Circular RNAs in Epithelial Ovarian Cancer: From Biomarkers to Therapeutic Targets
Abstract
Simple Summary
Abstract
1. Introduction
2. Overview of EOC
2.1. Histological Subtypes of EOC
2.2. Stages of EOC
2.3. Diagnosis and Treatment
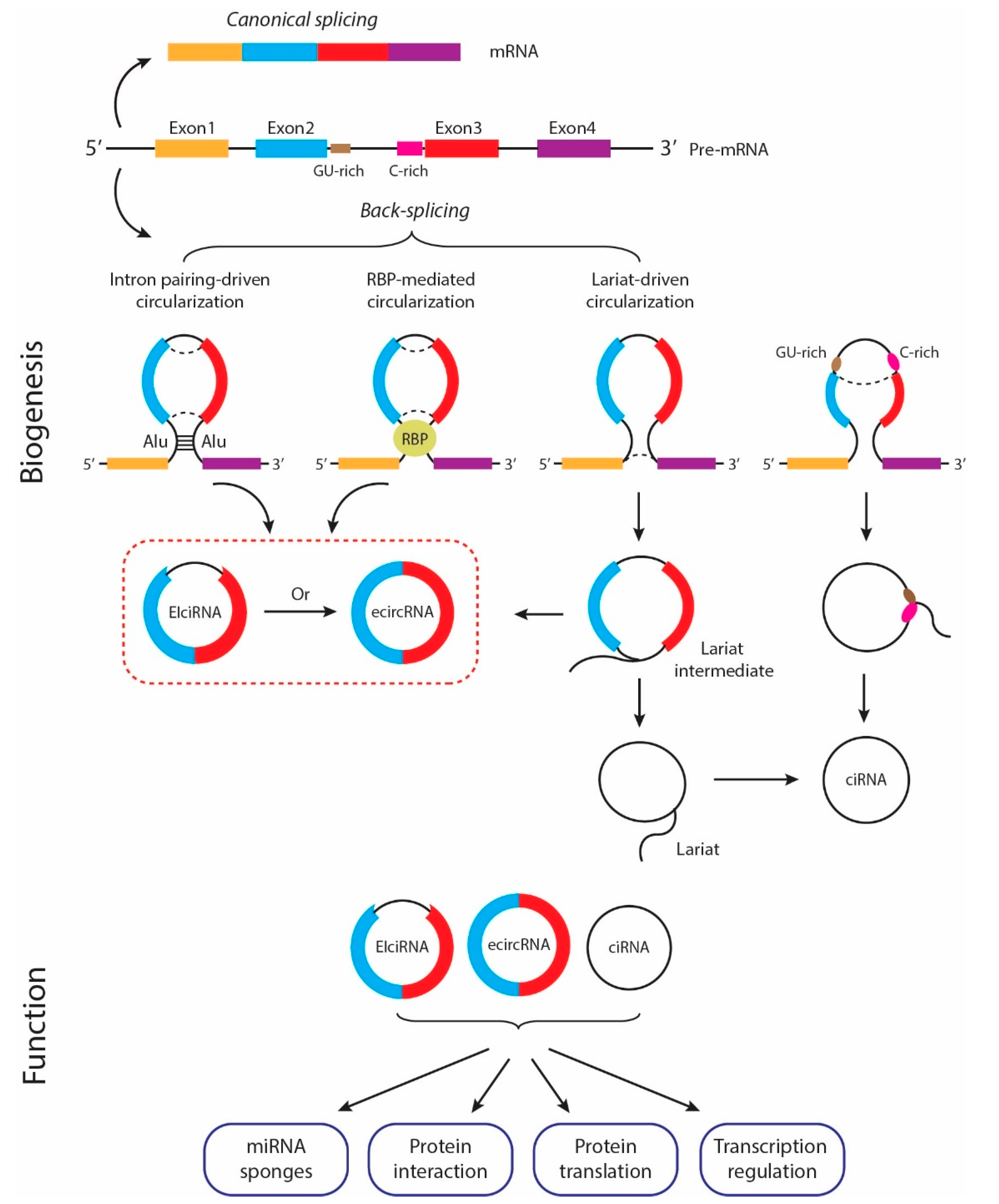
3. Overview of Circular RNA
3.1. Biogenesis of circRNAs
3.2. Mechanisms of circRNA Functions
4. CircRNAs as Potential Biomarkers in EOC
4.1. CircRNAs as Potential Prognostic Biomarkers
4.2. Circulating circRNAs as Potential Diagnostic Biomarkers
5. circRNAs as Potential Therapeutic Targets
5.1. circRNAs That Exert Tumor-Promoting Effects in EOC
5.2. circRNAs That Exert Tumor-Suppressing Effects in EOC
6. Concluding Remarks and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ovarian Cancers: Evolving Paradigms in Research and Care; National Academies Press (US): Washington, DC, USA, 2016.
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.; Talhouk, A.; McAlpine, J.N.; Law, M.R.; Hanley, G.E. Long-term mortality among women with epithelial ovarian cancer: A population-based study in British Columbia, Canada. BMC Cancer 2018, 18, 1039. [Google Scholar] [CrossRef]
- Bach, D.H.; Lee, S.K.; Sood, A.K. Circular RNAs in Cancer. Mol. Ther. Nucleic Acids 2019, 16, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.S.; Ai, Y.; Wilusz, J.E. Biogenesis and Functions of Circular RNAs Come into Focus. Trends Cell Biol. 2020, 30, 226–240. [Google Scholar] [CrossRef]
- He, A.T.; Liu, J.; Li, F.; Yang, B.B. Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal Transduct. Target. Ther. 2021, 6, 185. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, F.; He, A.T.; Yang, B.B. Circular RNAs: Expression, localization, and therapeutic potentials. Mol. Ther. 2021, 29, 1683–1702. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Zhang, C.; Yang, W.; Yong, T.; Awan, F.M.; Yang, B.B. Identifying and Characterizing circRNA-Protein Interaction. Theranostics 2017, 7, 4183–4191. [Google Scholar] [CrossRef]
- Verduci, L.; Tarcitano, E.; Strano, S.; Yarden, Y.; Blandino, G. CircRNAs: Role in human diseases and potential use as biomarkers. Cell Death Dis. 2021, 12, 468. [Google Scholar] [CrossRef]
- Yang, X.; Mei, J.; Wang, H.; Gu, D.; Ding, J.; Liu, C. The emerging roles of circular RNAs in ovarian cancer. Cancer Cell Int. 2020, 20, 265. [Google Scholar] [CrossRef]
- Foruzandeh, Z.; Zeinali-Sehrig, F.; Nejati, K.; Rahmanpour, D.; Pashazadeh, F.; Seif, F.; Alivand, M.R. CircRNAs as potent biomarkers in ovarian cancer: A systematic scoping review. Cell Mol. Biol. Lett. 2021, 26, 41. [Google Scholar] [CrossRef]
- Li, F.; Yang, Q.; He, A.T.; Yang, B.B. Circular RNAs in cancer: Limitations in functional studies and diagnostic potential. Semin Cancer Biol. 2021, 75, 49–61. [Google Scholar] [CrossRef]
- Prat, J.; D’Angelo, E.; Espinosa, I. Ovarian carcinomas: At least five different diseases with distinct histological features and molecular genetics. Hum. Pathol. 2018, 80, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Kobel, M.; Kang, E.Y. The Evolution of Ovarian Carcinoma Subclassification. Cancers 2022, 14, 416. [Google Scholar] [CrossRef] [PubMed]
- De Leo, A.; Santini, D.; Ceccarelli, C.; Santandrea, G.; Palicelli, A.; Acquaviva, G.; Chiarucci, F.; Rosini, F.; Ravegnini, G.; Pession, A.; et al. What Is New on Ovarian Carcinoma: Integrated Morphologic and Molecular Analysis Following the New 2020 World Health Organization Classification of Female Genital Tumors. Diagnostics 2021, 11, 697. [Google Scholar] [CrossRef]
- Adhikari, H.; Hassell, L.A. WHO Classification. Available online: https://www.pathologyoutlines.com/topic/ovarytumorwhoclassif.html (accessed on 11 November 2022).
- Kim, J.; Park, E.Y.; Kim, O.; Schilder, J.M.; Coffey, D.M.; Cho, C.H.; Bast, R.C., Jr. Cell Origins of High-Grade Serous Ovarian Cancer. Cancers 2018, 10, 433. [Google Scholar] [CrossRef]
- Kurman, R.J.; Carcangiu, M.L.; Herrington, C.S.; Young, R.H. WHO Classification of Tumours of Female Reproductive Organs, 4th ed.; International Agency for Research on Cancer: Lyon, France, 2014. [Google Scholar]
- Matulonis, U.A.; Sood, A.K.; Fallowfield, L.; Howitt, B.E.; Sehouli, J.; Karlan, B.Y. Ovarian cancer. Nat. Rev. Dis. Primers 2016, 2, 16061. [Google Scholar] [CrossRef]
- Li, J.; Abushahin, N.; Pang, S.; Xiang, L.; Chambers, S.K.; Fadare, O.; Kong, B.; Zheng, W. Tubal origin of ‘ovarian’ low-grade serous carcinoma. Mod. Pathol. 2011, 24, 1488–1499. [Google Scholar] [CrossRef] [PubMed]
- Vang, R.; Shih Ie, M.; Kurman, R.J. Fallopian tube precursors of ovarian low- and high-grade serous neoplasms. Histopathology 2013, 62, 44–58. [Google Scholar] [CrossRef] [PubMed]
- Ahn, G.; Folkins, A.K.; McKenney, J.K.; Longacre, T.A. Low-grade Serous Carcinoma of the Ovary: Clinicopathologic Analysis of 52 Invasive Cases and Identification of a Possible Noninvasive Intermediate Lesion. Am. J. Surg. Pathol. 2016, 40, 1165–1176. [Google Scholar] [CrossRef]
- Moujaber, T.; Balleine, R.L.; Gao, B.; Madsen, I.; Harnett, P.R.; DeFazio, A. New therapeutic opportunities for women with low-grade serous ovarian cancer. Endocr. Relat. Cancer 2021, 29, R1–R16. [Google Scholar] [CrossRef] [PubMed]
- Santillan, A.; Kim, Y.W.; Zahurak, M.L.; Gardner, G.J.; Giuntoli, R.L., 2nd; Shih, I.M.; Bristow, R.E. Differences of chemoresistance assay between invasive micropapillary/low-grade serous ovarian carcinoma and high-grade serous ovarian carcinoma. Int. J. Gynecol. Cancer 2007, 17, 601–606. [Google Scholar] [CrossRef]
- Babaier, A.; Mal, H.; Alselwi, W.; Ghatage, P. Low-Grade Serous Carcinoma of the Ovary: The Current Status. Diagnostics 2022, 12, 458. [Google Scholar] [CrossRef] [PubMed]
- Peres, L.C.; Cushing-Haugen, K.L.; Kobel, M.; Harris, H.R.; Berchuck, A.; Rossing, M.A.; Schildkraut, J.M.; Doherty, J.A. Invasive Epithelial Ovarian Cancer Survival by Histotype and Disease Stage. J. Natl. Cancer Inst. 2019, 111, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.R.; Shih Ie, M. Ovarian cancer. Annu. Rev. Pathol. 2009, 4, 287–313. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mang, M.; Wang, Y.; Wang, L.; Klein, R.; Kong, B.; Zheng, W. Tubal origin of ovarian endometriosis and clear cell and endometrioid carcinoma. Am. J. Cancer Res. 2015, 5, 869–879. [Google Scholar] [PubMed]
- Chen, S.; Li, Y.; Qian, L.; Deng, S.; Liu, L.; Xiao, W.; Zhou, Y. A Review of the Clinical Characteristics and Novel Molecular Subtypes of Endometrioid Ovarian Cancer. Front. Oncol. 2021, 11, 668151. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Xu, Z.; Zhang, T.; Qian, L.; Xiao, W.; Wei, H.; Jin, T.; Zhou, Y. Updates of Pathogenesis, Diagnostic and Therapeutic Perspectives for Ovarian Clear Cell Carcinoma. J. Cancer 2021, 12, 2295–2316. [Google Scholar] [CrossRef]
- Iida, Y.; Okamoto, A.; Hollis, R.L.; Gourley, C.; Herrington, C.S. Clear cell carcinoma of the ovary: A clinical and molecular perspective. Int. J. Gynecol. Cancer 2021, 31, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Oseledchyk, A.; Leitao, M.M., Jr.; Konner, J.; O’Cearbhaill, R.E.; Zamarin, D.; Sonoda, Y.; Gardner, G.J.; Long Roche, K.; Aghajanian, C.A.; Grisham, R.N.; et al. Adjuvant chemotherapy in patients with stage I endometrioid or clear cell ovarian cancer in the platinum era: A Surveillance, Epidemiology, and End Results Cohort Study, 2000-2013. Ann. Oncol. 2017, 28, 2985–2993. [Google Scholar] [CrossRef]
- Babaier, A.; Ghatage, P. Mucinous Cancer of the Ovary: Overview and Current Status. Diagnostics 2020, 10, 52. [Google Scholar] [CrossRef]
- Koshiyama, M.; Matsumura, N.; Konishi, I. Recent concepts of ovarian carcinogenesis: Type I and type II. Biomed. Res. Int. 2014, 2014, 934261. [Google Scholar] [CrossRef]
- Prat, J. FIGO’s staging classification for cancer of the ovary, fallopian tube, and peritoneum: Abridged republication. J. Gynecol. Oncol. 2015, 26, 87–89. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhu, Y.; Zhang, W.; Lang, J.; Ning, L. Utility Of Plasma circBNC2 As A Diagnostic Biomarker In Epithelial Ovarian Cancer. Onco. Targets Ther. 2019, 12, 9715–9723. [Google Scholar] [CrossRef] [PubMed]
- Bast, R.C., Jr.; Badgwell, D.; Lu, Z.; Marquez, R.; Rosen, D.; Liu, J.; Baggerly, K.A.; Atkinson, E.N.; Skates, S.; Zhang, Z.; et al. New tumor markers: CA125 and beyond. Int. J. Gynecol. Cancer 2005, 15 (Suppl. 3), 274–281. [Google Scholar] [CrossRef] [PubMed]
- Sroczynski, G.; Gogollari, A.; Conrads-Frank, A.; Hallsson, L.R.; Pashayan, N.; Widschwendter, M.; Siebert, U. Cost-Effectiveness of Early Detection and Prevention Strategies for Endometrial Cancer-A Systematic Review. Cancers 2020, 12, 1874. [Google Scholar] [CrossRef]
- Dochez, V.; Caillon, H.; Vaucel, E.; Dimet, J.; Winer, N.; Ducarme, G. Biomarkers and algorithms for diagnosis of ovarian cancer: CA125, HE4, RMI and ROMA, a review. J. Ovarian Res. 2019, 12, 28. [Google Scholar] [CrossRef]
- Sandri, M.T.; Bottari, F.; Franchi, D.; Boveri, S.; Candiani, M.; Ronzoni, S.; Peiretti, M.; Radice, D.; Passerini, R.; Sideri, M. Comparison of HE4, CA125 and ROMA algorithm in women with a pelvic mass: Correlation with pathological outcome. Gynecol. Oncol. 2013, 128, 233–238. [Google Scholar] [CrossRef]
- Ferraro, S.; Schiumarini, D.; Panteghini, M. Human epididymis protein 4: Factors of variation. Clin. Chim. Acta 2015, 438, 171–177. [Google Scholar] [CrossRef]
- Moore, R.G.; Miller, M.C.; Eklund, E.E.; Lu, K.H.; Bast, R.C., Jr.; Lambert-Messerlian, G. Serum levels of the ovarian cancer biomarker HE4 are decreased in pregnancy and increase with age. Am. J. Obstet Gynecol. 2012, 206, 349 e341–e347. [Google Scholar] [CrossRef]
- Ning, L.; Lang, J.; Wu, L. Plasma circN4BP2L2 is a promising novel diagnostic biomarker for epithelial ovarian cancer. BMC Cancer 2022, 22, 6. [Google Scholar] [CrossRef]
- Coleridge, S.L.; Bryant, A.; Kehoe, S.; Morrison, J. Chemotherapy versus surgery for initial treatment in advanced ovarian epithelial cancer. Cochrane Database Syst. Rev. 2021, 2, CD005343. [Google Scholar] [CrossRef] [PubMed]
- Matulonis, U.A. Ovarian Cancer; Elsevier: Amsterdam, The Netherlands, 2018; Volume 32. [Google Scholar]
- Sapiezynski, J.; Taratula, O.; Rodriguez-Rodriguez, L.; Minko, T. Precision targeted therapy of ovarian cancer. J. Control Release 2016, 243, 250–268. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Xia, B.R.; Zhang, Z.C.; Zhang, Y.J.; Lou, G.; Jin, W.L. Immunotherapy for Ovarian Cancer: Adjuvant, Combination, and Neoadjuvant. Front. Immunol. 2020, 11, 577869. [Google Scholar] [CrossRef] [PubMed]
- Kurnit, K.C.; Fleming, G.F.; Lengyel, E. Updates and New Options in Advanced Epithelial Ovarian Cancer Treatment. Obstet. Gynecol. 2021, 137, 108–121. [Google Scholar] [CrossRef]
- Murphy, A.D.; Morgan, R.D.; Clamp, A.R.; Jayson, G.C. The role of vascular endothelial growth factor inhibitors in the treatment of epithelial ovarian cancer. Br. J. Cancer 2022, 126, 851–864. [Google Scholar] [CrossRef]
- Bonello, M.; Sims, A.H.; Langdon, S.P. Human epidermal growth factor receptor targeted inhibitors for the treatment of ovarian cancer. Cancer Biol. Med. 2018, 15, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.W.Y.; Dand, S.; Doig, L.; Papenfuss, A.T.; Scott, C.L.; Ho, G.; Ooi, J.D. T-Cell Receptor Therapy in the Treatment of Ovarian Cancer: A Mini Review. Front. Immunol. 2021, 12, 672502. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Zhao, X.; Cai, Y.; Xu, J. Circular RNAs: Biogenesis, Mechanism, and Function in Human Cancers. Int. J. Mol. Sci. 2019, 20, 3926. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Jia, Y.; Zhang, Y.; Shi, L.; Li, Q.; Zang, A.; Wang, H. Circular RNA: Biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinogens. Epigenomics 2020, 12, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Xiao, Y.; Ma, J.; Tang, Y.; Tian, B.; Zhang, Y.; Li, X.; Wu, Z.; Yang, D.; Zhou, Y.; et al. Circular RNAs in Cancer: Emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 2019, 18, 90. [Google Scholar] [CrossRef]
- Huang, A.; Zheng, H.; Wu, Z.; Chen, M.; Huang, Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics 2020, 10, 3503–3517. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Huang, Y.; Zhu, Q. Mechanisms Regulating Abnormal Circular RNA Biogenesis in Cancer. Cancers 2021, 13, 4185. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Sinha, T.; Panigrahi, C.; Das, D.; Chandra Panda, A. Circular RNA translation, a path to hidden proteome. Wiley Interdiscip. Rev. RNA 2022, 13, e1685. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Lei, K.; Bai, H.; Wei, Z.; Xie, C.; Wang, J.; Li, J.; Chen, Q. The mechanism and function of circular RNAs in human diseases. Exp. Cell Res. 2018, 368, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Jakobsen, T.; Hager, H.; Kjems, J. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 2022, 19, 188–206. [Google Scholar] [CrossRef]
- Beilerli, A.; Gareev, I.; Beylerli, O.; Yang, G.; Pavlov, V.; Aliev, G.; Ahmad, A. Circular RNAs as biomarkers and therapeutic targets in cancer. Semin Cancer Biol. 2022, 83, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef]
- Zhang, F.; Xu, Y.; Ye, W.; Jiang, J.; Wu, C. Circular RNA S-7 promotes ovarian cancer EMT via sponging miR-641 to up-regulate ZEB1 and MDM2. Biosci. Rep. 2020, 40, BSR20200825. [Google Scholar] [CrossRef]
- Wu, H.; Zhao, X.; Wang, J.; Jiang, X.; Cheng, Y.; He, Y.; Sun, L.; Zhang, G. Circular RNA CDR1as Alleviates Cisplatin-Based Chemoresistance by Suppressing MiR-1299 in Ovarian Cancer. Front. Genet. 2021, 12, 815448. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Ji, M.; Wang, Q.; He, N.; Li, Y. Circular RNA Cdr1as Upregulates SCAI to Suppress Cisplatin Resistance in Ovarian Cancer via miR-1270 Suppression. Mol. Ther. Nucleic Acids 2019, 18, 24–33. [Google Scholar] [CrossRef]
- Chen, H.; Mao, M.; Jiang, J.; Zhu, D.; Li, P. Circular RNA CDR1as acts as a sponge of miR-135b-5p to suppress ovarian cancer progression. Onco Targets Ther 2019, 12, 3869–3879. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhang, J.; Zhang, L.Y.; Wang, L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3713–3718. [Google Scholar] [CrossRef]
- Zhang, M.; Xia, B.; Xu, Y.; Zhang, Y.; Xu, J.; Lou, G. Circular RNA (hsa_circ_0051240) promotes cell proliferation, migration and invasion in ovarian cancer through miR-637/KLK4 axis. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1224–1233. [Google Scholar] [CrossRef]
- Li, Q.H.; Liu, Y.; Chen, S.; Zong, Z.H.; Du, Y.P.; Sheng, X.J.; Zhao, Y. circ-CSPP1 promotes proliferation, invasion and migration of ovarian cancer cells by acting as a miR-1236-3p sponge. Biomed. Pharmacother. 2019, 114, 108832. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wu, W.; Li, Q.H.; Xie, B.M.; Shen, F.; Du, Y.P.; Zong, Z.H.; Wang, L.L.; Wei, X.Q.; Zhao, Y. Circ-NOLC1 promotes epithelial ovarian cancer tumorigenesis and progression by binding ESRP1 and modulating CDK1 and RhoA expression. Cell Death Discov. 2021, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Zong, Z.H.; Liu, Y.; Chen, S.; Wang, L.L.; Zhao, Y. circPUM1 Promotes Tumorigenesis and Progression of Ovarian Cancer by Sponging miR-615-5p and miR-6753-5p. Mol. Ther. Nucleic Acids 2019, 18, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Zong, Z.H.; Liu, Y.; Guan, X.; Chen, S.; Zhao, Y. CircRhoC promotes tumorigenicity and progression in ovarian cancer by functioning as a miR-302e sponge to positively regulate VEGFA. J. Cell Mol. Med. 2019, 23, 8472–8481. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Han, Y.; Qiao, H.; Han, Y.; Lu, X.; Lu, Y.; Wang, X.; Kai, H.; Zheng, Y. Hsa_circ_0063804 enhances ovarian cancer cells proliferation and resistance to cisplatin by targeting miR-1276/CLU axis. Aging (Albany NY) 2022, 14, 4699–4713. [Google Scholar] [CrossRef]
- Wang, S.; Li, Z.; Zhu, G.; Hong, L.; Hu, C.; Wang, K.; Cui, K.; Hao, C. RNA-binding protein IGF2BP2 enhances circ_0000745 abundancy and promotes aggressiveness and stemness of ovarian cancer cells via the microRNA-3187-3p/ERBB4/PI3K/AKT axis. J. Ovar. Res. 2021, 14, 154. [Google Scholar] [CrossRef]
- Cao, Y.; Xie, X.; Li, M.; Gao, Y. CircHIPK2 Contributes to DDP Resistance and Malignant Behaviors of DDP-Resistant Ovarian Cancer Cells Both in vitro and in vivo Through circHIPK2/miR-338-3p/CHTOP ceRNA Pathway. Oncol. Targets Ther. 2021, 14, 3151–3165. [Google Scholar] [CrossRef]
- Li, Y.; Lin, S.; An, N. Hsa_circ_0009910: Oncogenic circular RNA targets microRNA-145 in ovarian cancer cells. Cell Cycle 2020, 19, 1857–1868. [Google Scholar] [CrossRef]
- Zhao, Y.; Hu, Y.; Shen, Q.; Chen, Q.; Zhu, X.J.; Jiang, S.S.; Zhang, Q. CircRNA_MYLK promotes malignant progression of ovarian cancer through regulating microRNA-652. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 5281–5291. [Google Scholar] [CrossRef]
- Gan, X.; Zhu, H.; Jiang, X.; Obiegbusi, S.C.; Yong, M.; Long, X.; Hu, J. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol. Cancer 2020, 19, 45. [Google Scholar] [CrossRef]
- Sun, D.; Liu, J.; Zhou, L. Upregulation of circular RNA circFAM53B predicts adverse prognosis and accelerates the progression of ovarian cancer via the miR646/VAMP2 and miR647/MDM2 signaling pathways. Oncol. Rep. 2019, 42, 2728–2737. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, X.; Yang, L.; Li, M.; Zhang, Y.; Zhang, J. CircASH2L Promotes Ovarian Cancer Tumorigenesis, Angiogenesis, and Lymphangiogenesis by Regulating the miR-665/VEGFA Axis as a Competing Endogenous RNA. Front. Cell Dev. Biol. 2020, 8, 595585. [Google Scholar] [CrossRef] [PubMed]
- Pei, C.; Wang, H.; Shi, C.; Zhang, C.; Wang, M. CircRNA hsa_circ_0013958 may contribute to the development of ovarian cancer by affecting epithelial-mesenchymal transition and apoptotic signaling pathways. J. Clin. Lab. Anal. 2020, 34, e23292. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ye, X.; Xia, X.; Lin, X. Circular RNA ABCB10 correlates with advanced clinicopathological features and unfavorable survival, and promotes cell proliferation while reduces cell apoptosis in epithelial ovarian cancer. Cancer Biomark 2019, 26, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Zhao, Z.; Wu, Y.; Wang, Y.; Zhao, Y.; Wang, J. Circular RNA circTNPO3 Regulates Paclitaxel Resistance of Ovarian Cancer Cells by miR-1299/NEK2 Signaling Pathway. Mol. Ther. Nucleic Acids 2020, 21, 780–791. [Google Scholar] [CrossRef]
- Liang, Y.X.; Zhang, L.L.; Yang, L. circANKRD17(has_circ_0007883) confers paclitaxel resistance of ovarian cancer via interacting with FUS to stabilize FOXR2. Mol. Cell Biochem. 2022. [Google Scholar] [CrossRef]
- Yong, M.; Hu, J.; Zhu, H.; Jiang, X.; Gan, X.; Hu, L. Circ-EEF2 facilitated autophagy via interaction with mir-6881-3p and ANXA2 in EOC. Am. J. Cancer Res. 2020, 10, 3737–3751. [Google Scholar]
- Song, W.; Zeng, Z.; Zhang, Y.; Li, H.; Cheng, H.; Wang, J.; Wu, F. CircRNF144B/miR-342-3p/FBXL11 axis reduced autophagy and promoted the progression of ovarian cancer by increasing the ubiquitination of Beclin-1. Cell Death Dis. 2022, 13, 857. [Google Scholar] [CrossRef]
- Wu, D.; Liu, J.; Yu, L.; Wu, S.; Qiu, X. Circular RNA hsa_circ_0000144 aggravates ovarian Cancer progression by regulating ELK3 via sponging miR-610. J. Ovar. Res. 2022, 15, 113. [Google Scholar] [CrossRef]
- Zhou, X.; Jiang, J.; Guo, S. Hsa_circ_0004712 downregulation attenuates ovarian cancer malignant development by targeting the miR-331-3p/FZD4 pathway. J. Ovar. Res. 2021, 14, 118. [Google Scholar] [CrossRef]
- Ma, L.; Liu, W.; Li, M. Circ_0061140 Contributes to Ovarian Cancer Progression by Targeting miR-761/LETM1 Signaling. Biochem. Genet. 2022. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Huang, H.Y.; Long, Y.; Ma, Y.; Kamalibaike, M.; Dawuti, R.; Li, L. circ_C20orf11 enhances DDP resistance by inhibiting miR-527/YWHAZ through the promotion of extracellular vesicle-mediated macrophage M2 polarization in ovarian cancer. Cancer Biol. Ther. 2021, 22, 440–454. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Luo, F.; Jiang, X.; Zhang, W.; Xiang, T.; Pan, Q.; Cai, L.; Zhao, J.; Weng, D.; Li, Y.; et al. CircITGB6 promotes ovarian cancer cisplatin resistance by resetting tumor-associated macrophage polarization toward the M2 phenotype. J. Immunother. Cancer 2022, 10, e004029. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, J.; Zhang, X.; Liu, G. Serum circSETDB1 is a promising biomarker for predicting response to platinum-taxane-combined chemotherapy and relapse in high-grade serous ovarian cancer. Oncol. Targets Ther. 2019, 12, 7451–7457. [Google Scholar] [CrossRef]
- Luo, Y.; Gui, R. Circulating exosomal circFoxp1 confers cisplatin resistance in epithelial ovarian cancer cells. J. Gynecol. Oncol. 2020, 31, e75. [Google Scholar] [CrossRef]
- Wang, X.; Yao, Y.; Jin, M. Circ-0001068 is a novel biomarker for ovarian cancer and inducer of PD1 expression in T cells. Aging (Albany NY) 2020, 12, 19095–19106. [Google Scholar] [CrossRef]
- Lin, C.; Xu, X.; Yang, Q.; Liang, L.; Qiao, S. Circular RNA ITCH suppresses proliferation, invasion, and glycolysis of ovarian cancer cells by up-regulating CDH1 via sponging miR-106a. Cancer Cell Int. 2020, 20, 336. [Google Scholar] [CrossRef]
- Luo, L.; Gao, Y.; Sun, X. Circ-ITCH correlates with small tumor size, decreased FIGO stage and prolonged overall survival, and it inhibits cells proliferation while promotes cells apoptosis in epithelial ovarian cancer. Cancer Biomark 2018, 23, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lin, S.; Mo, Z.; Jiang, J.; Tang, H.; Wu, C.; Song, J. CircRNA_100395 inhibits cell proliferation and metastasis in ovarian cancer via regulating miR-1228/p53/epithelial-mesenchymal transition (EMT) axis. J. Cancer 2020, 11, 599–609. [Google Scholar] [CrossRef]
- Zou, T.; Wang, P.L.; Gao, Y.; Liang, W.T. Circular RNA_LARP4 is lower expressed and serves as a potential biomarker of ovarian cancer prognosis. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7178–7182. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, Z.; Yang, S.; Wang, Y.; Luan, Z. CircEXOC6B Suppresses the Proliferation and Motility and Sensitizes Ovarian Cancer Cells to Paclitaxel Through miR-376c-3p/FOXO3 Axis. Cancer Biother. Radiopharm. 2020, 37, 9. [Google Scholar] [CrossRef]
- Ning, L.; Long, B.; Zhang, W.; Yu, M.; Wang, S.; Cao, D.; Yang, J.; Shen, K.; Huang, Y.; Lang, J. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int. J. Oncol. 2018, 53, 2637–2646. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; Zhou, J.H.; Shen, F.R.; Shi, X.; Chen, Y.G. CircATRNL1 activates Smad4 signaling to inhibit angiogenesis and ovarian cancer metastasis via miR-378. Mol. Oncol. 2021, 15, 1217–1233. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Wang, H. Circ_0078607 inhibits the progression of ovarian cancer via regulating the miR-32-5p/SIK1 network. J. Ovar. Res. 2022, 15, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Yang, Z.; Jin, Y.; Cheng, S.; Yang, J.; Wang, Y. Low Expression of Circular RNA hsa_circ_0078607 Predicts Poor Prognosis in High-Grade Serous Ovarian Cancer. Cancer Manag. Res. 2021, 13, 2877–2883. [Google Scholar] [CrossRef]
- Fu, Y.; Sun, H. The molecular mechanism of circRHOBTB3 inhibits the proliferation and invasion of epithelial ovarian cancer by serving as the ceRNA of miR-23a-3p. J. Ovar. Res. 2022, 15, 66. [Google Scholar] [CrossRef]
- Wu, X.; Liu, D.; Wang, S.; Liu, J. Circ_0007444 Inhibits the Progression of Ovarian Cancer via Mediating the miR-570-3p/PTEN Axis. Oncol. Targets Ther. 2021, 14, 97–110. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, C.; Liu, Y.; Wang, M. Circular RNA profiling reveals circRNA1656 as a novel biomarker in high grade serous ovarian cancer. Biosci. Trends 2019, 13, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhou, J.; Li, Y.; Shi, X.; Shen, F.; Chen, M.; Chen, Y.; Wang, J. Hsa_circ_0001445 works as a cancer suppressor via miR-576-5p/SFRP1 axis regulation in ovarian cancer. Cancer Med. 2022. [Google Scholar] [CrossRef]
- Wu, M.; Qiu, Q.; Zhou, Q.; Li, J.; Yang, J.; Zheng, C.; Luo, A.; Li, X.; Zhang, H.; Cheng, X.; et al. circFBXO7/miR-96-5p/MTSS1 axis is an important regulator in the Wnt signaling pathway in ovarian cancer. Mol. Cancer 2022, 21, 137. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, Q.; Qiu, Q.; Hou, L.; Wu, M.; Li, J.; Li, X.; Lu, B.; Cheng, X.; Liu, P.; et al. CircPLEKHM3 acts as a tumor suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol. Cancer 2019, 18, 144. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Yuan, L.; Zou, X. Circular RNA circ-BNC2 (hsa_circ_0008732) inhibits the progression of ovarian cancer through microRNA-223-3p/ FBXW7 axis. J. Ovar. Res. 2022, 15, 95. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Sun, Y.; Shi, Y.; Liu, G.; Teng, F.; Geng, Z.; Chen, X.; Xu, H.; Xu, J.; Jia, X. Plasma circRNA microarray profiling identifies novel circRNA biomarkers for the diagnosis of ovarian cancer. J. Ovar. Res. 2022, 15, 58. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, R.; Wu, Z.; Bai, P. Circular RNA ITCH suppressed prostate cancer progression by increasing HOXB13 expression via spongy miR-17-5p. Cancer Cell Int. 2019, 19, 328. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, Y.Z.; Xu, L.; Jia, R. Circular RNA ITCH: A novel tumor suppressor in multiple cancers. Life Sci. 2020, 254, 117176. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, H.; Zheng, R.; Wu, P.; Sun, Z.; Chen, J.; Zhang, L.; Zhang, C.; Qian, H.; Jiang, J.; et al. Circular RNA ITCH suppresses metastasis of gastric cancer via regulating miR-199a-5p/Klotho axis. Cell Cycle 2021, 20, 522–536. [Google Scholar] [CrossRef] [PubMed]
- Kurman, R.J.; Shih Ie, M. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer—Shifting the paradigm. Hum. Pathol. 2011, 42, 918–931. [Google Scholar] [CrossRef]
- Ionescu, F.; Zhang, J.; Wang, L. Clinical Applications of Liquid Biopsy in Prostate Cancer: From Screening to Predictive Biomarker. Cancers 2022, 14, 1728. [Google Scholar] [CrossRef]
- Shabaninejad, Z.; Vafadar, A.; Movahedpour, A.; Ghasemi, Y.; Namdar, A.; Fathizadeh, H.; Pourhanifeh, M.H.; Savardashtaki, A.; Mirzaei, H. Circular RNAs in cancer: New insights into functions and implications in ovarian cancer. J. Ovar. Res. 2019, 12, 84. [Google Scholar] [CrossRef]
- Najafi, S. The emerging roles and potential applications of circular RNAs in ovarian cancer: A comprehensive review. J. Cancer Res. Clin. Oncol. 2022. [Google Scholar] [CrossRef]
- Zhu, J.; Luo, J.E.; Chen, Y.; Wu, Q. Circ_0061140 knockdown inhibits tumorigenesis and improves PTX sensitivity by regulating miR-136/CBX2 axis in ovarian cancer. J. Ovar. Res. 2021, 14, 136. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, J.; He, Y.; Wang, Y. hsa_circ_0061140 Knockdown Reverses FOXM1-Mediated Cell Growth and Metastasis in Ovarian Cancer through miR-370 Sponge Activity. Mol. Ther. Nucleic Acids 2018, 13, 55–63. [Google Scholar] [CrossRef]
- Zhang, Y.; Di, Q.; Chen, J.; Chang, M.; Ma, Y.; Yu, J. Circ_0061140 Contributes to the Malignant Progression in Ovarian Cancer Cells by Mediating the RAB1A Level Through Sponging miR-361-5p. Biochem. Genet. 2022, 60, 1946–1962. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Qi, L.; Wang, L. Overexpression of circ_CELSR1 facilitates paclitaxel resistance of ovarian cancer by regulating miR-149-5p/SIK2 axis. Anticancer Drugs 2021, 32, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cheng, J.; Quan, C.; Wen, H.; Feng, Z.; Hu, Q.; Zhu, J.; Huang, Y.; Wu, X. circCELSR1 (hsa_circ_0063809) Contributes to Paclitaxel Resistance of Ovarian Cancer Cells by Regulating FOXR2 Expression via miR-1252. Mol. Ther. Nucleic Acids 2020, 19, 718–730. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.Y.; Yuan, J.; Wang, C.; Zeng, D.; Yong, J.H.; Jiang, X.Y.; Lan, H.; Xiao, S.S. circCELSR1 facilitates ovarian cancer proliferation and metastasis by sponging miR-598 to activate BRD4 signals. Mol. Med. 2020, 26, 70. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Yan, L.; Zhong, J.; Hong, L.; Zhang, N.; Luo, X. Circ_0025033 deficiency suppresses paclitaxel resistance and malignant development of paclitaxel-resistant ovarian cancer cells by modulating the miR-532-3p/FOXM1 network. Immunopharmacol. Immunotoxicol. 2022, 44, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Zhang, Y. Circ_0025033 promotes the progression of ovarian cancer by activating the expression of LSM4 via targeting miR-184. Pathol. Res. Pract. 2021, 217, 153275. [Google Scholar] [CrossRef]
- Gao, Y.; Huang, Y. Circ_0007841 knockdown confers c.cisplatin sensitivity to ovarian cancer cells by down-regulation of NFIB expression in a miR-532-5p-dependent manner. J. Chemother. 2022, 2022, 1–14. [Google Scholar] [CrossRef]
- Huang, K.; Liu, D.; Su, C. Circ_0007841 accelerates ovarian cancer development through facilitating MEX3C expression by restraining miR-151-3p activity. Aging (Albany NY) 2021, 13, 12058–12066. [Google Scholar] [CrossRef]
- Yang, H.; Guo, Y.; Zhang, Y.; Wang, D.; Zhang, G.; Hou, J.; Yang, J. Circ_MUC16 attenuates the effects of Propofol to promote the aggressive behaviors of ovarian cancer by mediating the miR-1182/S100B signaling pathway. BMC Anesthesiol. 2021, 21, 297. [Google Scholar] [CrossRef]
- Lin, X.; Chen, Y.; Ye, X.; Xia, X. Circular RNA ABCB10 promotes cell proliferation and invasion, but inhibits apoptosis via regulating the microRNA1271mediated Capn4/Wnt/betacatenin signaling pathway in epithelial ovarian cancer. Mol. Med. Rep. 2021, 23, 387. [Google Scholar] [CrossRef]
- Sheng, M.; Wei, N.; Yang, H.Y.; Yan, M.; Zhao, Q.X.; Jing, L.J. CircRNA UBAP2 promotes the progression of ovarian cancer by sponging microRNA-144. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7283–7294. [Google Scholar] [CrossRef]
- Xu, Q.; Deng, B.; Li, M.; Chen, Y.; Zhuan, L. circRNA-UBAP2 promotes the proliferation and inhibits apoptosis of ovarian cancer though miR-382-5p/PRPF8 axis. J. Ovar. Res. 2020, 13, 81. [Google Scholar] [CrossRef]
- Li, M.; Chi, C.; Zhou, L.; Chen, Y.; Tang, X. Circular PVT1 regulates cell proliferation and invasion via miR-149-5p/FOXM1 axis in ovarian cancer. J. Cancer 2021, 12, 611–621. [Google Scholar] [CrossRef]
- Sun, X.; Luo, L.; Gao, Y. Circular RNA PVT1 enhances cell proliferation but inhibits apoptosis through sponging microRNA-149 in epithelial ovarian cancer. J. Obstet Gynaecol. Res. 2020, 46, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Wang, Q.; Guo, N.; Wang, H.; Chen, H.; Ni, G.; Li, P. CircRNA circ_0072995 promotes the progression of epithelial ovarian cancer by modulating miR-147a/CDK6 axis. Aging (Albany NY) 2020, 12, 17209–17223. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Luo, Y.; Li, X. Circ_0072995 Promotes Ovarian Cancer Progression Through Regulating miR-122-5p/SLC1A5 Axis. Biochem. Genet. 2022, 60, 153–172. [Google Scholar] [CrossRef]
- Liang, Y.; Meng, K.; Qiu, R. Circular RNA Circ_0013958 Functions as a Tumor Promoter in Ovarian Cancer by Regulating miR-637/PLXNB2 Axis. Front. Genet. 2021, 12, 644451. [Google Scholar] [CrossRef]
- Li, M.; Cai, J.; Han, X.; Ren, Y. Downregulation of circNRIP1 Suppresses the Paclitaxel Resistance of Ovarian Cancer via Regulating the miR-211-5p/HOXC8 Axis. Cancer Manag. Res. 2020, 12, 9159–9171. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Liu, L.U.; He, C.; Zhao, M.; Ni, R.; Zhang, Z.; Shui, C. Circ_0002711 knockdown suppresses cell growth and aerobic glycolysis by modulating miR-1244/ROCK1 axis in ovarian cancer. J. Biosci. 2021, 46, 21. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Li, S.; Zhao, X.; Yuan, Y.; Zhang, B.; Guan, Y. Knockdown of Circular RNA Hsa_circ_0000714 Can Regulate RAB17 by Sponging miR-370-3p to Reduce Paclitaxel Resistance of Ovarian Cancer Through CDK6/RB Pathway. Oncol. Targets Ther. 2020, 13, 13211–13224. [Google Scholar] [CrossRef] [PubMed]
- Ying, H.; Zhao, R.; Yu, Q.; Zhang, K.; Deng, Q. CircATL2 enhances paclitaxel resistance of ovarian cancer via impacting miR-506-3p/NFIB axis. Drug. Dev. Res. 2022, 83, 512–524. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yin, Z.; Wu, Y.; Zhan, Q.; Huang, H.; Fan, J. Circular RNA lysophosphatidic acid receptor 3 (circ-LPAR3) enhances the cisplatin resistance of ovarian cancer. Bioengineered 2022, 13, 3739–3750. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Ni, M.; Li, J.; Cheng, J.; Zhao, H.; Zhao, J.; Huang, S.; Wu, X. Circ0004390 promotes cell proliferation through sponging miR-198 in ovarian cancer. Biochem. Biophys. Res. Commun. 2020, 526, 14–20. [Google Scholar] [CrossRef]
- Ji, J.; Li, C.; Wang, J.; Wang, L.; Huang, H.; Li, Y.; Fang, J. Hsa_circ_0001756 promotes ovarian cancer progression through regulating IGF2BP2-mediated RAB5A expression and the EGFR/MAPK signaling pathway. Cell Cycle 2022, 21, 685–696. [Google Scholar] [CrossRef]
- Yin, Y.; Li, J.; Rong, J.; Zhang, B.; Wang, X.; Han, H. Circ_0067934 reduces JNK phosphorylation through a microRNA-545-3p/PPA1 axis to enhance tumorigenesis and cisplatin resistance in ovarian cancer. Immunopharmacol. Immunotoxicol. 2022, 44, 261–274. [Google Scholar] [CrossRef]
- Fu, L.; Zhang, D.; Yi, N.; Cao, Y.; Wei, Y.; Wang, W.; Li, L. Circular RNA circPBX3 promotes cisplatin resistance of ovarian cancer cells via interacting with IGF2BP2 to stabilize ATP7A mRNA expression. Hum. Cell 2022, 35, 1560–1576. [Google Scholar] [CrossRef]
- Qu, D.; Zou, X.; Liu, Z. Propofol modulates glycolysis reprogramming of ovarian tumor via restraining circular RNA-zinc finger RNA-binding protein/microRNA-212-5p/superoxide dismutase 2 axis. Bioengineered 2022, 13, 11881–11892. [Google Scholar] [CrossRef]
- Zhang, M.; Xu, Y.; Zhang, Y.; Li, B.; Lou, G. Circular RNA circE2F2 promotes malignant progression of ovarian cancer cells by upregulating the expression of E2F2 protein via binding to HuR protein. Cell Signal. 2021, 84, 110014. [Google Scholar] [CrossRef]
- Wu, S.G.; Zhou, P.; Chen, J.X.; Lei, J.; Hua, L.; Dong, Y.; Hu, M.; Lian, C.L.; Yang, L.C.; Zhou, J. circ-PTK2 (hsa_circ_0008305) regulates the pathogenic processes of ovarian cancer via miR-639 and FOXC1 regulatory cascade. Cancer Cell Int. 2021, 21, 277. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J.; Li, H.; Sun, Y.; Tong, X. Downregulation of hsa_circ_0026123 suppresses ovarian cancer cell metastasis and proliferation through the miR1243p/EZH2 signaling pathway. Int. J. Mol. Med. 2021, 47, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhu, H.; Hu, J. CircRAB11FIP1 promoted autophagy flux of ovarian cancer through DSC1 and miR-129. Cell Death Dis. 2021, 12, 219. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Dong, Z.N.; Qiu, B.Q.; Hu, M.; Liang, X.Q.; Dai, X.; Hong, D.; Sun, Y.F. CircRNA FGFR3 induces epithelial-mesenchymal transition of ovarian cancer by regulating miR-29a-3p/E2F1 axis. Aging (Albany NY) 2020, 12, 14080–14091. [Google Scholar] [CrossRef] [PubMed]
- An, Q.; Liu, T.; Wang, M.Y.; Yang, Y.J.; Zhang, Z.D.; Lin, Z.J.; Yang, B. circKRT7-miR-29a-3p-COL1A1 Axis Promotes Ovarian Cancer Cell Progression. Oncol. Targets Ther. 2020, 13, 8963–8976. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, X.; Qiao, L.; Wang, H. CircRNA circ_0000554 promotes ovarian cancer invasion and proliferation by regulating miR-567. Environ. Sci. Pollut. Res. Int. 2022, 29, 19072–19080. [Google Scholar] [CrossRef]
- Luo, N.; Sulaiman, Z.; Wang, C.; Ding, J.; Chen, Y.; Liu, B.; Cheng, Z.; Liu, S. Hsa_circ_0000497 and hsa_circ_0000918 contributed to peritoneal metastasis of ovarian cancer via ascites. J. Transl. Med. 2022, 20, 201. [Google Scholar] [CrossRef]
- Xie, J.; Wang, S.; Li, G.; Zhao, X.; Jiang, F.; Liu, J.; Tan, W. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J. Cell Mol. Med. 2019, 23, 3597–3602. [Google Scholar] [CrossRef]
- Liu, J.; Yu, F.; Wang, S.; Zhao, X.; Jiang, F.; Xie, J.; Deng, M. circGFRA1 Promotes Ovarian Cancer Progression By Sponging miR-449a. J. Cancer 2019, 10, 3908–3913. [Google Scholar] [CrossRef]
- Sun, Y.; Li, X.; Chen, A.; Shi, W.; Wang, L.; Yi, R.; Qiu, J. circPIP5K1A serves as a competitive endogenous RNA contributing to ovarian cancer progression via regulation of mi.iR-661/IGFBP5 signaling. J. Cell Biochem. 2019, 120, 19406–19414. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, H.; Li, P. Upregulation of hsa_circRNA_102958 Indicates Poor Prognosis and Promotes Ovarian Cancer Progression Through miR-1205/SH2D3A Axis. Cancer Manag. Res. 2020, 12, 4045–4053. [Google Scholar] [CrossRef]
- Li, B.; Zhang, L. CircSETDB1 knockdown inhibits the malignant progression of serous ovarian cancer through miR-129-3p-dependent regulation of MAP3K3. J. Ovar. Res. 2021, 14, 160. [Google Scholar] [CrossRef]
- Zhang, C.; Li, Y.; Zhao, W.; Liu, G.; Yang, Q. Circ-PGAM1 promotes malignant progression of epithelial ovarian cancer through regulation of the miR-542-3p/CDC5L/PEAK1 pathway. Cancer Med. 2020, 9, 3500–3521. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, J.; Lu, C. Circular RNA Foxo3 enhances progression of ovarian carcinoma cells. Aging (Albany NY) 2021, 13, 22432–22443. [Google Scholar] [CrossRef]
- Ma, R.; Ye, X.; Cheng, H.; Cui, H.; Chang, X. Tumor-derived exosomal circRNA051239 promotes proliferation and migration of epithelial ovarian cancer. Am. J. Transl. Res. 2021, 13, 1125–1139. [Google Scholar] [PubMed]
- Zong, Z.H.; Du, Y.P.; Guan, X.; Chen, S.; Zhao, Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J. Exp. Clin. Cancer Res. 2019, 38, 437. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Liu, X.; Zhang, S.; Chen, S.; Guan, X.; Li, Q.; Chen, X.; Zhao, Y. CircCRIM1 promotes ovarian cancer progression by working as ceRNAs of CRIM1 and targeting miR-383-5p/ZEB2 axis. Reprod. Biol. Endocrinol. 2021, 19, 176. [Google Scholar] [CrossRef]
- Lu, H.; Zheng, G.; Gao, X.; Chen, C.; Zhou, M.; Zhang, L. Propofol suppresses cell viability, cell cycle progression and motility and induces cell apoptosis of ovarian cancer cells through suppressing MEK/ERK signaling via targeting circVPS13C/miR-145 axis. J. Ovar. Res. 2021, 14, 30. [Google Scholar] [CrossRef]
- Cai, W.; Zhang, Q. The transcription factor ZEB1 mediates the progression of epithelial ovarian cancer by promoting the transcription of CircANKRD17. J. Biochem. Mol. Toxicol. 2022, 36, e23086. [Google Scholar] [CrossRef]
- Du, Z.; Wang, L.; Xia, Y. Circ_0015756 promotes the progression of ovarian cancer by regulating miR-942-5p/CUL4B pathway. Cancer Cell Int. 2020, 20, 572. [Google Scholar] [CrossRef]
- Sheng, S.; Hu, Y.; Yu, F.; Tong, W.; Wang, S.; Cai, Y.; Zhu, J. circKIF4A sponges miR-127 to promote ovarian cancer progression. Aging (Albany NY) 2020, 12, 17921–17929. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, W.; Fang, J.; Xie, P.; Miao, M.; Yang, H. Circular RNA circEXOC6B Inhibits the Progression of Ovarian Cancer by Sponging miR-421 and Regulating RUS1 Expression. Oncol. Targets Ther. 2020, 13, 8233–8243. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Fang, H. Curcumin inhibits ovarian cancer progression by regulating circ-PLEKHM3/miR-320a/SMG1 axis. J. Ovar. Res. 2021, 14, 158. [Google Scholar] [CrossRef] [PubMed]
- Yalan, S.; Yanfang, L.; He, C.; Yujie, T. Circular RNA circRHOBTB3 inhibits ovarian cancer progression through PI3K/AKT signaling pathway. Panminerva Med. 2020. [Google Scholar] [CrossRef]
- Lyu, M.; Li, X.; Shen, Y.; Lu, J.; Zhang, L.; Zhong, S.; Wang, J. CircATRNL1 and circZNF608 Inhibit Ovarian Cancer by Sequestering miR-152-5p and Encoding Protein. Front. Genet. 2022, 13, 784089. [Google Scholar] [CrossRef]
- Hu, J.; Wang, L.; Chen, J.; Gao, H.; Zhao, W.; Huang, Y.; Jiang, T.; Zhou, J.; Chen, Y. The circular RNA circ-ITCH suppresses ovarian carcinoma progression through targeting miR-145/RASA1 signaling. Biochem. Biophys. Res. Commun. 2018, 505, 222–228. [Google Scholar] [CrossRef]
- Luo, L.; Gao, Y.Q.; Sun, X.F. Circular RNA ITCH suppresses proliferation and promotes apoptosis in human epithelial ovarian cancer cells by sponging miR-10a-alpha. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8119–8126. [Google Scholar] [CrossRef]
- Yan, H.; Xiang, H.; Sun, B.; Feng, F.; Chen, P. Circular RNA-ITCH Inhibits the Proliferation of Ovarian Carcinoma by Downregulating lncRNA HULC. Reprod. Sci. 2020, 27, 375–379. [Google Scholar] [CrossRef]
- Zhang, N.; Jin, Y.; Hu, Q.; Cheng, S.; Wang, C.; Yang, Z.; Wang, Y. Circular RNA hsa_circ_0078607 suppresses ovarian cancer progression by regulating miR-518a-5p/Fas signaling pathway. J. Ovar. Res. 2020, 13, 64. [Google Scholar] [CrossRef]
- Lin, W.; Ye, H.; You, K.; Chen, L. Up-regulation of circ_LARP4 suppresses cell proliferation and migration in ovarian cancer by regulating miR-513b-5p/LARP4 axis. Cancer Cell Int. 2020, 20, 5. [Google Scholar] [CrossRef]
- Gong, J.; Xu, X.; Zhang, X.; Zhou, Y. Circular RNA-9119 suppresses in ovarian cancer cell viability via targeting the microRNA-21-5p-PTEN-Akt pathway. Aging (Albany NY) 2020, 12, 14314–14328. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Cao, Q.X.; Tian, J.; Ren, L.; Cheng, H.L.; Yang, S.Q. Circular RNA MTO1 Inhibits the Proliferation and Invasion of Ovarian Cancer Cells Through the miR-182-5p/KLF15 Axis. Cell Transplant. 2020, 29, 963689720943613. [Google Scholar] [CrossRef]
- Li, L.; Yu, P.; Zhang, P.; Wu, H.; Chen, Q.; Li, S.; Wang, Y. Upregulation of hsa_circ_0007874 suppresses the progression of ovarian cancer by regulating the miR-760/SOCS3 pathway. Cancer Med. 2020, 9, 2491–2499. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.; Xu, J.; Zhang, M.; Liu, S.; Gu, Y.; Zhang, M.; Wang, X.; Ni, J.; Qian, B.; Shen, R.; et al. Comprehensive circular RNA expression profiles and the tumor-suppressive function of circHIPK3 in ovarian cancer. Int. J. Biochem. Cell Biol. 2019, 112, 8–17. [Google Scholar] [CrossRef]
- Li, X.; Jiang, X.; Lu, J.; Lin, Y.; Jiang, L.; Li, Y.; Wan, F.; Wang, C. CircCERS6 Suppresses the Development of Epithelial Ovarian Cancer Through Mediating miR-630/RASSF8. Biochem. Genet. 2022, 60, 2611–2629. [Google Scholar] [CrossRef] [PubMed]
- Ford, C.E.; Werner, B.; Hacker, N.F.; Warton, K. The untapped potential of ascites in ovarian cancer research and treatment. Br. J. Cancer 2020, 123, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.H.L.; Yue, C.; Du, K.Y.; Salem, M.; O’Brien, J.; Peng, C. The Role of microRNAs in Epithelial Ovarian Cancer Metastasis. Int. J. Mol. Sci. 2020, 21, 7093. [Google Scholar] [CrossRef]
- Rickard, B.P.; Conrad, C.; Sorrin, A.J.; Ruhi, M.K.; Reader, J.C.; Huang, S.A.; Franco, W.; Scarcelli, G.; Polacheck, W.J.; Roque, D.M.; et al. Malignant Ascites in Ovarian Cancer: Cellular, Acellular, and Biophysical Determinants of Molecular Characteristics and Therapy Response. Cancers 2021, 13, 4318. [Google Scholar] [CrossRef]
- Yousefi, M.; Dehghani, S.; Nosrati, R.; Ghanei, M.; Salmaninejad, A.; Rajaie, S.; Hasanzadeh, M.; Pasdar, A. Current insights into the metastasis of epithelial ovarian cancer—Hopes and hurdles. Cell Oncol. 2020, 43, 515–538. [Google Scholar] [CrossRef]
- Pascual-Anton, L.; Cardenes, B.; Sainz de la Cuesta, R.; Gonzalez-Cortijo, L.; Lopez-Cabrera, M.; Cabanas, C.; Sandoval, P. Mesothelial-to-Mesenchymal Transition and Exosomes in Peritoneal Metastasis of Ovarian Cancer. Int. J. Mol. Sci. 2021, 22, 11496. [Google Scholar] [CrossRef]
- Majidpoor, J.; Mortezaee, K. Angiogenesis as a hallmark of solid tumors—Clinical perspectives. Cell Oncol. 2021, 44, 715–737. [Google Scholar] [CrossRef] [PubMed]
- Zada, S.; Hwang, J.S.; Ahmed, M.; Lai, T.H.; Pham, T.M.; Elashkar, O.; Kim, D.R. Cross talk between autophagy and oncogenic signaling pathways and implications for cancer therapy. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188565. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.W.; Lee, S.H. The Roles of Autophagy in Cancer. Int. J. Mol. Sci. 2018, 19, 3466. [Google Scholar] [CrossRef] [PubMed]
- White, E.; DiPaola, R.S. The double-edged sword of autophagy modulation in cancer. Clin. Cancer Res. 2009, 15, 5308–5316. [Google Scholar] [CrossRef] [PubMed]
- Das, C.K.; Mandal, M.; Kogel, D. Pro-survival autophagy and cancer cell resistance to therapy. Cancer Metastasis Rev. 2018, 37, 749–766. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, S.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Liang, X.; Lv, X.; Cheng, Y.; Du, J.; Liu, C.; Yang, Y. New insights into the role of circular RNAs in ovarian cancer. Pathol. Res. Pract. 2022, 238, 154073. [Google Scholar] [CrossRef]
- Chong, X.; Chen, J.; Zheng, N.; Zhou, Z.; Hai, Y.; Chen, S.; Zhang, Y.; Yu, Q.; Yu, S.; Chen, Z.; et al. PIK3CA mutations-mediated downregulation of circLHFPL2 inhibits colorectal cancer progression via upregulating PTEN. Mol. Cancer 2022, 21, 118. [Google Scholar] [CrossRef]
- Zhou, H.; Du, Y.; Wei, X.; Song, C.; Song, J.; Xu, N.; Huang, W.; Chen, L.; Yao, F.; Du, D.; et al. DDX56 transcriptionally activates MIST1 to facilitate tumorigenesis of HCC through PTEN-AKT signaling. Theranostics 2022, 12, 6069–6087. [Google Scholar] [CrossRef]
- Bi, X.; Lv, X.; Liu, D.; Guo, H.; Yao, G.; Wang, L.; Liang, X.; Yang, Y. METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. 2021, 28, 335–349. [Google Scholar] [CrossRef]
- Zhu, X.; Shen, H.; Yin, X.; Yang, M.; Wei, H.; Chen, Q.; Feng, F.; Liu, Y.; Xu, W.; Li, Y. Macrophages derived exosomes deliver miR-223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J. Exp. Clin. Cancer Res. 2019, 38, 81. [Google Scholar] [CrossRef] [PubMed]
- Martins, F.C.; Santiago, I.; Trinh, A.; Xian, J.; Guo, A.; Sayal, K.; Jimenez-Linan, M.; Deen, S.; Driver, K.; Mack, M.; et al. Combined image and genomic analysis of high-grade serous ovarian cancer reveals PTEN loss as a common driver event and prognostic classifier. Genome Biol. 2014, 15, 526. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.G.; McMeekin, D.S.; Brown, A.K.; DiSilvestro, P.; Miller, M.C.; Allard, W.J.; Gajewski, W.; Kurman, R.; Bast, R.C., Jr.; Skates, S.J. A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol. Oncol. 2009, 112, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.G.; Miller, M.C.; Disilvestro, P.; Landrum, L.M.; Gajewski, W.; Ball, J.J.; Skates, S.J. Evaluation of the diagnostic accuracy of the risk of ovarian malignancy algorithm in women with a pelvic mass. Obstet. Gynecol. 2011, 118, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.G.; Brown, A.K.; Miller, M.C.; Skates, S.; Allard, W.J.; Verch, T.; Steinhoff, M.; Messerlian, G.; DiSilvestro, P.; Granai, C.O.; et al. The use of multiple novel tumor biomarkers for the detection of ovarian carcinoma in patients with a pelvic mass. Gynecol. Oncol. 2008, 108, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Dahl, M.; Daugaard, I.; Andersen, M.S.; Hansen, T.B.; Gronbaek, K.; Kjems, J.; Kristensen, L.S. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab. Invest. 2018, 98, 1657–1669. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S. Profiling of circRNAs using an enzyme-free digital counting method. Methods 2021, 196, 11–16. [Google Scholar] [CrossRef]
- Gnant, M.; Filipits, M.; Greil, R.; Stoeger, H.; Rudas, M.; Bago-Horvath, Z.; Mlineritsch, B.; Kwasny, W.; Knauer, M.; Singer, C.; et al. Predicting distant recurrence in receptor-positive breast cancer patients with limited clinicopathological risk: Using the PAM50 Risk of Recurrence score in 1478 postmenopausal patients of the ABCSG-8 trial treated with adjuvant endocrine therapy alone. Ann. Oncol. 2014, 25, 339–345. [Google Scholar] [CrossRef]
- Dowsett, M.; Sestak, I.; Lopez-Knowles, E.; Sidhu, K.; Dunbier, A.K.; Cowens, J.W.; Ferree, S.; Storhoff, J.; Schaper, C.; Cuzick, J. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J. Clin. Oncol. 2013, 31, 2783–2790. [Google Scholar] [CrossRef]
- Ye, F.; Chen, C.; Qin, J.; Liu, J.; Zheng, C. Genetic profiling reveals an alarming rate of cross-contamination among human cell lines used in China. FASEB J. 2015, 29, 4268–4272. [Google Scholar] [CrossRef]
- Bian, X.; Yang, Z.; Feng, H.; Sun, H.; Liu, Y. A Combination of Species Identification and STR Profiling Identifies Cross-contaminated Cells from 482 Human Tumor Cell Lines. Sci. Rep. 2017, 7, 9774. [Google Scholar] [CrossRef]
- Furlong, M.T.; Hough, C.D.; Sherman-Baust, C.A.; Pizer, E.S.; Morin, P.J. Evidence for the colonic origin of ovarian cancer cell line SW626. J. Natl. Cancer Inst. 1999, 91, 1327–1328. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nielsen, A.F.; Bindereif, A.; Bozzoni, I.; Hanan, M.; Hansen, T.B.; Irimia, M.; Kadener, S.; Kristensen, L.S.; Legnini, I.; Morlando, M.; et al. Best practice standards for circular RNA research. Nat. Methods 2022, 19, 1208–1220. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, X.; Xue, W.; Zhang, L.; Yang, L.Z.; Cao, S.M.; Lei, Y.N.; Liu, C.X.; Guo, S.K.; Shan, L.; et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat. Methods 2021, 18, 51–59. [Google Scholar] [CrossRef]
- Zhang, Y.; Nguyen, T.M.; Zhang, X.O.; Wang, L.; Phan, T.; Clohessy, J.G.; Pandolfi, P.P. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs. Genome Biol. 2021, 22, 41. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Wang, Y.; He, L.; Zhang, J.; Zhu, X.; Liu, N.; Wang, J.; Lu, T.; He, L.; Tian, Y.; et al. Circular RNA circIPO11 drives self-renewal of liver cancer initiating cells via Hedgehog signaling. Mol. Cancer 2021, 20, 132. [Google Scholar] [CrossRef]
- Han, K.; Wang, F.W.; Cao, C.H.; Ling, H.; Chen, J.W.; Chen, R.X.; Feng, Z.H.; Luo, J.; Jin, X.H.; Duan, J.L.; et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol. Cancer 2020, 19, 60. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Li, X.; Awan, F.M.; Yang, Z.; Fang, L.; Lyu, J.; Li, F.; Peng, C.; Krylov, S.N.; et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene 2018, 37, 5829–5842. [Google Scholar] [CrossRef]
- Lu, M.; Wu, Y.; Zeng, B.; Sun, J.; Li, Y.; Luo, J.; Wang, L.; Yi, Z.; Li, H.; Ren, G. CircEHMT1 inhibits metastatic potential of breast cancer cells by modulating miR-1233-3p/KLF4/MMP2 axis. Biochem. Biophys. Res. Commun. 2020, 526, 306–313. [Google Scholar] [CrossRef]
| circRNA (circBase ID) | Level in OC | Detection Methods, Cohort Size/Subtype | Sample Source | Clinicopathologic Features | Prognosis | Diagnosis | Refs |
|---|---|---|---|---|---|---|---|
| circCDR1as (ciRS-7, circ_0001946) | Up | qRT-PCR: OC vs. adjacent normal tissues (n = 40) | Tissues | TNM stage, metastasis | OS | - | [69] |
| Down | qRT-PCR: OC vs. adjacent normal tissues (n = 6) | Tissues | - | - | - | [70] | |
| Down | Microarray: DDP-resistant vs. DDP-sensitive SOC (n = 5). qRT-PCR: DDP-resistant SOC (tissues and serum exosome, n = 36), DDP-sensitive SOC (tissues and serum exosome, n = 30), adjacent normal tissues (n = 66). | Tissues, serum | DDP resistance | - | - | [71] | |
| Down | qRT-PCR: OC (n = 65), normal ovarian epithelial tissues (n = 37) | Tissues | - | - | - | [72] | |
| circHIPK3 * | Up | qRT-PCR: EOC vs. adjacent non-cancerous tissues (n = 69) | Tissues | FIGO stage, metastasis | OS, DFS | - | [73] |
| circCEACAM5 (circ_0051240) | Up | Microarray: OC vs. adjacent normal tissues (n = 10) qRT-PCR: OC vs. adjacent non-cancerous tissues (n = 33) | Tissues | FIGO stage, metastasis | OS | - | [74] |
| circCSPP1 (circ_0001806) | Up | qRT-PCR: EOC (n = 117), borderline tumors (n = 12), normal ovarian tissues (n = 15), benign ovarian tumors (n = 12) | Tissues | FIGO stage, differentiation | - | - | [75] |
| circNOLC1 (circ_0000257) | Up | qRT-PCR: EOC (86 serous, 32 others), normal ovarian tissues (n = 15), benign ovarian tumors (n = 11), borderline ovarian tumors (n = 11) | Tissues | FIGO stage, differentiation, serum CA125 level | - | - | [76] |
| circPUM1 (circ_0000043) | Up | qRT-PCR: EOC (n = 62), normal ovarian tissues (n = 13) | Tissues | FIGO stage | - | - | [77] |
| circRHOC (circ_0013549) | Up | qRT-PCR: OC (100 serous carcinomas, 27 others), normal ovarian tissues (n = 24) | Tissues | FIGO stage, differentiation | - | - | [78] |
| circCELSR1 (circ_0063804) | Up | qRT-PCR: OC vs. adjacent normal tissues (n = 108) | Tissues | FIGO stage, tumor grade, tumor size | OS | - | [79] |
| circSPECC1 (circ_0000745) | Up | qRT-PCR: OC (35 EOC, 9 germ cell tumors, 6 others), para-cancerous tissues (n = 50) | Tissues | FIGO stage, tumor grade | - | - | [80] |
| circHIPK2 (circ_0001756) | Up | qRT-PCR: OC (26 DDP-sensitive, 20 DDP-resistant), adjacent normal tissues (n = 46) | Tissues | FIGO stage, tumor size, metastasis, DDP resistance | - | - | [81] |
| circMFN2 (circ_0009910) | Up | qRT-PCR: OC (n = 50), normal ovarian tissues (n = 50) | Tissues | FIGO stage, metastasis | OS | - | [82] |
| circMYLK * | Up | qRT-PCR: OC vs. adjacent normal tissues (n = 46) | Tissues | TNM stage | OS | - | [83] |
| circMUC16 (circ_0049116) | Up | RNA-seq: EOC (n = 3), healthy ovarian tissues (n = 4). qRT-PCR: EOC (n = 70), normal ovarian tissues (n = 30); serum of EOC patients (n = 70), serum of healthy subjects (n = 30) | Tissues, serum | FIGO stage, tumor grade | - | - | [84] |
| circFAM53B (circ_0000267) | Up | qRT-PCR: EOC (36 serous, 18 mucinous), paired non-cancerous tissues (n = 54) | Tissues | FIGO stage, tumor size, metastasis | OS | - | [85] |
| circASH2L (circ_0006302) | Up | qRT-PCR: OC vs. adjacent normal tissues (n = 50) | Tissues | FIGO stage, tumor size, metastasis | - | - | [86] |
| circACP6 (circ_0013958) | Up | qRT-PCR: serous ovarian carcinomas (SOC, n = 45), normal ovarian tissues (n = 45) | Tissues | FIGO stage, metastasis | - | AUC = 0.912 | [87] |
| circABCB10 * | Up | qRT-PCR: EOC (67 serous, 36 others), adjacent tissues (n = 53) | Tissues | FIGO stage, differentiation, tumor size | OS | AUC = 0.766 | [88] |
| circTNPO3 (circ_0001741) | Up | Microarray: PTX-sensitive vs. PTX-resistant OC (n = 3) qRT-PCR: OC (20 PTX-sensitive, 28 PTX-resistant), adjacent normal tissues (n = 48) | Tissues | FIGO stage, PTX resistance | OS | AUC = 0.910 | [89] |
| circANKRD17 (circ_0007883) | Up | RNA-seq: EOC (n = 3), healthy ovarian tissues (n = 4) qRT-PCR: EOC (28 serous, 20 mucinous; 28 PTX resistant, 20 PTX sensitive), adjacent normal tissues (n = 48) | Tissues | PTX resistance | OS | - | [90] |
| circEEF2 (circ_0048559) | Up | FISH: EOC (40 serous, 9 mucinous, 8 clear cell), normal ovarian tissues (n = 12) | Tissues | FIGO stage, tumor grade | - | - | [91] |
| circRNF144B (circ_0075797) | Up | RNA-seq: OC tissues with high and low LC3 dots (n =3) qRT-PCR: OC vs. adjacent non-cancerous tissues (n = 36) | Tissues | Metastasis | OS | - | [92] |
| circSLAMF6 (circ_0000144) | Up | qRT-PCR: OC tissues vs. adjacent normal tissues (n = 60); serum of OC patients vs. serum of healthy (n = 60) | Tissues, serum | - | OS, DFS | - | [93] |
| circPDE7B (circ_0004712) | Up | qRT-PCR: EOC (20 serous, 10 mucinous) vs. adjacent non-cancerous tissues (n = 30) | Tissues | Tumor grade, metastasis | OS, PFS | - | [94] |
| circC20orf11 (circ_0061140) | Up | qRT-PCR: OC vs. adjacent normal tissues (n = 55) | Tissues | TNM stage, metastasis, tumor size | - | - | [95] |
| circC20orf11 * | Up | qRT-PCR: OC (47 serous, 13 others; 26 DDP-sensitive, 34 DDP-resistant) | Serum | FIGO stage, tumor grade, metastasis, DDP resistance | OS | - | [96] |
| circITGB6 (circ_0056856) | Up | RNA-seq: DDP-sensitive vs. resistant OC tissues (n = 5) qRT-PCR: Serum of OC (20 DDP-resistant, 20 DDP-sensitive), serum of normal controls (n = 15); OC tissues (20 DDP-resistant, 20 DDP-sensitive). ISH: EOC tissues (95 SOC, 18 MC, 3 EC, 2 CCC, 1 other) | Serum, tissue | FIGO stage, tumor recurrence, DDP resistance | OS, RFS | - | [97] |
| circSETDB1 (circ_0006352) | Up | qRT-PCR: SOC (18 primary chemoresistance, 42 primary chemosensitive), healthy (n = 60). | Serum | FIGO stage, metastasis, chemosensitivity | PFS | SOC vs. healthy: AUC = 0.803 Chemoresistant vs. chemosensitive SOC: AUC = 0.811 | [98] |
| circFOXP1 (circ_0001320) | Up | qRT-PCR: EOC (46 DDP-resistant, 76 DDP-sensitive), healthy (n = 82) | Serum | FIGO stage, metastasis, tumor size, DDP resistance | OS, DFS | AUC = 0.914 | [99] |
| circMGAT5 (circ_0001068) | Up | Microarray: OC vs. healthy (n = 4). qRT-PCR: (1) training set: OC (n = 10), healthy (n = 10); (2) larger cohort: OC (n = 85), healthy (n = 43). | Serum | - | - | AUC = 0.970 | [100] |
| circITCH * | Down | qRT-PCR: OC vs. adjacent normal tissues (n = 45) | Tissues | FIGO stage, tumor size | OS | - | [101] |
| circITCH * | Down | qRT-PCR: EOC (48 serous, 29 others) vs. adjacent normal tissues (n = 77) | Tissues | FIGO stage, tumor size | OS | - | [102] |
| circRNA_100395 (circ_0015278) # | Down | qRT-PCR: OC vs. adjacent normal tissues (n = 60) | Tissues | FIGO stage, metastasis | OS | - | [103] |
| circLARP4 * | Down | qRT-PCR: EOC vs. adjacent normal tissues (n = 78) | Tissues | FIGO stage, metastasis. | OS, DFS | - | [104] |
| circEXOC6B (circ_0009043) | Down | qRT-PCR: OC vs. adjacent normal tissues (n = 60) | Tissues | TNM stage, metastasis | OS | - | [105] |
| circEXOC6B (circ_0009043) | Down | RNA-seq: EOC vs. normal ovarian tissues (n = 4) qRT-PCR: EOC vs. normal ovarian tissues (n = 54) | Tissues | Metastasis | OS | - | [106] |
| circATRNL1 (circ_0020093) | Down | qRT-PCR: OC (30 serous, 26 others), adjacent non-cancerous tissues (n = 56) | Tissues | FIGO stage, metastasis | OS | - | [107] |
| circSLC22A3 (circ_0078607) | Down | qRT-PCR: OC vs. adjacent normal tissues (n = 43) | Tissues | FIGO stage, metastasis | - | - | [108] |
| qRT-PCR: HGSC vs. adjacent non-cancerous tissues (n = 49) | FIGO stage, serum CA125 level | OS, PFS | [109] | ||||
| circRHOBTB3 (circ_0007444) | Down | qRT-PCR: EOC (n = 40), normal ovarian tissues (n = 20) | Tissues | FIGO stage, metastasis | OS | - | [110] |
| qRT-PCR: OC vs. adjacent normal tissues (n = 87) | FIGO stage, grade, tumor size | OS | [111] | ||||
| circRNA1656 (circ_0002755) | Down | RNA-seq: HGSC (n = 3), benign ovarian diseases (n = 3). qRT-PCR: HGSC (n = 60), ovarian benign diseases (n = 60) | Tissues | FIGO stage | - | - | [112] |
| circSMARCA5 (circ_0001445) | Down | qRT-PCR: OC (36 serous, 14 others), normal ovarian tissues | Tissues | FIGO stage, metastasis | OS | - | [113] |
| circFBXO7 (circ_0001222) | Down | RNA-seq: EOC (n = 27), normal tissues(n = 26) qRT-PCR: OC vs. normal tissues (n = 12) BaseScope assay: EOC (n = 77) | Tissues | - | OS, RFS | - | [114] |
| circPLEKHM3 (circ_0001095) | Down | RNA-seq: OC vs. normal ovarian tissues (n = 5) qRT-PCR: OC vs. normal ovarian tissues (n = 12); primary OC vs. matched peritoneal metastatic OC (n = 26) BaseScope assay: OC (n = 86) | Tissues | Metastasis | OS, RFS | - | [115] |
| circBNC2 (circ_0008732) | Down | qRT-PCR: EOC vs. benign ovarian cysts vs. healthy (n = 83) | Plasma | Tumor grade, subtype, metastasis | - | EOC vs. benign: AUC = 0.879 EOC vs. healthy: AUC = 0.923 Early stage EOC vs. benign: AUC = 0.864 Early stage EOC vs. healthy: AUC = 0.908 | [36] |
| qRT-PCR: OC vs. adjacent tissues (n = 40) | Tissues | FIGO stage, metastasis | OS | - | [116] | ||
| circN4BP2L2 * | Down | qRT-PCR: Stage I + II EOC (n = 36), stage III + IV EOC (n = 90), benign ovarian cysts (n = 126), healthy (n = 126) | Plasma | FIGO stage, tumor grade, metastasis | - | EOC vs. benign: AUC = 0.82 EOC vs. normal: AUC = 0.90 Early-stage EOC vs. benign: AUC = 0.81 Early stage EOC vs. normal: AUC = 0.90 | [43] |
| circN4BP2L2 (circ_0000471) | Down | RNA-seq: EOC vs. normal ovarian tissues (n = 4) qRT-PCR: EOC vs. normal ovarian tissues (n = 54) | Tissues | FIGO stage, metastasis | PFS | - | [106] |
| circFAM120A (circ_0003972) circTOM1L1 (circ_0007288) | Down | Microarray: OC (n = 4), vs. uterine myoma patients (n = 4) qRT-PCR: OC (n = 60: 48 EOC) vs. benign tumors (n = 60) | Plasma | Metastasis (circ_0007288) | - | circ_0003972: AUC = 0.724 circ_0007288: AUC = 0.790 | [117] |
| circRNA (circBase ID) | Experimental Models | Functions | Molecular Mechanisms | Refs |
|---|---|---|---|---|
| circC20orf11 (circ_0061140) | SKOV3/PTX, HEYA8/PTX Female BALB/c mice s.c. injected with SKOV3/PTX | In vitro: ↑ proliferation/migration/invasion/PTX resistance; ↓ apoptosis In vivo: ↑ tumor growth/PTX resistance | miR-136/CBX2 | [125] |
| SKOV3, A2780 Male BALB/c mice s.c. injected with SKOV3 | In vitro: ↑ proliferation/migration/EMT In vivo: ↑ tumor growth | miR-370/FOXM1 | [126] | |
| SKOV3, A2780 BALB/c mice s.c. injected with SKOV3 | In vitro: ↑ proliferation/migration/invasion; ↑ angiogenesis; ↓ apoptosis In vivo: ↑ tumor growth | miR-761/LETM1 | [95] | |
| SKOV3, CAOV3 BALB/c female mice s.c. injected with SKOV3 | In vitro: ↑ proliferation/migration/invasion/EMT; ↑ angiogenesis; ↓ apoptosis In vivo: ↑ tumor growth | miR-361-5p/RAB1A | [127] | |
| circC20orf11 * | SKOV3, A2780, SKOV3/DDP, A2780/DDP Male BALB/c mice s.c. injected with A2780/DDP and SKOV3/DDP | In vitro: ↑ proliferation/DDP resistance; ↓ apoptosis In vivo: ↑ tumor DDP resistance | miR-527/YWHAZ | [96] |
| circCELSR1 * | SKOV3/PTX, A2780/PTX BALB/c mice inoculated with SKOV3/PTX | In vitro: ↑ PTX resistance/viability/proliferation; ↓ apoptosis In vivo: ↑ tumor PTX resistance | miR-149-5p/SIK2 | [128] |
| circCELSR1 (circ_0063809) | SKOV3, HEYA8, SKOV3/PTX, HEYA8/PTX Female BALB/c mice s.c. injected with SKOV3/PTX | In vitro: ↑ PTX resistance In vivo: ↑ tumor PTX resistance | miR-1252/FOXR2 | [129] |
| SKOV3, A2780 Athymic female nude mice i.p. injected with SKOV3 | In vitro: ↑ proliferation/migration/invasion/EMT; ↓ apoptosis In vivo: ↑ tumor growth/metastasis | miR-598/BRD4 | [130] | |
| circCELSR1 (circ_0063804) | OVCAR3, SKOV3 Nude mice s.c. injected with OVCAR3 | In vitro: ↑ proliferation/DDP resistance; ↓ apoptosis. In vivo: ↑ tumor growth/DDP resistance | miR-1276/CLU | [79] |
| circFOXM1 (circ_0025033) | SKOV3/PTX, HEYA8/PTX | In vitro: ↑ PTX resistance/migration/invasion; ↓ apoptosis | miR-532-3p/FOXM1 | [131] |
| SKOV3, A2780 Female BALB/c mice s.c. injected with SKOV3 | In vitro: ↑ proliferation/migration/invasion/glycolysis. In vivo: ↑ tumor growth | miR-184/LSM4 | [132] | |
| circSEC61A1 (circ_0007841) | A2780/DDP, SKOV3/DDP Female BALB/c mice s.c. injected with SKOV3/DDP | In vitro: ↑viability/proliferation/invasion/migration/DDP resistance; ↓ apoptosis. In vivo: ↑ tumor growth/DDP resistance | miR-532-5p/NFIB | [133] |
| SKOV3, OVCAR3 BALB/c mice s.c. injected with SKOV3 | In vitro: ↑ proliferation/migration/invasion In vivo: ↑ tumor growth | miR-151-3p/MEX3C | [134] | |
| circMUC16 (circ_0049116) | SKOV3, A2780 BALB/c mice i.p. injected with SKOV3 | In vitro: ↑ autophagy/proliferation/invasion In vivo: ↑ tumor metastasis | miR-199a-5p/Beclin1 & RUNX1; ATG13 protein | [84] |
| A2780, SKOV3 Female BALB/c mice s.c. injected with A2780 | In vitro: ↓ anti-tumor effects of Propofol In vivo: ↑ tumor growth, ↓ anti-tumor effects of Propofol | miR-1182/S100B | [135] | |
| circABCB10 * | SKOV3, UWB1.289 | In vitro: ↑ proliferation/invasion; ↓ apoptosis | miR-1271/Wnt/β-catenin | [136] |
| In vitro: ↑ proliferation; ↓ apoptosis. | miR-1271, miR-1252, miR-203 | [88] | ||
| circUBAP2 * | OVCAR3, HO8910 | In vitro: ↑ proliferation/migration | miRNA-144/CHD2 | [137] |
| circUBAP2 * | OVCAR3, ES-2 | In vitro: ↑ proliferation; ↓ apoptosis. | miR-382-5p/PRPF8 | [138] |
| circPVT1 * | SKOV3, A2780 | In vitro: ↑ proliferation/migration/invasion | miR-149-5p/FOXM1 | [139] |
| circPVT1 * | CAOV3, SKOV3 | In vitro: ↑ proliferation; ↓ apoptosis. | miR-149 | [140] |
| circRGNEF (circ_0072995) | A2780, HO8910 BALB/c mice s.c. injected with HO8910 | In vitro: ↑ proliferation/migration; ↓ apoptosis. In vivo: ↑ tumor growth | miR-147a/CDK6 | [141] |
| OVCAR3, SKOV3 Female BALB/c mice s.c. injected with OVCAR3 | In vitro: ↑ proliferation/migration/invasion; ↓ apoptosis. In vivo: ↑ tumor growth | miR-122-5p/SLC1A5 | [142] | |
| circACP6 (circ_0013958) | SKOV3, CAOV3 Female BALB/c mice s.c. injected with SKOV3 | In vitro: ↑ proliferation/migration/invasion; ↓ apoptosis. In vivo: ↑ tumor growth | miR-637/PLXNB2 | [143] |
| A2780, OVCAR3 | In vitro: ↑ proliferation/migration/invasion/EMT; ↓ apoptosis | [87] | ||
| circNRIP1 (circ_0002711) | A2780/PTX, SKOV3/PTX BALB/c mice s.c. injected with SKOV3/PTX | In vitro: ↑ proliferation/migration/invasion/PTX resistance; ↓ apoptosis. In vivo: ↑ tumor PTX resistance | miR-211-5p/HOXC8 | [144] |
| SKOV3, OV90 Nude mice s.c. injected with SKOV3 | In vitro: ↑ viability/proliferation/glycolysis In vivo: ↑ tumor growth | miR-1244/ROCK1 | [145] | |
| circAARS (circ_0000714) | A2780/PTX | In vitro: ↑ PTX resistance/proliferation/cell cycle progression | miR-370-3p/RAB17 | [146] |
| circATL2 (circ_0000993) | HEYA8/PTX, SKOV3/PTX BALB/c mice s.c. infected with SKOV3/PTX | In vitro: ↑ PTX resistance/proliferation; ↓ cell cycle arrest/apoptosis. In vivo: ↑ tumor PTX resistance | miR-506-3p/NFIB | [147] |
| circTNPO3 (circ_0001741) | SKOV3, HEYA8, SKOV3/PTX, HEYA8/PTX Nude mice s.c. injected with SKOV3/PTX | In vitro: ↑ PTX resistance/cell cycle progression; ↓ apoptosis. In vivo: ↑ tumor PTX resistance | miR-1299/NEK2 | [89] |
| circLPAR3 (circ_0004390) | SKOV3/DDP, A2780/DDP BALB/c mice s.c. injected with SKOV3/DDP | In vitro: ↑ DDP resistance resistances/proliferation/migration/invasion; ↓ apoptosis. In vivo: ↑ tumor growth/DDP resistance | miR-634/PDK1 | [148] |
| SKOV3, HEYA8, OVCAR429 | In vitro: ↑ proliferation | miR-198/MET | [149] | |
| circHIPK2 (circ_0001756) | SKOV3/DDP, A2780/DDP Female BALB/c mice s.c. injected with A2780/DDP | In vitro: ↑ DDP resistance/proliferation/cell cycle progression/migration/invasion; ↓ apoptosis In vivo: ↑ tumor DDP resistance/growth | miR-338-3p/CHTOP | [81] |
| SKOV3, A2780 Female BALB/c mice i.v. injected with SKOV3 | In vitro: ↑ proliferation/invasion/EMT In vivo: ↑ tumor growth | IGF2BP2/EGFR/MAPK | [150] | |
| circFOXP1 (circ_0001320) | COC1, SKOV3, SKOV3/DDP Nude mice s.c. injected with SKOV3/DDP | In vitro: ↑ proliferation/DDP resistance In vivo: ↑ tumor growth/DDP resistance | miR-22 & miR-150-3p/CEBPG & FMNL3 | [99] |
| circPRKCI (circ_0067934) | A2780/DDP Female BALB/c mice s.c. injected with A2780/DDP | In vitro: ↑ DDP resistance/proliferation/invasion; ↓ apoptosis In vivo: ↑ tumor growth | miR-545-3p/PPA1/JNK | [151] |
| circPBX3 (circ_0004804) | OV90, SKOV3; OV90-Res; SKOV3-Res Female nude mice s.c. injected with OV90 | In vitro: ↑ DDP resistance In vivo: ↑ tumor DDP resistance | IGF2BP2/ATP7A | [152] |
| circZFR* | A2780 | In vitro: ↑ proliferation/migration/invasion/glycolysis; ↓ apoptosis | miR-212-5p/SOD2 | [153] |
| circE2F2 (circ_0000030) | OVCAR3, SKOV3 Female BALB/c mice s.c. injected with OVCAR3 | In vitro: ↑ proliferation/migration/invasion/ glucose. In vivo: ↑ tumor growth | HuR/E2F2. | [154] |
| circPTK2 (circ_0008305) | SKOV3, OVCAR3 Female nude mice i.p. injected with SKOV3 | In vitro: ↑ migration/invasion/EMT; ↑ angiogenesis In vivo: ↑ tumor growth | miR-639/FOXC1 | [155] |
| circASH2L (circ_0006302) | A2780, SKOV3 Female BALB/c mice s.c. injected with SKOV3 | In vitro: ↑ invasion/proliferation; ↑angiogenesis In vivo: ↑ tumor angiogenesis/lymphangiogenesis/growth | miR-665/VEGFA | [86] |
| circITGB6 (circ_0056856) | OVCAR3, CAOV3, ID8 (mice) Female C57BL/6 mice i.p. injected with ID8 | In vitro: ↑ DDP resistance In vivo: ↑ M2 macrophage-dependent DDP resistance of tumor | IGF2BP2/FGF9 | [97] |
| circMGAT5 (circ_0001068) | A2780 | In vitro: ↑ PD1 expression in T cells, ↑ T cell exhaustion | miR-28-5p/PD1 | [100] |
| circSPECC1 (circ_0000745) | ES-2, SKOV3 | In vitro: ↑ proliferation/migration/invasion/EMT/stemness | miR-3187-3p/ERBB4/PI3K/AKT | [80] |
| circTUBA1B (circ_0026123) | SKOV3 Female BALB/c mice s.c. injected with SKOV3 | In vitro: ↑ migration/proliferation/cancer stem cell differentiation In vivo: ↑ tumor growth | miR-124-3p/EZH2 | [156] |
| circRAB11FIP1 (circ_0005630) | SKOV3, A2780 BALB/c mice i.p. injected with SKOV3 | In vitro: ↑ autophagic flux/proliferation/invasion In vivo: ↑ tumor metastasis | miR-129/ATG14 & ATG7; DSC1/ATG101 | [157] |
| circFGFR3 * | SKOV3, A2780 Mice inoculated with SKOV3 | In vitro: ↑ proliferation/migration/invasion/EMT In vivo: ↑ tumor growth/metastasis | miR-29a-3p/E2F1 | [158] |
| circCSPP1 (circ_0001806) | OVCAR3, A2780, CAOV3 | In vitro: ↑ proliferation/migration/invasion/EMT | miR-1236-3p/ZEB1 | [75] |
| circKRT7 (circ_0026360) | ES-2, SKOV3 BALB/c mice s.c. injected with ES-2 | In vitro: ↑ proliferation/migration/invasion/EMT In vivo: ↑ tumor growth | miR-29a-3p/COL1A1 | [159] |
| circLIN52 (circ_0000554) | HO8910 | In vitro: ↑ proliferation/invasion/EMT | miR-567 | [160] |
| circSLAIN1 (circ_0000497) circGMIP (circ_0000918) | SKOV3, OVCAR3 | In vitro: ↑ migration/invasion/EMT | predicted miRNAs/mRNAs | [161] |
| circEPSTI1 (circ_0000479) | OV119, A2780 BALB/c mice injected (s.c. and i.v.) with OV119 | In vitro: ↑ proliferation/invasion; ↓ apoptosis In vivo: ↑ tumor growth/metastasis | miR-942/EPSTI1 | [162] |
| circGFRA1 * | OV119, A2780 BALB/c mice injected (s.c. and i.v.) with OV119 | In vitro: ↑ proliferation/invasion In vivo: ↑ tumor growth/metastasis | miR-449a/GFRA1 | [163] |
| circMFN2 (circ_0009910) | SKOV3 | In vitro: ↑ proliferation/migration/invasion | miR-145/NF-κB & Notch | [82] |
| circCEACAM5 (circ_0051240) | OVCAR3, HO8910 Male nude mice, s.c. injected with OVCAR3 | In vitro: ↑ proliferation/migration/invasion In vivo: ↑ tumorigenesis | miR-637/KLK4 | [74] |
| circPIP5K1A (circ_0014130) | SKOV3, A2780 BALB/c mice s.c. injected with SKOV3 | In vitro: ↑ proliferation/migration/invasion In vivo: ↑ tumor growth | miR-661/IGFBP5 | [164] |
| circRNA_102958 (circ_0003854) # | SKOV3, OVCAR3 | In vitro: ↑ proliferation/migration/invasion | miR-1205/SH2D3A | [165] |
| circFAM53B (circ_0000267) | A2780, HO8910 | In vitro: ↑ proliferation/migration/invasion; ↓ apoptosis | miRNA-646/VAMP2, miRNA-647/MDM2 | [85] |
| circSETDB1 (circ_0006352) | A2780, SKOV3 Female BALB/c mice i.p. injected with SKOV3 | In vitro: ↑ proliferation/invasion/migration; ↓ apoptosis In vivo: ↑ tumor growth | miR-129-3p/MAP3K3 | [166] |
| circPDE7B (circ_0004712) | OVCAR3, SKOV3 Female BALB/c mice s.c. injected with SKOV3 | In vitro: ↑ proliferation/invasion/migration; ↓ apoptosis In vivo: ↑ tumor growth | miR-331-3p/FZD4 | [94] |
| circMYLK * | A2780, CAOV3 | In vitro: ↑ proliferation | miR-652 | [83] |
| circPGAM1 (circ_0019340) | CAOV3, OVCAR3 Nude mice s.c. injected with CAOV3 and OVCAR3 | In vitro: ↑ proliferation/migration/invasion; ↓ apoptosis In vivo: ↑ tumor growth | miR-542-3p/CDC5L/PEAK1 | [167] |
| circFOXO3 * | SKOV3 | In vitro: ↑ proliferation/migration/invasion | miR-422a/PLP2 | [168] |
| circRNA_051239 (circ_0051239) # | SKOV3, SKOV3.ip | In vitro: ↑ proliferation/migration/invasion | miR-509-5p/PRSS3 | [169] |
| circPUM1 (circ_0000043) | A2780, CAOV3 Female BALB/c mice i.p. injected with A2780 and CAOV3 | In vitro: ↑ proliferation/migration/invasion; ↓ apoptosis; ↑ MMT of peritoneal mesothelial cells In vivo: ↑ tumor growth/metastasis | miR-615-5p/NF-κB, miR-6753-5p/MMP2 | [77] |
| circWHSC1 (circ_0001387) | CAOV3, OVCAR3 Female BALB/c mice injected (s.c. or i.p.) with CAOV3. | In vitro: ↑ proliferation/migration/invasion; ↓ apoptosis; ↑ MMT of peritoneal mesothelial cells In vivo: ↑ tumor growth/metastasis | miR-145 & miR-1182/MUC1 & hTERT | [170] |
| circRHOC (circ_0013549) | A2780, CAOV3 Female BALB/c mice i.p. injected with A2780 | In vitro: ↑ viability/migration/invasion In vivo: ↑ tumor metastasis | miR-302e/VEGFA; VEGFA | [78] |
| circNOLC1 (circ_0000257) | CAOV3, A2780 Female BALB/c mice s.c. injected with A2780 | In vitro: ↑ proliferation/migration/invasion; ↓ apoptosis In vivo: ↑ tumor growth | ESRP1/CDK1/RhoA | [76] |
| circCRIM1 (circ_0002346) | OVCAR3, CAOV3 Female BALB/c mice s.c. injected with CAOV3 | In vitro: ↑ viability/migration/invasion; ↓ apoptosis In vivo: ↑ tumor growth | miR-145-5p/CRIM1, miR-383-5p/ZEB2; encoding a 188aa protein | [171] |
| circVPS13C* | A2780, SKOV3 | In vitro: ↑ proliferation/migration/invasion/cell cycle progression; ↓ apoptosis | miR-145/MEK/ERK | [172] |
| circSLAMF6 (circ_0000144) | SKOV3, ES-2 Female BALB/c mice s.c. injected with SKOV3 | In vitro: ↑ proliferation/invasion/migration In vivo: ↑ tumor growth | miR-610/ELK3 | [93] |
| circRNF144B (circ_0075797) | SKOV3, OVCAR3 Mice s.c. injected (s.c. and i.v.) with SKOV3 | In vitro: ↓ autophagy; ↑ proliferation/migration/invasion In vivo: ↑ tumor growth/metastasis | miR-342-3p/FBXL11/Beclin-1 | [92] |
| circANKRD17 (circ_0007883) | A2780, SKOV3, A2780/PTX, SKOV3/PTX BALB/c female mice s.c. injected with SKOV3/PTX | In vitro: ↑ PTX chemoresistance In vivo: ↑ tumor growth/PTX chemoresistance | FUS/FOXR2 | [90] |
| A2780, SKOV3 | In vitro: ↑ proliferation/invasion/migration/EMT; ↓ apoptosis | ZEB1/circANKRD17 | [173] | |
| circEEF2 (circ_0048559) | SKOV3, A2780 BALB/c mice s.c. injected with SKOV3, A2780 and i.p. injected with SKOV3 | In vitro: ↑ autophagy/proliferation/invasion In vivo: ↑ tumor growth/metastasis | miR-6881-3p/ATG5 & ATG7, ANXA2/p-mTOR | [91] |
| circCFH (circ_0015756) | OV90, SKOV3 Female BALB/c mice s.c. injected with SKOV3 | In vitro: ↑ proliferation/migration/invasion; ↓ apoptosis In vivo: ↑ tumor growth | miR-942-5p/CUL4B | [174] |
| circKIF4A (circ_0007255) | CAOV3, SKOV3 Female BALB/c mice injected (s.c. and i.v.) with EOC cells | In vitro: ↑ proliferation/invasion In vivo: ↑ tumor growth/metastasis | miR-127/JAM3 | [175] |
| circEXOC6B (circ_0009043) | A2780, SKOV3 BALB/c mice s.c. injected with SKOV3 | In vitro: ↓ proliferation/migration/invasion/PTX resistance In vivo: ↓ tumor PTX resistance | miR-376c-3p/FOXO3 | [105] |
| circEXOC6B* | A2870, SKOV3 | In vitro: ↓ proliferation/invasion; ↑ apoptosis | miR-421/RUS1 | [176] |
| circPLEKHM3 (circ_0001095) | A2780, MDAH2274, OV90 Female BALB/c mice s.c. injected with A2780 | In vitro: ↓ proliferation/migration/EMT In vivo: ↓ tumor growth | miR-9/BRCA1/DNAJB6/KLF4/AKT | [115] |
| SKOV3, A2780 Female BALB/c mice s.c. injected with A2780 | In vitro: ↓ proliferation; ↑ apoptosis; ↑ anti-tumor effect of curcumin In vivo: ↓ tumor growth; ↑ anti-tumor effect of curcumin | miR-320a/SMG1 | [177] | |
| circCDR1as (ciRS-7, circ_0001946) | SKOV3, A2780 Male BALB/c mice s.c. injected with SKOV3 | In vitro: ↑ proliferation/migration/invasion In vivo: ↑ tumor growth | miR-641/ZEB1 & MDM2 | [69] |
| SKOV3, SKOV3/CDDP, HO8910, HO8910/CDDP Female BALB/c mice s.c. injected with SKOV3 | In vitro: ↓ DDP resistance resistance/proliferation/migration/invasion; ↑ apoptosis In vivo: ↓ tumor growth/DDP resistance | miR-1299/PPP1R12B; AKT/mTOR | [70] | |
| A2780, SKOV3, A2780-DDP, SKOV3-DDP BALB/c female mice s.c. injected with SKOV3 | In vitro: ↓ DDP resistance/proliferation/migration; ↑ apoptosis In vivo: ↓ tumor growth/DDP resistance | miR-1270/SCAI | [71] | |
| A2780, HO8910 | In vitro: ↓ proliferation/invasion/migration | miR-135b-5p/HIF1AN | [72] | |
| circRHOBTB3 * | A2780, OV90 Female BALB/mice s.c. injected with A2780 | In vitro: ↓ proliferation/migration/invasion/glycolysis/EMT In vivo: ↓ tumor growth/metastasis | PI3K/AKT | [178] |
| circRHOBTB3 (circ_0007444) | SKOV3, OVCAR8 BALB/c mice s.c. injected with SKOV3 | In vitro: ↓ proliferation/G1/S transition/invasion; ↑ apoptosis In vivo: ↓ tumor growth | miR-23a-3p/PTEN/AKT | [110] |
| SKOV3, OVCAR3 Female nude mice s.c. injected with SKOV3, OVCAR3 | In vitro: ↓ proliferation/migration/invasion; ↑ apoptosis. In vivo: ↓ tumor growth | miR-570-3p/PTEN | [111] | |
| circATRNL1 (circ_0020093) | SKOV3, CAOV3 Nude mice i.p. injected with SKOV3 and CAOV3 | In vitro: ↓ proliferation/migration/invasion; ↓ angiogenesis; ↑ apoptosis In vivo: ↓ tumor growth/metastasis | miR-378/SMAD4 | [107] |
| A2780, SKOV3, SW626 | In vitro: ↓ proliferation/migration/invasion | encoding a 131 aa protein | [179] | |
| circITCH * | SKOV3, CAOV3 Female BALB/c mice s.c. injected with SKOV3 | In vitro: ↓ proliferation/migration/invasion In vivo: ↓ tumor growth | miR-145/RASA1 | [180] |
| circITCH * | SKOV3 | In vitro: ↓ proliferation; ↑ apoptosis | miR-10a | [181] |
| circITCH * | UWB1.289 + BRCA1, UWB1.289 | In vitro: ↓ proliferation | lncRNA HULC | [182] |
| circITCH * | A2780, OVCAR3 BALB/c mice s.c. injected with OVCAR3 | In vitro: ↓ proliferation/invasion/glycolysis; ↑ apoptosis In vivo: ↓ tumor growth | miR-106a/CDH1 | [101] |
| circSLC22A3 (circ_0078607) | HEY, ES-2 Female BALB/c mice s.c. injected with HEY | In vitro: ↓ proliferation/migration/invasion; ↑ apoptosis In vivo: ↓ tumor growth | miR-32-5p/SIK1 | [108] |
| SKOV3, A2780 | In vitro: ↓ proliferation; ↑ apoptosis | miR-518a-5p/Fas | [183] | |
| circRNA_100395 (circ_0015278) # | SKOV3, ES-2 | In vitro: ↓ proliferation/migration/invasion/EMT | miR-1228/p53 | [103] |
| circLARP4 * | SKOV3, A2780 | In vitro: ↓ proliferation/migration/invasion; ↑ apoptosis | miR-513b-5p/LARP4 | [184] |
| circFBXO7 (circ_0001222) | A2780, MDAH2774, SKOV3, OV90 Female BALB/c mice s.c. injected with MDAH2774, NOD/SCID mice i.p. injected with MDAH2774 | In vitro: ↓ proliferation/migration/invasion In vivo: ↓ tumor growth/metastasis | miR-96-5p/MTSS1/Wnt/β-catenin | [114] |
| circBNC2 (circ_0008732) | SKOV3, HO8910 | In vitro: ↓ proliferation/migration/invasion | miR-223-3p/FBXW7 | [116] |
| circ_9119 * | ES-2, SKOV3 Female BALB/c mice s.c. injected with SKOV3 | In vitro: ↓ proliferation; ↑ apoptosis In vivo: ↓ tumor growth | miR-21/PTEN/AKT | [185] |
| circZNF608 (circ_0001523) | A2780, SKOV3, SW626 | In vitro: ↓ proliferation/migration/invasion | miR-152-5p | [179] |
| circMTO1 * | SKOV3, OVCAR3 | In vitro: ↓ proliferation/invasion | miR-182-5p/KLF15 | [186] |
| circMTO1 (circ_0007874) | A2780, SKOV3 BALB/c mice s.c. injected with A2780 | In vitro: ↓ migration/proliferation In vivo: ↓ tumor growth | miR-760/SOCS3 | [187] |
| circHIPK3 (circ_0000284) | A2780, SKOV3 | In vitro: ↓ proliferation/migration/invasion; ↑ apoptosis | predicted miRNAs/mRNAs | [188] |
| circSMARCA5 (circ_0001445) | SKOV3, HO8910 Female BALB/c mice injected (s.c. and i.p.) with HO8910 | In vitro: ↓ proliferation/invasion/migration; ↑ apoptosis In vivo: ↓ tumor growth/metastasis | miR-576-5p/SFRP1/WNT/β-catenin | [113] |
| circCERS6 (circ_0007024) | SKOV3, OVCAR3 Nude mice s.c. injected with SKOV3 | In vitro: ↓ proliferation/invasion/migration/EMT In vivo: ↓ tumor growth | miR-630/RASSF8 | [189] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, Y.; Chen, Y.; Agbede, O.; Eshaghi, E.; Peng, C. Circular RNAs in Epithelial Ovarian Cancer: From Biomarkers to Therapeutic Targets. Cancers 2022, 14, 5711. https://doi.org/10.3390/cancers14225711
Qiu Y, Chen Y, Agbede O, Eshaghi E, Peng C. Circular RNAs in Epithelial Ovarian Cancer: From Biomarkers to Therapeutic Targets. Cancers. 2022; 14(22):5711. https://doi.org/10.3390/cancers14225711
Chicago/Turabian StyleQiu, Yumin, Yan Chen, Oluwatobi Agbede, Esra Eshaghi, and Chun Peng. 2022. "Circular RNAs in Epithelial Ovarian Cancer: From Biomarkers to Therapeutic Targets" Cancers 14, no. 22: 5711. https://doi.org/10.3390/cancers14225711
APA StyleQiu, Y., Chen, Y., Agbede, O., Eshaghi, E., & Peng, C. (2022). Circular RNAs in Epithelial Ovarian Cancer: From Biomarkers to Therapeutic Targets. Cancers, 14(22), 5711. https://doi.org/10.3390/cancers14225711






