment of early-stage breast cancer. NPJ Breast Cancer 2022, 8, 94. [Google Scholar] [CrossRef]
- Tyanova, S.; Albrechtsen, R.; Kronqvist, P.; Cox, J.; Mann, M.; Geiger, T. Proteomic maps of breast cancer subtypes. Nat. Commun. 2016, 7, 10259. [Google Scholar] [CrossRef] [PubMed]
- Johansson, H.J.; Socciarelli, F.; Vacanti, N.M.; Haugen, M.H.; Zhu, Y.; Siavelis, I.; Fernandez-Woodbridge, A.; Aure, M.R.; Sennblad, B.; Vesterlund, M.; et al. Breast cancer quantitative proteome and proteogenomic landscape. Nat. Commun. 2019, 10, 1600. [Google Scholar] [CrossRef] [PubMed]
- De Marchi, T.; Pyl, P.T.; Sjostrom, M.; Klasson, S.; Sartor, H.; Tran, L.; Pekar, G.; Malmstrom, J.; Malmstrom, L.; Nimeus, E. Proteogenomic Workflow Reveals Molecular Phenotypes Related to Breast Cancer Mammographic Appearance. J. Proteome Res. 2021, 20, 2983–3001. [Google Scholar] [CrossRef]
- Bouchal, P.; Schubert, O.T.; Faktor, J.; Capkova, L.; Imrichova, H.; Zoufalova, K.; Paralova, V.; Hrstka, R.; Liu, Y.; Ebhardt, H.A.; et al. Breast Cancer Classification Based on Proteotypes Obtained by SWATH Mass Spectrometry. Cell Rep. 2019, 28, 832–843.e837. [Google Scholar] [CrossRef]
- Gotti, C.; Roux-Dalvai, F.; Joly-Beauparlant, C.; Mangnier, L.; Leclercq, M.; Droit, A. Extensive and Accurate Benchmarking of DIA Acquisition Methods and Software Tools Using a Complex Proteomic Standard. J. Proteome Res. 2021, 20, 4801–4814. [Google Scholar] [CrossRef]
- Searle, B.C.; Pino, L.K.; Egertson, J.D.; Ting, Y.S.; Lawrence, R.T.; MacLean, B.X.; Villen, J.; MacCoss, M.J. Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat. Commun. 2018, 9, 5128. [Google Scholar] [CrossRef]
- Demichev, V.; Messner, C.B.; Vernardis, S.I.; Lilley, K.S.; Ralser, M. DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 2020, 17, 41–44. [Google Scholar] [CrossRef]
- Ryden, L.; Loman, N.; Larsson, C.; Hegardt, C.; Vallon-Christersson, J.; Malmberg, M.; Lindman, H.; Ehinger, A.; Saal, L.H.; Borg, A. Minimizing inequality in access to precision medicine in breast cancer by real-time population-based molecular analysis in the SCAN-B initiative. Br. J. Surg. 2018, 105, e158–e168. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Sun, Z.; Campbell, D.S.; Binz, P.A.; Farrah, T.; Shteynberg, D.; Mendoza, L.; Omenn, G.S.; Moritz, R.L. Tiered Human Integrated Sequence Search Databases for Shotgun Proteomics. J. Proteome Res. 2016, 15, 4091–4100. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Searle, B.C.; Swearingen, K.E.; Barnes, C.A.; Schmidt, T.; Gessulat, S.; Kuster, B.; Wilhelm, M. Generating high quality libraries for DIA MS with empirically corrected peptide predictions. Nat. Commun. 2020, 11, 1548. [Google Scholar] [CrossRef] [PubMed]
- Willforss, J.; Chawade, A.; Levander, F. NormalyzerDE: Online Tool for Improved Normalization of Omics Expression Data and High-Sensitivity Differential Expression Analysis. J. Proteome Res. 2019, 18, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Taverner, T.; Karpievitch, Y.V.; Polpitiya, A.D.; Brown, J.N.; Dabney, A.R.; Anderson, G.A.; Smith, R.D. DanteR: An extensible R-based tool for quantitative analysis of -omics data. Bioinformatics 2012, 28, 2404–2406. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef]
- Gillet, L.C.; Leitner, A.; Aebersold, R. Mass Spectrometry Applied to Bottom-Up Proteomics: Entering the High-Throughput Era for Hypothesis Testing. Annu. Rev. Anal. Chem. 2016, 9, 449–472. [Google Scholar] [CrossRef]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Oshi, M.; Endo, I.; Takabe, K. MYC Targets Scores Are Associated with Cancer Aggressiveness and Poor Survival in ER-Positive Primary and Metastatic Breast Cancer. Int. J. Mol. Sci. 2020, 21, 8127. [Google Scholar] [CrossRef]
- Oshi, M.; Takahashi, H.; Tokumaru, Y.; Yan, L.; Rashid, O.M.; Matsuyama, R.; Endo, I.; Takabe, K. G2M Cell Cycle Pathway Score as a Prognostic Biomarker of Metastasis in Estrogen Receptor (ER)-Positive Breast Cancer. Int. J. Mol. Sci. 2020, 21, 2921. [Google Scholar] [CrossRef]
- Murthy, V.; Oshi, M.; Tokumaru, Y.; Endo, I.; Takabe, K. Increased apoptosis is associated with robust immune cell infiltration and cytolytic activity in breast cancer. Am. J. Cancer Res. 2021, 11, 3674–3687. [Google Scholar] [PubMed]
- Savas, P.; Salgado, R.; Denkert, C.; Sotiriou, C.; Darcy, P.K.; Smyth, M.J.; Loi, S. Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat. Rev. Clin. Oncol. 2016, 13, 228–241. [Google Scholar] [CrossRef]
- Salmans, M.L.; Zhao, F.; Andersen, B. The estrogen-regulated anterior gradient 2 (AGR2) protein in breast cancer: A potential drug target and biomarker. Breast Cancer Res. BCR 2013, 15, 204. [Google Scholar] [CrossRef]
- Barraclough, D.L.; Platt-Higgins, A.; de Silva Rudland, S.; Barraclough, R.; Winstanley, J.; West, C.R.; Rudland, P.S. The metastasis-associated anterior gradient 2 protein is correlated with poor survival of breast cancer patients. Am. J. Pathol. 2009, 175, 1848–1857. [Google Scholar] [CrossRef]
- Obacz, J.; Brychtova, V.; Podhorec, J.; Fabian, P.; Dobes, P.; Vojtesek, B.; Hrstka, R. Anterior gradient protein 3 is associated with less aggressive tumors and better outcome of breast cancer patients. OncoTargets Ther. 2015, 8, 1523–1532. [Google Scholar] [CrossRef][Green Version]
- Segers, V.F.M.; Dugaucquier, L.; Feyen, E.; Shakeri, H.; De Keulenaer, G.W. The role of ErbB4 in cancer. Cell. Oncol. 2020, 43, 335–352. [Google Scholar] [CrossRef]
- Sundvall, M.; Iljin, K.; Kilpinen, S.; Sara, H.; Kallioniemi, O.P.; Elenius, K. Role of ErbB4 in breast cancer. J. Mammary Gland. Biol. Neoplasia 2008, 13, 259–268. [Google Scholar] [CrossRef]
- Nicolini, A.; Ferrari, P.; Duffy, M.J. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin. Cancer Biol. 2018, 52, 56–73. [Google Scholar] [CrossRef] [PubMed]
- Tarighati, E.; Keivan, H.; Mahani, H. A review of prognostic and predictive biomarkers in breast cancer. Clin. Exp. Med. 2022. [Google Scholar] [CrossRef] [PubMed]
- Naresh, A.; Long, W.; Vidal, G.A.; Wimley, W.C.; Marrero, L.; Sartor, C.I.; Tovey, S.; Cooke, T.G.; Bartlett, J.M.; Jones, F.E. The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein promoting apoptosis of breast cancer cells. Cancer Res. 2006, 66, 6412–6420. [Google Scholar] [CrossRef]
- Cohen, N.; Shani, O.; Raz, Y.; Sharon, Y.; Hoffman, D.; Abramovitz, L.; Erez, N. Fibroblasts drive an immunosuppressive and growth-promoting microenvironment in breast cancer via secretion of Chitinase 3-like 1. Oncogene 2017, 36, 4457–4468. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, S.; Wang, Q.; Zhang, X. Tumor-recruited M2 macrophages promote gastric and breast cancer metastasis via M2 macrophage-secreted CHI3L1 protein. J. Hematol. Oncol. 2017, 10, 36. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.; Warden, C.D.; Huang, Y.; Loera, S.; Xue, L.; Zhang, S.; Chu, P.; Zheng, S.; Yen, Y. Prognostic and therapeutic significance of ribonucleotide reductase small subunit M2 in estrogen-negative breast cancers. BMC Cancer 2014, 14, 664. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Mahfouz, M.; Habashy, H.O. RRM2 expression in different molecular subtypes of breast cancer and its prognostic significance. Diagn. Pathol. 2022, 17, 1. [Google Scholar] [CrossRef]
- Ben-David, A. Comparison of classification accuracy using Cohen’s Weighted Kappa. Expert Syst. Appl. 2008, 34, 825–832. [Google Scholar] [CrossRef]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Lantz, B. Machine Learning with R: Learn How to Use R to Apply Powerful Machine Learning Methods and Gain an Insight into Real-World Applications; Packt Publishing: Birmingham, UK, 2013. [Google Scholar]
| Experiment | Proteins 1 | Common Proteins 2 |
|---|---|---|
| DDA Match Between Runs | 6826 | 1820 |
| DDA no Match Between Runs | 6445 | 817 |
| DIA-NN | 9902 | 3205 |
| EncyclopeDIA | 5066 | 1920 |
| Experiment | Correlation RNA-Seq | Pairs | Proteins 5% FDR * Spearman | Median Spearman Score | Proteins 5% FDR * Pearson | Median Pearson Score |
|---|---|---|---|---|---|---|
| DIA-NN * | Overall | 8537 | 6113 | 0.503 | 6235 | 0.516 |
| Low variance removed | 3109 | 2330 | 0.622 | 2355 | 0.644 | |
| MQ DDA MBR * | Overall | 5520 | 3403 | 0.444 | 3400 | 0.452 |
| Low variance removed | 1965 | 1258 | 0.536 | 1242 | 0.546 | |
| MQ DDA no MBR * | Overall | 5211 | 2808 | 0.457 | 2804 | 0.470 |
| Low variance removed | 1733 | 1072 | 0.600 | 1066 | 0.640 | |
| EncyclopeDIA * | Overall | 4128 | 2878 | 0.436 | 2882 | 0.441 |
| Low variance removed | 1359 | 1091 | 0.574 | 1118 | 0.591 |
| Predicted | Basal | Her2 | LumA | LumB | Normal | |
|---|---|---|---|---|---|---|
| Reference | ||||||
| Basal | 4 | 0 | 0 | 0 | 1 | |
| Her2 | 0 | 3 | 0 | 2 | 0 | |
| LumA | 0 | 1 | 7 | 0 | 0 | |
| LumB | 0 | 1 | 0 | 2 | 0 | |
| Normal | 0 | 2 | 1 | 0 | 0 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mosquim Junior, S.; Siino, V.; Rydén, L.; Vallon-Christersson, J.; Levander, F. Choice of High-Throughput Proteomics Method Affects Data Integration with Transcriptomics and the Potential Use in Biomarker Discovery. Cancers 2022, 14, 5761. https://doi.org/10.3390/cancers14235761
Mosquim Junior S, Siino V, Rydén L, Vallon-Christersson J, Levander F. Choice of High-Throughput Proteomics Method Affects Data Integration with Transcriptomics and the Potential Use in Biomarker Discovery. Cancers. 2022; 14(23):5761. https://doi.org/10.3390/cancers14235761
Chicago/Turabian StyleMosquim Junior, Sergio, Valentina Siino, Lisa Rydén, Johan Vallon-Christersson, and Fredrik Levander. 2022. "Choice of High-Throughput Proteomics Method Affects Data Integration with Transcriptomics and the Potential Use in Biomarker Discovery" Cancers 14, no. 23: 5761. https://doi.org/10.3390/cancers14235761
APA StyleMosquim Junior, S., Siino, V., Rydén, L., Vallon-Christersson, J., & Levander, F. (2022). Choice of High-Throughput Proteomics Method Affects Data Integration with Transcriptomics and the Potential Use in Biomarker Discovery. Cancers, 14(23), 5761. https://doi.org/10.3390/cancers14235761









