Histone Chaperones and Digestive Cancer: A Review of the Literature
Abstract
Simple Summary
Abstract
1. Introduction
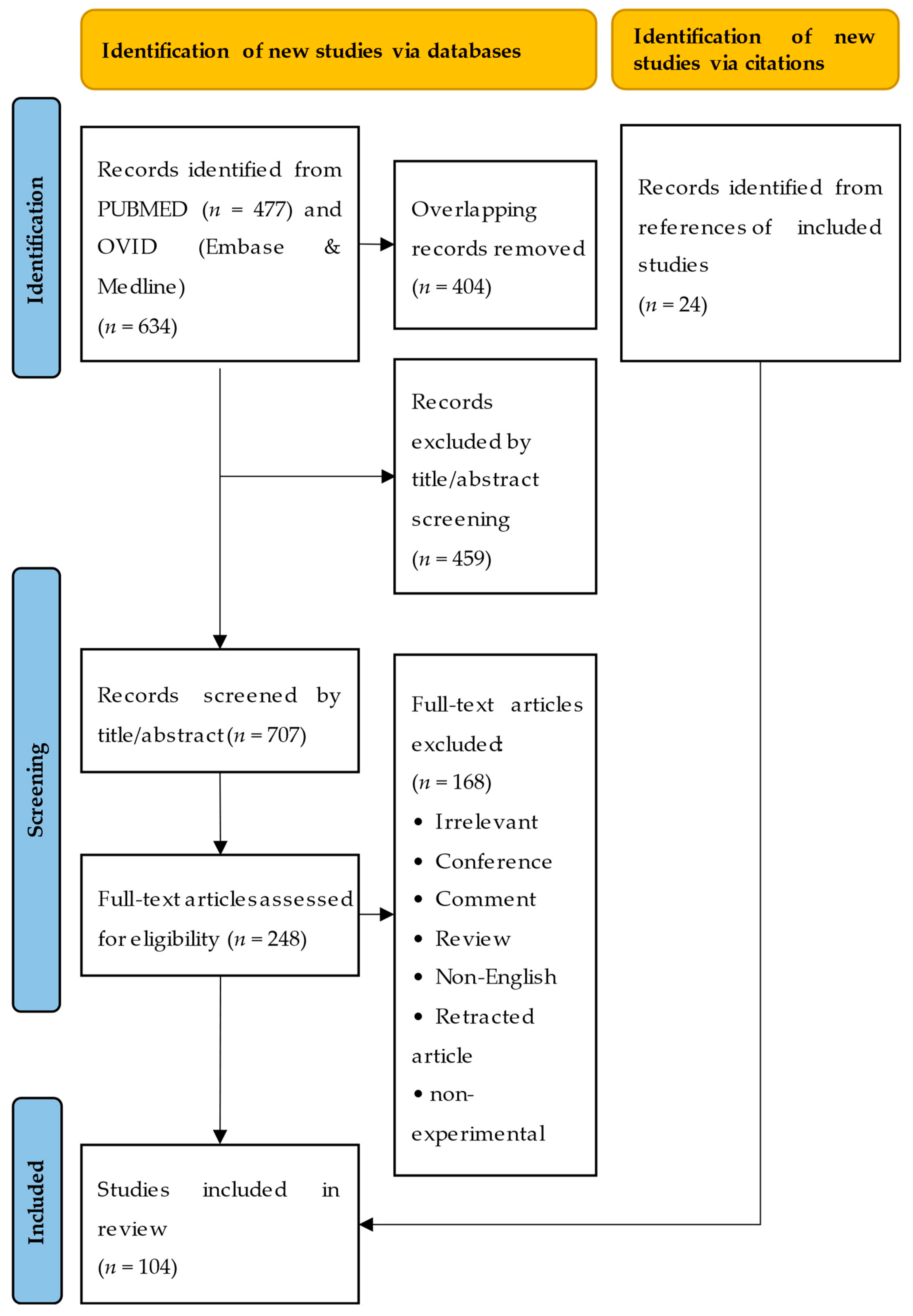
2. Literature Search
2.1. Search Process
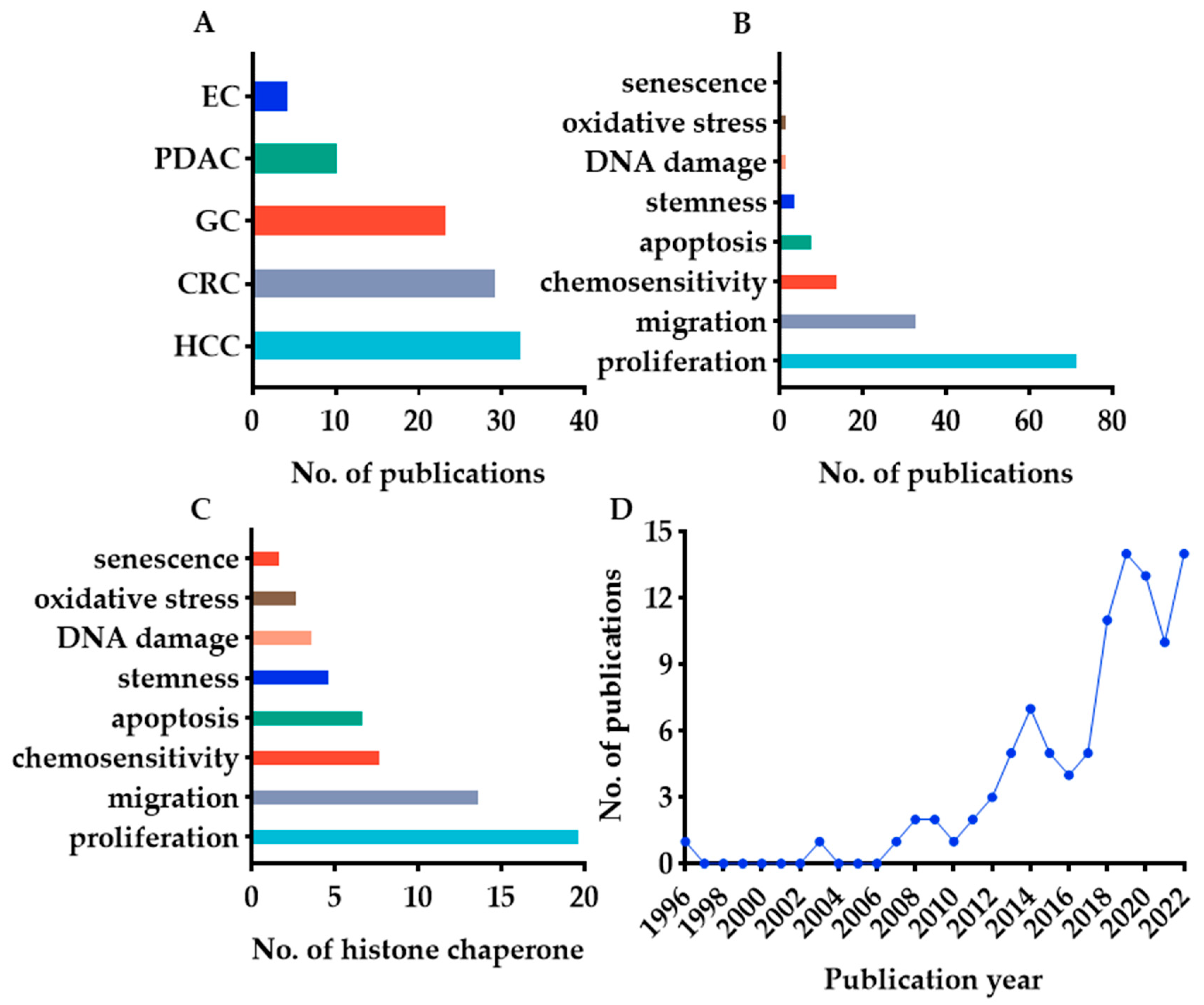
2.2. Search Results
3. Histone Chaperones in Digestive Cancers
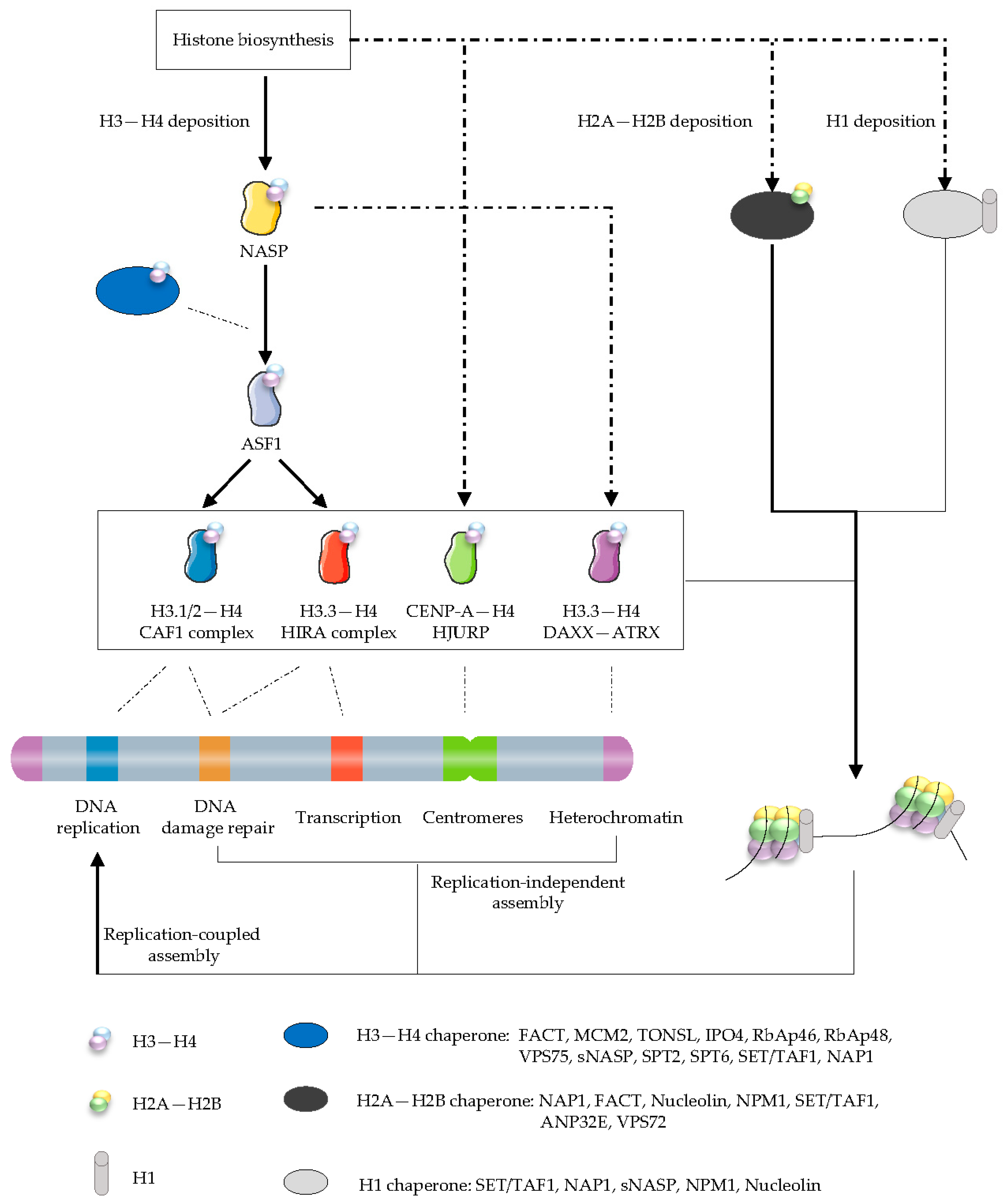
3.1. The Histone Chaperone Network
3.2. Clinical Prognostic Value of Histone Chaperones in Digestive Cancer—General Information
3.3. Phenotype and Molecular Mechanisms of Histone Chaperones in Digestive Cancer—General Information
3.4. ASF1A/B in Digestive Cancer
3.5. FACT Complex in Digestive Cancer
3.6. HJURP in Digestive Cancer
3.7. CHAF1A/B in Digestive Cancer
3.8. NPM1 in Digestive Cancer
3.9. NCL in Digestive Cancer
3.10. NAP1L1 in Digestive Cancer
3.11. DAXX and ATRX in Digestive Cancer
3.12. Other Histone Chaperones in Digestive Cancer
3.13. Histone Chaperone-Targeted Cancer Therapeutics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Adam, R.S.; Blomberg, I.; Hoorn, S.T.; Bijlsma, M.F.; Vermeulen, L. The recurring features of molecular subtypes in distinct gastrointestinal malignancies—A systematic review. Crit. Rev. Oncol. Hematol. 2021, 164, 103428. [Google Scholar] [CrossRef] [PubMed]
- Hammond, C.M.; Strømme, C.B.; Huang, H.; Patel, D.J.; Groth, A. Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 2017, 18, 141–158. [Google Scholar] [CrossRef]
- Laskey, R.A.; Honda, B.M.; Mills, A.D.; Finch, J.T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 1978, 275, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, F.; Gurard-Levin, Z.A.; Rojas-Villalobos, C.; Vassias, I.; Quatrini, R.; Almouzni, G.; Loyola, A. JMJD1B, a novel player in histone H3 and H4 processing to ensure genome stability. Epigenet. Chromatin 2020, 13, 6. [Google Scholar] [CrossRef]
- Hogan, A.; Sathyan, K.; Willis, A.; Khurana, S.; Srivastava, S.; Zasadzińska, E.; Lee, A.; Bailey, A.; Gaynes, M.; Huang, J.; et al. UBR7 acts as a histone chaperone for post-nucleosomal histone H3. EMBO J. 2021, 40, e108307. [Google Scholar] [CrossRef]
- Renaud-Pageot, C.; Quivy, J.P.; Lochhead, M.; Almouzni, G. CENP-A Regulation and Cancer. Front. Cell Dev. Biol. 2022, 10, 907120. [Google Scholar] [CrossRef]
- Ray-Gallet, D.; Almouzni, G. H3-H4 histone chaperones and cancer. Curr. Opin. Genet. Dev. 2022, 73, 101900. [Google Scholar] [CrossRef]
- Chen, T.; Huang, H.; Zhou, Y.; Geng, L.; Shen, T.; Yin, S.; Zhou, L.; Zheng, S. HJURP promotes hepatocellular carcinoma proliferation by destabilizing p21 via the MAPK/ERK1/2 and AKT/GSK3β signaling pathways. J. Exp. Clin. Cancer Res. 2018, 37, 193. [Google Scholar] [CrossRef]
- Chen, T.; Tu, Y.; Lv, D.; Lin, K.; Tang, H.; Huang, W. Vacuolar protein sorting-associated protein 72 homolog (VPS72) binding to lysine acetyltransferase 5 (KAT5) promotes the proliferation, invasion and migration of hepatocellular carcinoma through regulating phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) signaling pathway. Bioengineered 2022, 13, 9197–9210. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, W.; Pu, L.; Zhang, L.; Han, G.; Zuo, X.; Zhang, Y.; Li, X.; Shen, H.; Wu, J.; et al. PRDM8 exhibits antitumor activities toward hepatocellular carcinoma by targeting NAP1L1. Hepatology 2018, 68, 994–1009. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Chen, M.; Lee, D.; Law, C.T.; Wei, L.; Tsang, F.H.; Chin, D.W.; Cheng, C.L.; Lee, J.M.; Ng, I.O.; et al. Histone chaperone FACT complex mediates oxidative stress response to promote liver cancer progression. Gut 2020, 69, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Yuan, X.; Yu, J.; Wu, Y.; Li, K.; Sun, C.; Li, S.; Shen, L.; Kong, F.; Jia, J.; et al. Histone Chaperone ASF1A Predicts Poor Outcomes for Patients With Gastrointestinal Cancer and Drives Cancer Progression by Stimulating Transcription of β-Catenin Target Genes. eBioMedicine 2017, 21, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, X.; Yu, J.; Björkholm, M.; Xu, D. ASF1a inhibition induces p53-dependent growth arrest and senescence of cancer cells. Cell Death Dis. 2019, 10, 76. [Google Scholar] [CrossRef]
- Qiu, F.; Wang, Y.; Chu, X.; Wang, J. ASF1A regulates H4Y72 phosphorylation and promotes autophagy in colon cancer cells via a kinase activity. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2754–2763. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, L.; Feng, J.; Tan, D.; Zhu, Y.; Hou, J.; Li, W.; Lv, K.; Wang, W.; Jiang, L.; et al. ASF1B is a Promising Prognostic Biomarker and Correlates With Immunotherapy Efficacy in Hepatocellular Carcinoma. Front. Genet. 2022, 13, 842351. [Google Scholar] [CrossRef]
- Zhan, T.; Gao, X.; Wang, G.; Li, F.; Shen, J.; Lu, C.; Xu, L.; Li, Y.; Zhang, J. Construction of Novel lncRNA-miRNA-mRNA Network Associated With Recurrence and Identification of Immune-Related Potential Regulatory Axis in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 626663. [Google Scholar] [CrossRef]
- Ouyang, X.; Lv, L.; Zhao, Y.; Zhang, F.; Hu, Q.; Li, Z.; Zhu, D.; Li, L. ASF1B Serves as a Potential Therapeutic Target by Influencing Cell Cycle and Proliferation in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 801506. [Google Scholar] [CrossRef]
- Chen, C.; Bao, H.; Lin, W.; Chen, X.; Huang, Y.; Wang, H.; Yang, Y.; Liu, J.; Lv, X.; Teng, L. ASF1b is a novel prognostic predictor associated with cell cycle signaling pathway in gastric cancer. J. Cancer 2022, 13, 1985–2000. [Google Scholar] [CrossRef]
- Kim, J.H.; Youn, Y.; Lee, J.C.; Kim, J.; Ryu, J.K.; Hwang, J.H. Downregulation of ASF1B inhibits tumor progression and enhances efficacy of cisplatin in pancreatic cancer. Cancer Biomark. 2022, 34, 647–659. [Google Scholar] [CrossRef]
- Wang, K.; Hao, Z.; Fu, X.; Li, W.; Jiao, A.; Hua, X. Involvement of elevated ASF1B in the poor prognosis and tumorigenesis in pancreatic cancer. Mol. Cell. Biochem. 2022, 477, 1945–1957. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; He, K.; Luo, T.; Deng, Y.; Wang, H.; Liu, H.; Zhang, J.; Chen, K.; Xiao, J.; Duan, X.; et al. SSRP1 Contributes to the Malignancy of Hepatocellular Carcinoma and Is Negatively Regulated by miR-497. Mol. Ther. 2016, 24, 903–914. [Google Scholar] [CrossRef]
- Min, J.; Jin, D.; Zhang, F.; Kang, Y.; Qi, Y.; Du, P. DLG1-AS1 is activated by MYC and drives the proliferation and migration of hepatocellular carcinoma cells through miR-497-5p/SSRP1 axis. Cancer Cell Int. 2021, 21, 16. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Guo, Y.; Dai, L.; Liang, Z.; Yang, Q.; Yi, S. Long intergenic noncoding RNA01134 accelerates hepatocellular carcinoma progression by sponging microRNA-4784 and downregulating structure specific recognition protein 1. Bioengineered 2020, 11, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Zhao, R.; Zhang, J.; Cao, T.; Tang, T. SSRP1 affects the growth and apoptosis of gastric cancer cells through AKT pathway. J. Med. Biochem. 2022, 41, 100–107. [Google Scholar] [CrossRef]
- Song, H.; Zeng, J.; Roychoudhury, S.; Biswas, P.; Mohapatra, B.; Ray, S.; Dowlatshahi, K.; Wang, J.; Band, V.; Talmon, G.; et al. Targeting Histone Chaperone FACT Complex Overcomes 5-Fluorouracil Resistance in Colon Cancer. Mol. Cancer Ther. 2020, 19, 258–269. [Google Scholar] [CrossRef]
- Wang, Q.; Jia, S.; Jiao, Y.; Xu, L.; Wang, D.; Chen, X.; Hu, X.; Liang, H.; Wen, N.; Zhang, S.; et al. SSRP1 influences colorectal cancer cell growth and apoptosis via the AKT pathway. Int. J. Med. Sci. 2019, 16, 1573–1582. [Google Scholar] [CrossRef]
- Wu, W.; He, K.; Guo, Q.; Chen, J.; Zhang, M.; Huang, K.; Yang, D.; Wu, L.; Deng, Y.; Luo, X.; et al. SSRP1 promotes colorectal cancer progression and is negatively regulated by miR-28-5p. J. Cell. Mol. Med. 2019, 23, 3118–3129. [Google Scholar] [CrossRef]
- Huang, H.; Cai, L.; Li, R.; Ye, L.; Chen, Z. A novel lncRNA LOC101927746 accelerates progression of colorectal cancer via inhibiting miR-584-3p and activating SSRP1. Biochem. Biophys. Res. Commun. 2019, 509, 734–738. [Google Scholar] [CrossRef]
- Chen, T.; Zhou, L.; Zhou, Y.; Zhou, W.; Huang, H.; Yin, S.; Xie, H.; Zhou, L.; Zheng, S. HJURP Promotes Epithelial-to-Mesenchymal Transition via Upregulating SPHK1 in Hepatocellular Carcinoma. Int. J. Biol. Sci. 2019, 15, 1139–1147. [Google Scholar] [CrossRef]
- Hu, B.; Wang, Q.; Wang, Y.; Chen, J.; Li, P.; Han, M. Holliday junction-recognizing protein promotes cell proliferation and correlates with unfavorable clinical outcome of hepatocellular carcinoma. OncoTargets Ther. 2017, 10, 2601–2607. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, Y.; Yi, Q.; Liao, X.; Han, C.; Zheng, L.; Li, H.; Yu, Q.; Yan, X.; Chen, X.; Zhu, H.; et al. Hypomethylation-driven overexpression of HJURP promotes progression of hepatocellular carcinoma and is associated with poor prognosis. Biochem. Biophys. Res. Commun. 2021, 566, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.J.; Li, X.; Shi, P.; Ding, H.Y.; Liu, Y.P.; Li, T.; Lin, P.P.; Wang, Y.S.; Zhang, G.Q.; Cao, Y. Holliday junction recognition protein promotes pancreatic cancer growth and metastasis via modulation of the MDM2/p53 signaling. Cell Death Dis. 2020, 11, 386. [Google Scholar] [CrossRef]
- Kang, D.H.; Woo, J.; Kim, H.; Kim, S.Y.; Ji, S.; Jaygal, G.; Ahn, T.S.; Kim, H.J.; Kwak, H.J.; Kim, C.J.; et al. Prognostic Relevance of HJURP Expression in Patients with Surgically Resected Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 7928. [Google Scholar] [CrossRef]
- Zhang, D.; Wu, F.; Song, J.; Meng, M.; Fan, X.; Lu, C.; Weng, Q.; Fang, S.; Zheng, L.; Tang, B.; et al. A role for the NPM1/PTPN14/YAP axis in mediating hypoxia-induced chemoresistance to sorafenib in hepatocellular carcinoma. Cancer Cell Int. 2022, 22, 65. [Google Scholar] [CrossRef]
- Liu, X.; Liu, D.; Qian, D.; Dai, J.; An, Y.; Jiang, S.; Stanley, B.; Yang, J.; Wang, B.; Liu, X.; et al. Nucleophosmin (NPM1/B23) interacts with activating transcription factor 5 (ATF5) protein and promotes proteasome- and caspase-dependent ATF5 degradation in hepatocellular carcinoma cells. J. Biol. Chem. 2012, 287, 19599–19609. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, D.H.; Liu, F.; Liu, G.Y.; Lu, K.; Deng, X.L.; Li, Q.F.; Shi, S.L. Relocation of NPM Affects the Malignant Phenotypes of Hepatoma SMMC-7721 Cells. J. Cell. Biochem. 2017, 118, 3225–3236. [Google Scholar] [CrossRef]
- Ching, R.H.H.; Lau, E.Y.T.; Ling, P.M.T.; Lee, J.M.F.; Ma, M.K.F.; Cheng, B.Y.L.; Lo, R.C.L.; Ng, I.O.L.; Lee, T.K.W. Phosphorylation of Nucleophosmin at Threonine 234/237 is associated with HCC metastasis. Oncotarget 2015, 6, 43483–43495. [Google Scholar] [CrossRef]
- Lo, S.J.; Fan, L.C.; Tsai, Y.F.; Lin, K.Y.; Huang, H.L.; Wang, T.H.; Liu, H.; Chen, T.C.; Huang, S.F.; Chang, C.J.; et al. A novel interaction of nucleophosmin with BCL2-associated X protein regulating death evasion and drug sensitivity in human hepatoma cells. Hepatology 2013, 57, 1893–1905. [Google Scholar] [CrossRef]
- Ch, N.P.L.; Sivagnanam, A.; Raja, S.; Mahalingam, S. Molecular basis for RASSF10/NPM/RNF2 feedback cascade-mediated regulation of gastric cancer cell proliferation. J. Biol. Chem. 2021, 297, 100935. [Google Scholar] [CrossRef]
- Zhu, Y.; Shi, M.; Chen, H.; Gu, J.; Zhang, J.; Shen, B.; Deng, X.; Xie, J.; Zhan, X.; Peng, C. NPM1 activates metabolic changes by inhibiting FBP1 while promoting the tumorigenicity of pancreatic cancer cells. Oncotarget 2015, 6, 21443–21451. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, G.; Abraham, A.G.; Morton, J.; Sampson, O.; Pefani, D.E.; Khoronenkova, S.; Grawenda, A.; Papaspyropoulos, A.; Jamieson, N.; McKay, C.; et al. AKT regulates NPM dependent ARF localization and p53mut stability in tumors. Oncotarget 2014, 5, 6142–6167. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chen, H.; Wong, C.C.; Liu, D.; Li, T.; Wang, X.; Ji, J.; Sung, J.J.; Fang, J.Y.; Yu, J. DEAD-box helicase 27 promotes colorectal cancer growth and metastasis and predicts poor survival in CRC patients. Oncogene 2018, 37, 3006–3021. [Google Scholar] [CrossRef]
- Wong, J.C.; Hasan, M.R.; Rahman, M.; Yu, A.C.; Chan, S.K.; Schaeffer, D.F.; Kennecke, H.F.; Lim, H.J.; Owen, D.; Tai, I.T. Nucleophosmin 1, upregulated in adenomas and cancers of the colon, inhibits p53-mediated cellular senescence. Int. J. Cancer 2013, 133, 1567–1577. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.C.Y.; Chern, Y.J.; Zhang, P.; Pasiliao, C.C.; Rahman, M.; Chang, G.; Ren, J.; Tai, I.T. Inhibition of nucleophosmin 1 suppresses colorectal cancer tumor growth of patient -derived xenografts via activation of p53 and inhibition of AKT. Cancer Biol. Ther. 2021, 22, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Grbčić, P.; Čupić, D.F.; Gamberi, T.; Kraljević Pavelić, S.; Sedić, M. Proteomic Profiling of BRAFV600E Mutant Colon Cancer Cells Reveals the Involvement of Nucleophosmin/c-Myc Axis in Modulating the Response and Resistance to BRAF Inhibition by Vemurafenib. Int. J. Mol. Sci. 2021, 22, 6174. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, F.; Zhang, X.F.; Qi, L.S.; Yang, L.; Guo, H.; Zhang, N. Expression of nucleophosmin/NPM1 correlates with migration and invasiveness of colon cancer cells. J. Biomed. Sci. 2012, 19, 53. [Google Scholar] [CrossRef]
- Wang, D.; Li, Y.; Liu, Y.; Cheng, S.; Liu, F.; Zuo, R.; Ding, C.; Shi, S.; Liu, G. NPM1 promotes cell proliferation by targeting PRDX6 in colorectal cancer. Int. J. Biochem. Cell Biol. 2022, 147, 106233. [Google Scholar] [CrossRef]
- Liu, Y.; Li, C.; Fang, L.; Wang, L.; Liu, H.; Tian, H.; Zheng, Y.; Fan, T.; He, J. Lipid metabolism-related lncRNA SLC25A21-AS1 promotes the progression of oesophageal squamous cell carcinoma by regulating the NPM1/c-Myc axis and SLC25A21 expression. Clin. Transl. Med. 2022, 12, e944. [Google Scholar] [CrossRef]
- Zhang, Y.W.; Chen, Q.; Li, B.; Li, H.Y.; Zhao, X.K.; Xiao, Y.Y.; Liu, S.; Zuo, S. NAP1L1 Functions as a Tumor Promoter via Recruiting Hepatoma-Derived Growth Factor/c-Jun Signal in Hepatocellular Carcinoma. Front. Cell Dev. Biol. 2021, 9, 659680. [Google Scholar] [CrossRef]
- Le, Y.; Kan, A.; Li, Q.J.; He, M.K.; Chen, H.L.; Shi, M. NAP1L1 is a prognostic biomarker and contribute to doxorubicin chemotherapy resistance in human hepatocellular carcinoma. Cancer Cell Int. 2019, 19, 228. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xiang, B.; Liu, Y.; Wang, Y.; Kan, H. LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett. 2018, 437, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Schimmack, S.; Taylor, A.; Lawrence, B.; Alaimo, D.; Schmitz-Winnenthal, H.; Büchler, M.W.; Modlin, I.M.; Kidd, M. A mechanistic role for the chromatin modulator, NAP1L1, in pancreatic neuroendocrine neoplasm proliferation and metastases. Epigenet. Chromatin 2014, 7, 15. [Google Scholar] [CrossRef]
- Xu, M.; Jia, Y.; Liu, Z.; Ding, L.; Tian, R.; Gu, H.; Wang, Y.; Zhang, H.; Tu, K.; Liu, Q. Chromatin assembly factor 1, subunit A (P150) facilitates cell proliferation in human hepatocellular carcinoma. OncoTargets Ther. 2016, 9, 4023–4035. [Google Scholar] [CrossRef]
- Zheng, L.; Liang, X.; Li, S.; Li, T.; Shang, W.; Ma, L.; Jia, X.; Shao, W.; Sun, P.; Chen, C.; et al. CHAF1A interacts with TCF4 to promote gastric carcinogenesis via upregulation of c-MYC and CCND1 expression. eBioMedicine 2018, 38, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, X.; Shen, B.; Chen, X.; Shu, Y. Histone chaperone CHAF1A impacts the outcome of fluoropyrimidines-based adjuvant therapy in gastric cancer by regulating the expression of thymidylate synthetase. Gene 2019, 716, 144034. [Google Scholar] [CrossRef]
- Peng, X.; Fu, H.; Yin, J.; Zhao, Q. CHAF1B knockdown blocks migration in a hepatocellular carcinoma model. Oncol. Rep. 2018, 40, 405–413. [Google Scholar] [CrossRef]
- Qiu, W.; Wang, G.; Sun, X.; Ye, J.; Wei, F.; Shi, X.; Lv, G. The involvement of cell surface nucleolin in the initiation of CCR6 signaling in human hepatocellular carcinoma. Med. Oncol. 2015, 32, 75. [Google Scholar] [CrossRef]
- Chen, S.C.; Hu, T.H.; Huang, C.C.; Kung, M.L.; Chu, T.H.; Yi, L.N.; Huang, S.T.; Chan, H.H.; Chuang, J.H.; Liu, L.F.; et al. Hepatoma-derived growth factor/nucleolin axis as a novel oncogenic pathway in liver carcinogenesis. Oncotarget 2015, 6, 16253–16270. [Google Scholar] [CrossRef]
- De Lara, S.B.; Tran, D.D.H.; Allister, A.B.; Polenkowski, M.; Nashan, B.; Koch, M.; Tamura, T. C20orf204, a hepatocellular carcinoma-specific protein interacts with nucleolin and promotes cell proliferation. Oncogenesis 2021, 10, 31. [Google Scholar] [CrossRef]
- Devanand, P.; Oya, Y.; Sundaramoorthy, S.; Song, K.Y.; Watanabe, T.; Kobayashi, Y.; Shimizu, Y.; Hong, S.A.; Suganuma, M.; Lim, I.K. Inhibition of TNFα-interacting protein α (Tipα)-associated gastric carcinogenesis by BTG2/TIS21 via downregulating cytoplasmic nucleolin expression. Exp. Mol. Med. 2018, 50, e449. [Google Scholar] [CrossRef] [PubMed]
- Gilles, M.E.; Maione, F.; Cossutta, M.; Carpentier, G.; Caruana, L.; di Maria, S.; Houppe, C.; Destouches, D.; Shchors, K.; Prochasson, C.; et al. Nucleolin Targeting Impairs the Progression of Pancreatic Cancer and Promotes the Normalization of Tumor Vasculature. Cancer Res. 2016, 76, 7181–7193. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, Y.; Tu, Q.; Zhang, Z.; Chen, M.; Mwangi, J.; Li, Y.; Jin, Y.; Zhao, X.; Lai, R. Targeting surface nucleolin induces autophagy-dependent cell death in pancreatic cancer via AMPK activation. Oncogene 2019, 38, 1832–1844. [Google Scholar] [CrossRef]
- Wu, D.M.; Zhang, P.; Liu, R.Y.; Sang, Y.X.; Zhou, C.; Xu, G.C.; Yang, J.L.; Tong, A.P.; Wang, C.T. Phosphorylation and changes in the distribution of nucleolin promote tumor metastasis via the PI3K/Akt pathway in colorectal carcinoma. FEBS Lett. 2014, 588, 1921–1929. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Reyes, E.M.; Akiyama, S.K. Cell-surface nucleolin is a signal transducing P-selectin binding protein for human colon carcinoma cells. Exp. Cell Res. 2008, 314, 2212–2223. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Li, L.; Bai, Y.; Yu, B.; Xie, C.; Wu, H.; Zhang, Y.; Huang, L.; Yan, Y.; Li, X.; et al. The long noncoding RNA LUCAT1 promotes colorectal cancer cell proliferation by antagonizing Nucleolin to regulate MYC expression. Cell Death Dis. 2020, 11, 908. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Sun, X.; Xie, W.; Chen, S.; Hu, Y.; Xing, D.; Xu, J.; Chen, X.; Zhao, Z.; Han, Z.; et al. Opposing biological functions of the cytoplasm and nucleus DAXX modified by SUMO-2/3 in gastric cancer. Cell Death Dis. 2020, 11, 514. [Google Scholar] [CrossRef]
- Wu, C.; Ding, H.; Wang, S.; Li, Y.; Liu, S.B.; Wang, X.; Zheng, J.; Xue, T.; Amin, H.M.; Song, Y.H.; et al. DAXX inhibits cancer stemness and epithelial-mesenchymal transition in gastric cancer. Br. J. Cancer 2020, 122, 1477–1485. [Google Scholar] [CrossRef]
- Chen, Y.C.; Lee, T.H.; Tzeng, S.L. Reduced DAXX Expression Is Associated with Reduced CD24 Expression in Colorectal Cancer. Cells 2019, 8, 1242. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, F.; Zhu, X.; Guo, W.; Fu, T.; Wang, W. Death Domain-Associated Protein Promotes Colon Cancer Metastasis through Direct Interaction with ZEB1. J. Cancer 2020, 11, 750–758. [Google Scholar] [CrossRef]
- Huang, Y.S.; Wu, C.C.; Chang, C.C.; Huang, S.F.; Kuo, H.Y.; Shih, H.M. Reciprocal regulation of Daxx and PIK3CA promotes colorectal cancer cell growth. Cell. Mol. Life Sci. 2022, 79, 367. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, S.L.; Cheng, Y.W.; Li, C.H.; Lin, Y.S.; Hsu, H.C.; Kang, J.J. Physiological and functional interactions between Tcf4 and Daxx in colon cancer cells. J. Biol. Chem. 2006, 281, 15405–15411. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Oh, S.; Song, H.; Shin, S.; Zhang, B.; Freeman, W.M.; Janknecht, R. A potential common role of the Jumonji C domain-containing 1A histone demethylase and chromatin remodeler ATRX in promoting colon cancer. Oncol. Lett. 2018, 16, 6652–6662. [Google Scholar] [CrossRef]
- Young, C.C.; Baker, R.M.; Howlett, C.J.; Hryciw, T.; Herman, J.E.; Higgs, D.; Gibbons, R.; Crawford, H.; Brown, A.; Pin, C.L. The Loss of ATRX Increases Susceptibility to Pancreatic Injury and Oncogenic KRAS in Female But Not Male Mice. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 93–113. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.Y.; Salah, M.M.; Kassim, S.K.; Abdelaal, A.; Elayat, W.M.; Mohamed, D.A.; Fouly, A.E.; Abu-Zahra, F.A.E. Evaluation of the diagnostic and therapeutic roles of non-coding RNA and cell proliferation related gene association in hepatocellular carcinoma. Gene 2019, 706, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, H.; Zhang, J.; Cheng, A.S.L.; Yu, J.; To, K.F.; Kang, W. MCM family in gastrointestinal cancer and other malignancies: From functional characterization to clinical implication. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188415. [Google Scholar] [CrossRef]
- Wang, P.; Yang, X.; Zhao, L.; Liu, D.; Liu, J.; Ding, Y. A novel long non-coding RNA TONSL-AS1 regulates progression of gastric cancer via activating TONSL. Exp. Cell Res. 2019, 382, 111453. [Google Scholar] [CrossRef]
- Jin, X.; Jiang, R.; Xiang, Y.; Fan, Z.; Wu, Z.; Yang, B.; Yang, L.; Wei, S.; Yang, Y. Overexpression of retinoblastoma-binding protein 4 contributes to the radiosensitivity of AGS gastric cancer cells via phosphoinositide3-kinase/protein kinase B pathway suppression. Mol. Med. Rep. 2018, 18, 1571–1581. [Google Scholar] [CrossRef]
- Kang, X.; Feng, Y.; Gan, Z.; Zeng, S.; Guo, X.; Chen, X.; Zhang, Y.; Wang, C.; Liu, K.; Chen, X.; et al. NASP antagonize chromatin accessibility through maintaining histone H3K9me1 in hepatocellular carcinoma. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 3438–3448. [Google Scholar] [CrossRef]
- Yu, B.; Chen, X.; Li, J.; Gu, Q.; Zhu, Z.; Li, C.; Su, L.; Liu, B. microRNA-29c inhibits cell proliferation by targeting NASP in human gastric cancer. BMC Cancer 2017, 17, 109. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, X.; Xing, H.; Liu, Z.; Jia, J.; Jin, C.; Zhang, Y. Importin-4 functions as a driving force in human primary gastric cancer. J. Cell. Biochem. 2019, 120, 12638–12646. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Tang, Y.; Xiao, M.; Li, M.; Fu, Z.M.H.; Wang, Y.L. The role of histone chaperone spty2d1 in human colorectal cancer. Mol. Cell. Probes 2022, 64, 101832. [Google Scholar] [CrossRef] [PubMed]
- Diao, C.; Guo, P.; Yang, W.; Sun, Y.; Liao, Y.; Yan, Y.; Zhao, A.; Cai, X.; Hao, J.; Hu, S.; et al. SPT6 recruits SND1 to co-activate human telomerase reverse transcriptase to promote colon cancer progression. Mol. Oncol. 2021, 15, 1180–1202. [Google Scholar] [CrossRef]
- Zhang, J.; Lan, Z.; Qiu, G.; Ren, H.; Zhao, Y.; Gu, Z.; Li, Z.; Feng, L.; He, J.; Wang, C. Over-expression of ANP32E is associated with poor prognosis of pancreatic cancer and promotes cell proliferation and migration through regulating β-catenin. BMC Cancer 2020, 20, 1065. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.S.; Yang, X.L.; Liu, Y.S.; Ding, H.; Wu, J.J.; Shi, Y.; Jia, C.Y.; Lu, G.X.; Zhang, D.D.; Wang, H.M.; et al. Long non-coding RNA NORAD promotes pancreatic cancer stem cell proliferation and self-renewal by blocking microRNA-202-5p-mediated ANP32E inhibition. J. Transl. Med. 2021, 19, 400. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zou, Y.; Wu, T.; Ni, J.; Tan, Q.; Wang, Q.; Zhang, M. ANP32E contributes to gastric cancer progression via NUF2 upregulation. Mol. Med. Rep. 2022, 26, 275. [Google Scholar] [CrossRef] [PubMed]
- Mbianda, J.; Bakail, M.; André, C.; Moal, G.; Perrin, M.E.; Pinna, G.; Guerois, R.; Becher, F.; Legrand, P.; Traoré, S.; et al. Optimal anchoring of a foldamer inhibitor of ASF1 histone chaperone through backbone plasticity. Sci. Adv. 2021, 7, eabd9153. [Google Scholar] [CrossRef]
- Gonzalez-Muñoz, E.; Arboleda-Estudillo, Y.; Otu, H.H.; Cibelli, J.B. Cell reprogramming. Histone chaperone ASF1A is required for maintenance of pluripotency and cellular reprogramming. Science 2014, 345, 822–825. [Google Scholar] [CrossRef]
- Corpet, A.; de Koning, L.; Toedling, J.; Savignoni, A.; Berger, F.; Lemaître, C.; O’Sullivan, R.J.; Karlseder, J.; Barillot, E.; Asselain, B.; et al. Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 2011, 30, 480–493. [Google Scholar] [CrossRef]
- Ye, X.; Zerlanko, B.; Zhang, R.; Somaiah, N.; Lipinski, M.; Salomoni, P.; Adams, P.D. Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 2007, 27, 2452–2465. [Google Scholar] [CrossRef]
- Battu, A.; Ray, A.; Wani, A.A. ASF1A and ATM regulate H3K56-mediated cell-cycle checkpoint recovery in response to UV irradiation. Nucleic Acids Res. 2011, 39, 7931–7945. [Google Scholar] [CrossRef] [PubMed]
- Devenport, S.N.; Shah, Y.M. Functions and Implications of Autophagy in Colon Cancer. Cells 2019, 8, 1349. [Google Scholar] [CrossRef] [PubMed]
- Chou, R.H.; Wang, Y.N.; Hsieh, Y.H.; Li, L.Y.; Xia, W.; Chang, W.C.; Chang, L.C.; Cheng, C.C.; Lai, C.C.; Hsu, J.L.; et al. EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev. Cell 2014, 30, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Li, J.; Zhao, P. Roles of ncRNAs as ceRNAs in Gastric Cancer. Genes 2021, 12, 1036. [Google Scholar] [CrossRef]
- Chen, Z.; Ou, D.; Huang, Z.; Shen, P. Identification of hsa_circ_0002024 as a prognostic competing endogenous RNA (ceRNA) through the hsa_miR_129-5p/Anti-Silencing Function 1B Histone Chaperone (ASF1B) axis in renal cell carcinoma. Bioengineered 2021, 12, 6579–6593. [Google Scholar] [CrossRef]
- Hayashi, R.; Goto, Y.; Tanaka, R.; Oonogi, K.; Hisasue, M.; Yoshida, K. Transcriptional regulation of human chromatin assembly factor ASF1. DNA Cell Biol. 2007, 26, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Miknis, G.F.; Stevens, S.J.; Smith, L.E.; Ostrov, D.A.; Churchill, M.E. Development of novel Asf1-H3/H4 inhibitors. Bioorg. Med. Chem. Lett. 2015, 25, 963–968. [Google Scholar] [CrossRef][Green Version]
- Bakail, M.; Gaubert, A.; Andreani, J.; Moal, G.; Pinna, G.; Boyarchuk, E.; Gaillard, M.C.; Courbeyrette, R.; Mann, C.; Thuret, J.Y.; et al. Design on a Rational Basis of High-Affinity Peptides Inhibiting the Histone Chaperone ASF1. Cell Chem. Biol. 2019, 26, 1573–1585.e10. [Google Scholar] [CrossRef]
- Orphanides, G.; Wu, W.H.; Lane, W.S.; Hampsey, M.; Reinberg, D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 1999, 400, 284–288. [Google Scholar] [CrossRef]
- Winkler, D.D.; Muthurajan, U.M.; Hieb, A.R.; Luger, K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 2011, 286, 41883–41892. [Google Scholar] [CrossRef]
- Panieri, E.; Pinho, S.A.; Afonso, G.J.M.; Oliveira, P.J.; Cunha-Oliveira, T.; Saso, L. NRF2 and Mitochondrial Function in Cancer and Cancer Stem Cells. Cells 2022, 11, 2401. [Google Scholar] [CrossRef] [PubMed]
- Hossan, T.; Nagarajan, S.; Baumgart, S.J.; Xie, W.; Magallanes, R.T.; Hernandez, C.; Chiaroni, P.M.; Indenbirken, D.; Spitzner, M.; Thomas-Chollier, M.; et al. Histone Chaperone SSRP1 is Essential for Wnt Signaling Pathway Activity During Osteoblast Differentiation. Stem Cells 2016, 34, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Dunleavy, E.M.; Roche, D.; Tagami, H.; Lacoste, N.; Ray-Gallet, D.; Nakamura, Y.; Daigo, Y.; Nakatani, Y.; Almouzni-Pettinotti, G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 2009, 137, 485–497. [Google Scholar] [CrossRef]
- Tachiwana, H.; Müller, S.; Blümer, J.; Klare, K.; Musacchio, A.; Almouzni, G. HJURP involvement in de novo CenH3(CENP-A) and CENP-C recruitment. Cell Rep. 2015, 11, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, J.H.; Rodríguez, M.G.; Martins, N.M.; Kimura, H.; Kelly, D.A.; Masumoto, H.; Larionov, V.; Jansen, L.E.; Earnshaw, W.C. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011, 30, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, D.; Gatto, A.; Podsypanina, K.; Renaud-Pageot, C.; Landete, R.P.; Bonneville, L.; Dumont, M.; Fachinetti, D.; Almouzni, G. CENP-A overexpression promotes distinct fates in human cells, depending on p53 status. Commun. Biol. 2021, 4, 417. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhang, H.; Hao, Y.; Xu, X.; Zhai, Y.; Wang, S.; Li, Y.; Ma, F.; Li, Y.; Wang, Z.; et al. A Non-Synonymous Single Nucleotide Polymorphism in the HJURP Gene Associated with Susceptibility to Hepatocellular Carcinoma among Chinese. PLoS ONE 2016, 11, e0148618. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.; Zhu, W.; Xiao, C.; Li, X.; Wang, Y.; Han, Y.; Zheng, J.; Li, Y.; Li, M.; Wen, X. HJURP promotes proliferation in prostate cancer cells through increasing CDKN1A degradation via the GSK3β/JNK signaling pathway. Cell Death Dis. 2021, 12, 583. [Google Scholar] [CrossRef]
- Mao, M.; Jia, Y.; Chen, Y.; Yang, J.; Xu, L.; Zhang, X.; Zhou, J.; Li, Z.; Chen, C.; Ju, S.; et al. HJURP regulates cell proliferation and chemo-resistance via YAP1/NDRG1 transcriptional axis in triple-negative breast cancer. Cell Death Dis. 2022, 13, 396. [Google Scholar] [CrossRef]
- Dou, Z.; Qiu, C.; Zhang, X.; Yao, S.; Zhao, C.; Wang, Z.; Chu, R.; Chen, J.; Chen, Z.; Li, R.; et al. HJURP Promotes Malignant Progression and Mediates Sensitivity to Cisplatin and WEE1-inhibitor in Serous Ovarian Cancer. Int. J. Biol. Sci. 2022, 18, 1188–1210. [Google Scholar] [CrossRef]
- Smith, S.; Stillman, B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 1989, 58, 15–25. [Google Scholar] [CrossRef]
- Kaufman, P.D.; Kobayashi, R.; Kessler, N.; Stillman, B. The p150 and p60 subunits of chromatin assembly factor I: A molecular link between newly synthesized histones and DNA replication. Cell 1995, 81, 1105–1114. [Google Scholar] [CrossRef]
- Sykaras, A.; Pergaris, A.; Theocharis, S. Challenging, Accurate and Feasible: CAF-1 as a Tumour Proliferation Marker of Diagnostic and Prognostic Value. Cancers 2021, 13, 2575. [Google Scholar] [CrossRef] [PubMed]
- Glinsky, G.V.; Glinskii, A.B.; Stephenson, A.J.; Hoffman, R.M.; Gerald, W.L. Gene expression profiling predicts clinical outcome of prostate cancer. J. Clin. Investig. 2004, 113, 913–923. [Google Scholar] [CrossRef]
- Peng, H.; Du, B.; Jiang, H.; Gao, J. Over-expression of CHAF1A promotes cell proliferation and apoptosis resistance in glioblastoma cells via AKT/FOXO3a/Bim pathway. Biochem. Biophys. Res. Commun. 2016, 469, 1111–1116. [Google Scholar] [CrossRef]
- Barbieri, E.; de Preter, K.; Capasso, M.; Chen, Z.; Hsu, D.M.; Tonini, G.P.; Lefever, S.; Hicks, J.; Versteeg, R.; Pession, A.; et al. Histone chaperone CHAF1A inhibits differentiation and promotes aggressive neuroblastoma. Cancer Res. 2014, 74, 765–774. [Google Scholar] [CrossRef]
- Dermani, F.K.; Khoei, S.G.; Afshar, S.; Amini, R. The potential role of nucleophosmin (NPM1) in the development of cancer. J. Cell. Physiol. 2021, 236, 7832–7852. [Google Scholar] [CrossRef]
- Falini, B.; Nicoletti, I.; Martelli, M.F.; Mecucci, C. Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): Biologic and clinical features. Blood 2007, 109, 874–885. [Google Scholar] [CrossRef]
- Yun, J.P.; Miao, J.; Chen, G.G.; Tian, Q.H.; Zhang, C.Q.; Xiang, J.; Fu, J.; Lai, P.B. Increased expression of nucleophosmin/B23 in hepatocellular carcinoma and correlation with clinicopathological parameters. Br. J. Cancer 2007, 96, 477–484. [Google Scholar] [CrossRef]
- Nozawa, Y.; van Belzen, N.; van der Made, A.C.; Dinjens, W.N.; Bosman, F.T. Expression of nucleophosmin/B23 in normal and neoplastic colorectal mucosa. J. Pathol. 1996, 178, 48–52. [Google Scholar] [CrossRef]
- Yang, Y.F.; Zhang, X.Y.; Yang, M.; He, Z.H.; Peng, N.F.; Xie, S.R.; Xie, Y.F. Prognostic role of nucleophosmin in colorectal carcinomas. Asian Pac. J. Cancer Prev. 2014, 15, 2021–2026. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, Y.; Sun, Z.; Liu, K.; Qiu, W.; Yao, R.; Feng, T.; Xin, C.; Yue, L. Prognostic significance of the co-expression of nucleophosmin and trefoil factor 3 in postoperative gastric cancer patients. Mol. Clin. Oncol. 2014, 2, 1055–1061. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, F.; Chen, E.; You, D.; Song, Y.; Sun, Z.; Yue, L. Both high expression of nucleophosmin/B23 and CRM1 predicts poorer prognosis in human gastric cancer. APMIS 2016, 124, 1046–1053. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Qiu, W.; Sun, L.; Xiang, J.; Sun, X.; Sui, A.; Ding, A.; Yue, L. Clinical significance of nucleophosmin/B23 and human epidermal growth factor receptor 2/neu expressions in gastric cancers. APMIS 2013, 121, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Nguyen, T.L.X.; Choi, J.W.; Lee, K.H.; Cho, S.W.; Liu, Z.; Ye, K.; Bae, S.S.; Ahn, J.Y. Nuclear Akt interacts with B23/NPM and protects it from proteolytic cleavage, enhancing cell survival. Proc. Natl. Acad. Sci. USA 2008, 105, 16584–16589. [Google Scholar] [CrossRef]
- Xu, D.H.; Liu, F.; Li, X.; Chen, X.F.; Jing, G.J.; Wu, F.Y.; Shi, S.L.; Li, Q.F. Regulatory role of nucleophosmin during the differentiation of human liver cancer cells. Int. J. Oncol. 2014, 45, 264–272. [Google Scholar] [CrossRef][Green Version]
- Guo, C.A.; Su, X.L.; Wang, W.J.; Xia, T.H.; Cao, X.M.; Yuan, S.B.; Wang, W.A.; Zhang, A.; Liu, H.B. NPM1 is a diagnostic and prognostic biomarker associated with the clinicopathological characteristics of gastric cancer. Neoplasma 2022, 20, 965–975. [Google Scholar] [CrossRef]
- Zhou, Y.; Fang, Y.; Zhou, J.; Liu, Y.; Wu, S.; Xu, B. NPM1 is a Novel Therapeutic Target and Prognostic Biomarker for Ewing Sarcoma. Front. Genet. 2021, 12, 771253. [Google Scholar] [CrossRef]
- Carvalho, L.S.; Gonçalves, N.; Fonseca, N.A.; Moreira, J.N. Cancer Stem Cells and Nucleolin as Drivers of Carcinogenesis. Pharmaceuticals 2021, 14, 60. [Google Scholar] [CrossRef]
- Angelov, D.; Bondarenko, V.A.; Almagro, S.; Menoni, H.; Mongélard, F.; Hans, F.; Mietton, F.; Studitsky, V.M.; Hamiche, A.; Dimitrov, S.; et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006, 25, 1669–1679. [Google Scholar] [CrossRef]
- Qi, J.; Li, H.; Liu, N.; Xing, Y.; Zhou, G.; Wu, Y.; Liu, Y.; Chen, W.; Yue, J.; Han, B.; et al. The implications and mechanisms of the extra-nuclear nucleolin in the esophageal squamous cell carcinomas. Med. Oncol. 2015, 32, 45. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Xiong, L.; Yu, L.; Li, R.; Wang, Z.; Ren, B.; Dong, J.; Li, B.; Wang, D. Increased level of nucleolin confers to aggressive tumor progression and poor prognosis in patients with hepatocellular carcinoma after hepatectomy. Diagn. Pathol. 2014, 9, 175. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Zhou, F.; Zhang, Q.; Sun, X.; Shi, X.; Liang, Y.; Wang, X.; Yue, L. Overexpression of nucleolin and different expression sites both related to the prognosis of gastric cancer. APMIS 2013, 121, 919–925. [Google Scholar] [CrossRef]
- Peng, L.; Liang, J.; Wang, H.; Song, X.; Rashid, A.; Gomez, H.F.; Corley, L.J.; Abbruzzese, J.L.; Fleming, J.B.; Evans, D.B.; et al. High levels of nucleolar expression of nucleolin are associated with better prognosis in patients with stage II pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2010, 16, 3734–3742. [Google Scholar] [CrossRef]
- Destouches, D.; Page, N.; Hamma-Kourbali, Y.; Machi, V.; Chaloin, O.; Frechault, S.; Birmpas, C.; Katsoris, P.; Beyrath, J.; Albanese, P.; et al. A simple approach to cancer therapy afforded by multivalent pseudopeptides that target cell-surface nucleoproteins. Cancer Res. 2011, 71, 3296–3305. [Google Scholar] [CrossRef] [PubMed]
- Raineri, F.; Bourgoin-Voillard, S.; Cossutta, M.; Habert, D.; Ponzo, M.; Houppe, C.; Vallée, B.; Boniotto, M.; Chalabi-Dchar, M.; Bouvet, P.; et al. Nucleolin Targeting by N6L Inhibits Wnt/β-Catenin Pathway Activation in Pancreatic Ductal Adenocarcinoma. Cancers 2021, 13, 2986. [Google Scholar] [CrossRef]
- Vasaturo, M.; Cotugno, R.; Fiengo, L.; Vinegoni, C.; Piaz, F.D.; de Tommasi, N. The anti-tumor diterpene oridonin is a direct inhibitor of Nucleolin in cancer cells. Sci. Rep. 2018, 8, 16735. [Google Scholar] [CrossRef]
- Goldshmit, Y.; Trangle, S.S.; Afergan, F.; Iram, T.; Pinkas-Kramarski, R. Nucleolin inhibitor GroA triggers reduction in epidermal growth factor receptor activation: Pharmacological implication for glial scarring after spinal cord injury. J. Neurochem. 2016, 138, 845–858. [Google Scholar] [CrossRef]
- Goldshmit, Y.; Trangle, S.S.; Kloog, Y.; Pinkas-Kramarski, R. Interfering with the interaction between ErbB1, nucleolin and Ras as a potential treatment for glioblastoma. Oncotarget 2014, 5, 8602–8613. [Google Scholar] [CrossRef]
- Luo, Q.; Wang, B.; Wu, Z.; Jiang, W.; Wang, Y.; Du, K.; Zhou, N.; Zheng, L.; Gan, J.; Shen, W.H.; et al. NAP1-Related Protein 1 (NRP1) has multiple interaction modes for chaperoning histones H2A-H2B. Proc. Natl. Acad. Sci. USA 2020, 117, 30391–30399. [Google Scholar] [CrossRef]
- Queiroz, C.J.S.; Song, F.; Reed, K.R.; Al-Khafaji, N.; Clarke, A.R.; Vimalachandran, D.; Miyajima, F.; Pritchard, D.M.; Jenkins, J.R. NAP1L1: A Novel Human Colorectal Cancer Biomarker Derived From Animal Models of Apc Inactivation. Front. Oncol. 2020, 10, 1565. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, Y.; Cui, S.; Song, D.; Li, B.; Chen, Q.; Yao, G.; Gong, B. NAP1L1 interacts with hepatoma-derived growth factor to recruit c-Jun inducing breast cancer growth. Cancer Cell Int. 2021, 21, 605. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xie, Y.; Luo, H.; Song, Y.; Que, T.; Hu, R.; Huang, H.; Luo, K.; Li, C.; Qin, C.; et al. NAP1L1 promotes proliferation and chemoresistance in glioma by inducing CCND1/CDK4/CDK6 expression through its interaction with HDGF and activation of c-Jun. Aging 2021, 13, 26180–26200. [Google Scholar] [CrossRef]
- Guidi, F.; Puglia, M.; Gabbiani, C.; Landini, I.; Gamberi, T.; Fregona, D.; Cinellu, M.A.; Nobili, S.; Mini, E.; Bini, L.; et al. 2D-DIGE analysis of ovarian cancer cell responses to cytotoxic gold compounds. Mol. Biosyst. 2012, 8, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Khosravi-Far, R.; Chang, H.Y.; Baltimore, D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 1997, 89, 1067–1076. [Google Scholar] [CrossRef]
- Gibbons, R.J.; Pellagatti, A.; Garrick, D.; Wood, W.G.; Malik, N.; Ayyub, H.; Langford, C.; Boultwood, J.; Wainscoat, J.S.; Higgs, D.R. Identification of acquired somatic mutations in the gene encoding chromatin-remodeling factor ATRX in the alpha-thalassemia myelodysplasia syndrome (ATMDS). Nat. Genet. 2003, 34, 446–449. [Google Scholar] [CrossRef]
- Hoelper, D.; Huang, H.; Jain, A.Y.; Patel, D.J.; Lewis, P.W. Structural and mechanistic insights into ATRX-dependent and -independent functions of the histone chaperone DAXX. Nat. Commun. 2017, 8, 1193. [Google Scholar] [CrossRef]
- Mahmud, I.; Liao, D. DAXX in cancer: Phenomena, processes, mechanisms and regulation. Nucleic Acids Res. 2019, 47, 7734–7752. [Google Scholar] [CrossRef]
- Jiao, Y.; Shi, C.; Edil, B.H.; de Wilde, R.F.; Klimstra, D.S.; Maitra, A.; Schulick, R.D.; Tang, L.H.; Wolfgang, C.L.; Choti, M.A.; et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 2011, 331, 1199–1203. [Google Scholar] [CrossRef]
- Hackeng, W.M.; Brosens, L.A.A.; Kim, J.Y.; O’Sullivan, R.; Sung, Y.N.; Liu, T.C.; Cao, D.; Heayn, M.; Brosnan-Cashman, J.; An, S.; et al. Non-functional pancreatic neuroendocrine tumours: ATRX/DAXX and alternative lengthening of telomeres (ALT) are prognostically independent from ARX/PDX1 expression and tumour size. Gut 2022, 71, 961–973. [Google Scholar] [CrossRef]
- Dyer, M.A.; Qadeer, Z.A.; Valle-Garcia, D.; Bernstein, E. ATRX and DAXX: Mechanisms and Mutations. Cold Spring Harb. Perspect. Med. 2017, 7, a026567. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.F.; Zhao, Z.G.; Ye, L.L.; Zhuge, W.; Han, Z.; Zhang, T.M.; Ye, S.S.; Chen, W.J.; Zhu, S.; Shi, L.; et al. Prognostic significance of Daxx NCR (Nuclear/Cytoplasmic Ratio) in gastric cancer. Cancer Med. 2017, 6, 2063–2075. [Google Scholar] [CrossRef] [PubMed]
- Ko, T.Y.; Kim, J.I.; Park, E.S.; Mun, J.M.; Park, S.D. The Clinical Implications of Death Domain-Associated Protein (DAXX) Expression. Korean J. Thorac. Cardiovasc. Surg. 2018, 51, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, J.H.; La, M.; Jang, M.J.; Chae, G.W.; Kim, S.B.; Tak, H.; Jung, Y.; Byun, B.; Ahn, J.K.; et al. Inhibition of NF-kappaB acetylation and its transcriptional activity by Daxx. J. Mol. Biol. 2007, 368, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Yang, Y.; Chen, W.; Li, E.; Liang, T. Demethylation at enhancer upregulates MCM2 and NUP37 expression predicting poor survival in hepatocellular carcinoma patients. J. Transl. Med. 2022, 20, 49. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Miyazaki, T.; Fukai, Y.; Nakajima, M.; Sohda, M.; Takita, J.; Masuda, N.; Fukuchi, M.; Manda, R.; Ojima, H.; et al. A new proliferation marker, minichromosome maintenance protein 2, is associated with tumor aggressiveness in esophageal squamous cell carcinoma. J. Surg. Oncol. 2003, 84, 24–30. [Google Scholar] [CrossRef]
- Yang, C.; Wen, Y.; Li, H.; Zhang, D.; Zhang, N.; Shi, X.; Jiang, B.; Ma, X.; Yang, P.; Tang, H.; et al. Overexpression of minichromosome maintenance 2 predicts poor prognosis in patients with gastric cancer. Oncol. Rep. 2012, 27, 135–142. [Google Scholar] [CrossRef]
- Giaginis, C.; Georgiadou, M.; Dimakopoulou, K.; Tsourouflis, G.; Gatzidou, E.; Kouraklis, G.; Theocharis, S. Clinical significance of MCM-2 and MCM-5 expression in colon cancer: Association with clinicopathological parameters and tumor proliferative capacity. Dig. Dis. Sci. 2009, 54, 282–291. [Google Scholar] [CrossRef]
- Gasparian, A.V.; Burkhart, C.A.; Purmal, A.A.; Brodsky, L.; Pal, M.; Saranadasa, M.; Bosykh, D.A.; Commane, M.; Guryanova, O.A.; Pal, S.; et al. Curaxins: Anticancer compounds that simultaneously suppress NF-κB and activate p53 by targeting FACT. Sci. Transl. Med. 2011, 3, 95ra74. [Google Scholar] [CrossRef]
- Song, H.; Xi, S.; Chen, Y.; Pramanik, S.; Zeng, J.; Roychoudhury, S.; Harris, H.; Akbar, A.; Elhag, S.S.; Coulter, D.W.; et al. Histone chaperone FACT complex inhibitor CBL0137 interferes with DNA damage repair and enhances sensitivity of medulloblastoma to chemotherapy and radiation. Cancer Lett. 2021, 520, 201–212. [Google Scholar] [CrossRef]
- De, S.; Lindner, D.J.; Coleman, C.J.; Wildey, G.; Dowlati, A.; Stark, G.R. The FACT inhibitor CBL0137 Synergizes with Cisplatin in Small-Cell Lung Cancer by Increasing NOTCH1 Expression and Targeting Tumor-Initiating Cells. Cancer Res. 2018, 78, 2396–2406. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yin, C.; Fedorov, A.; Qiao, L.; Bao, H.; Beknazarov, N.; Wang, S.; Gautam, A.; Williams, R.M.; Crawford, J.C.; et al. ADAR1 masks the cancer immunotherapeutic promise of ZBP1-driven necroptosis. Nature 2022, 606, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Huang, Q.; Luster, T.A.; Hu, H.; Zhang, H.; Ng, W.L.; Khodadadi-Jamayran, A.; Wang, W.; Chen, T.; Deng, J.; et al. In Vivo Epigenetic CRISPR Screen Identifies Asf1a as an Immunotherapeutic Target in Kras-Mutant Lung Adenocarcinoma. Cancer Discov. 2020, 10, 270–287. [Google Scholar] [CrossRef] [PubMed]

| Gene | Tumor | Role | Effect In Vitro | Effect In Vivo (Mouse Model) | Upstream Events | Downstream Events | Reference Number |
|---|---|---|---|---|---|---|---|
| ASF1A | HCC | promoter | cell senescence, DNA damage | NR | NR | p53/p21cip1 pathway | [14] |
| CRC | promoter | proliferation, migration, stemness | proliferation and migration (metastatic and subcutaneous CDX model) | NR | β-catenin | [13] | |
| CRC | promoter | NR | NR | NR | H4Y72ph/autophagy | [15] | |
| ASF1B | HCC | promoter | proliferation | NR | NR | NR | [16] |
| HCC | promoter | NR | NR | miRNA-214-3p | NR | [17] | |
| HCC | promoter | proliferation, migration | NR | NR | CDK9 stabilization | [18] | |
| GC | promoter | proliferation migration | proliferation (subcutaneous CDX model) | NR | PI3K/AKT/mTOR pathway | [19] | |
| PDAC | promoter | proliferation, migration, DNA damage, chemosensitivity | NR | NR | NR | [20] | |
| PDAC | promoter | proliferation, apoptosis | NR | NR | PI3K/AKT pathway | [21] | |
| SPT16 | HCC | promoter | proliferation, migration, oxidative stress chemosensitivity | proliferation (subcutaneous and orthotopic CDX model) | NRF2/KEAP1 pathway | NRF2 transcription elongation rate | [12] |
| SSRP1 | HCC | promoter | proliferation, migration, oxidative stress chemosensitivity | proliferation (subcutaneous and orthotopic CDX model) | NRF2/KEAP1 pathway | NRF2 transcription elongation rate | [12] |
| HCC | Promoter | proliferation, migration, apoptosis, chemosensitivity | proliferation and migration (metastatic and subcutaneous CDX model) | miR-497 | NR | [22] | |
| HCC | NR | NR | NR | DLG-AS1/miR-497-5p | NR | [23] | |
| HCC | promoter | proliferation | NR | LNC01134/miR-4784 | NR | [24] | |
| GC | promoter | proliferation, migration, apoptosis | NR | NR | AKT pathway | [25] | |
| CRC | promoter | DNA damage, chemosensitivity | chemosensitivity (subcutaneous CDX model) | NR | APE1 acetylation | [26] | |
| CRC | promoter | proliferation, migration | proliferation (subcutaneous CDX model) | NR | AKT pathway | [27] | |
| CRC | promoter | proliferation, migration, chemosensitivity | proliferation (subcutaneous CDX model) | miR-28-5p | MMP9 | [28] | |
| CRC | promoter | NR | NR | LOC101927746/miR-584-3p | NR | [29] | |
| HJURP | HCC | promoter | proliferation | proliferation (subcutaneous CDX model) | NR | MAPK/ERK1/2 and AKT/GSK3β pathways | [9] |
| HCC | promoter | migration | NR | NR | SPHK1 | [30] | |
| HCC | promoter | proliferation | NR | NR | NR | [31] | |
| HCC | promoter | proliferation, migration | NR | NR | NR | [32] | |
| PDAC | promoter | proliferation, migration | proliferation and migration (subcutaneous CDX model) | NR | H3K4me2/MDM2/p53 axis | [33] | |
| CRC | promoter | proliferation, migration | NR | NR | NR | [34] | |
| NPM1 | HCC | promoter | proliferation, chemosensitivity | NR | NR | PTPN14/YAP axis | [35] |
| HCC | promoter | proliferation | NR | NR | ATF5 degradation | [36] | |
| HCC | promoter | proliferation, migration | NR | NR | NR | [37] | |
| HCC | promoter | migration | NR | CDK1 | Rho/ROCK/LIMK pathway | [38] | |
| HCC | promoter | chemosensitivity | NR | NR | BAX mitochondria translocation and oligomerization | [39] | |
| GC | NR | NR | NR | RASSF10 | RNF2/RASSF10 feedback | [40] | |
| PDAC | promoter | proliferation | NR | NR | FBP1 | [41] | |
| PDAC | promoter | NR | NR | AKT | ARF localization | [42] | |
| CRC | promoter | proliferation | NR | DDX27 | NF-κB pathway | [43] | |
| CRC | promoter | proliferation, senescence | NR | NR | NR | [44] | |
| CRC | promoter | proliferation, chemosensitivity | proliferation (subcutaneous PDX model) | NA | PI3K/AKT pathway | [45] | |
| CRC | promoter | chemosensitivity | NR | NR | c-MYC | [46] | |
| CRC | promoter | proliferation, migration | NR | NR | NR | [47] | |
| CRC | promoter | proliferation, oxidative stress | proliferation (subcutaneous CDX model) | NR | CBX3/PRDX6 axis | [48] | |
| EC | NR | NR | NR | SLC25A21-AS1 | c-MYC | [49] | |
| NAP1L1 | HCC | promoter | proliferation | proliferation (subcutaneous CDX model) | NR | HDGF/c-Jun/CCND1 axis | [50] |
| HCC | promoter | proliferation | NR | PRDM8 | PI3K/AKT/mTOR pathway | [11] | |
| HCC | promoter | proliferation, chemosensitivity | proliferation and chemosensitivity (subcutaneous CDX model) | NR | NR | [51] | |
| HCC | promoter | proliferation | NR | Let-7c-5p | PI3K/AKT/mTOR pathway | [52] | |
| PDAC | promoter | proliferation | proliferation (orthotopic CDX model) | NR | NR | [53] | |
| CHAF1A | HCC | promoter | proliferation, apoptosis, | proliferation (subcutaneous CDX model) | NR | NR | [54] |
| GC | promoter | proliferation | proliferation (subcutaneous CDX model) | SP1 | TCF4/c-MYC/CCND1 axis | [55] | |
| GC | promoter | chemosensitivity | NR | NR | Thymidylate synthetase | [56] | |
| CHAF1B | HCC | promoter | proliferation, migration, apoptosis | proliferation (subcutaneous CDX model) | NR | NR | [57] |
| Nucleolin | HCC | promoter | migration | NR | NR | CCL20/CCR6 pathway | [58] |
| HCC | promoter | proliferation, migration | NR | HDGF | PI3K/Akt pathway | [59] | |
| HCC | NR | NR | NR | C20orf204-189AA | NR | [60] | |
| GC | promoter | NR | NR | BTG2/SP1 | TNF-α | [61] | |
| PDAC | promoter | migration | proliferation, migration, and angiogenesis (orthotopic CDA and CDX model) | NR | Endothelial cell activation, Ang-2 secretion | [62] | |
| PDAC | promoter | proliferation | proliferation (orthotopic CDX model) | NR | Autophagy via AMPK pathway | [63] | |
| CRC | promoter | NR | NR | VEGF/PI3K/AKT pathway | EMT pathway | [64] | |
| CRC | NR | NR | NR | P-Selectin Binding Protein | PI3K/p38/MAPK complex formation | [65] | |
| CRC | suppressor | proliferation | NR | LUCAT1 | MYC | [66] | |
| DAXX | GC | promoter | proliferation, migration | NR | RanBP2/RanGAP1 | NR | [67] |
| GC | suppressor | stemness, migration | NR | NR | HDAC-1/SNAIL3 axis | [68] | |
| CRC | suppressor | proliferation, migration | NR | NR | CD24 | [69] | |
| CRC | suppressor | migration | NR | NR | ZEB1 | [70] | |
| CRC | promoter | proliferation | proliferation (subcutaneous CDX model) | PI3KCA/AKT pathway | PI3KCA | [71] | |
| CRC | NR | NR | NR | NR | TCF4 | [72] | |
| ATRX | CRC | promoter | proliferation | NR | JMJD1A | NR | [73] |
| PDAC | suppressor | NR | KRASG12D tumorigenicity (inducible knockout mouse model) | NR | NR | [74] | |
| MCM2 | HCC | promoter | proliferation | NR | miR-34a-5p | NR | [75] |
| CRC | promoter | stemness | NR | miR-195-5p/497-5p | NR | [76] | |
| TONSL | GC | suppressor | proliferation, migration | NR | TONSL-AS1 | NR | [77] |
| RBAP48 | GC | promoter | apoptosis, radiosensitivity | NR | NR | PI3K/AKT pathway | [78] |
| sNASP | HCC | promoter | proliferation | proliferation (subcutaneous CDX model) | NR | H3K9me1 modification | [79] |
| GC | promoter | proliferation, apoptosis | NR | miR-29c | NR | [80] | |
| IPO4 | GC | promoter | proliferation, migration | NR | NR | NR | [81] |
| SPT2 | CRC | promoter | proliferation | NR | NR | NR | [82] |
| SPT6 | CRC | promoter | proliferation, migration, apoptosis, stemness, chemosensitivity | proliferation and migration (metastatic and subcutaneous CDX model) | NR | SND1/hTERT axis | [83] |
| ANP32E | PDAC | promoter | proliferation, migration | NR | NR | β-catenin | [84] |
| PDAC | promoter | proliferation, stemness | proliferation (subcutaneous CDX model) | miR-202-5p | NR | [85] | |
| GC | promoter | proliferation, apoptosis | NR | NR | NR | [86] | |
| VPS72 | HCC | promoter | proliferation, migration | NR | NR | KAT5/PI3K/AKT pathway | [10] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Cai, Z.; Jiang, T.; Han, J.; Zhang, B. Histone Chaperones and Digestive Cancer: A Review of the Literature. Cancers 2022, 14, 5584. https://doi.org/10.3390/cancers14225584
Zhao Z, Cai Z, Jiang T, Han J, Zhang B. Histone Chaperones and Digestive Cancer: A Review of the Literature. Cancers. 2022; 14(22):5584. https://doi.org/10.3390/cancers14225584
Chicago/Turabian StyleZhao, Zhou, Zhaolun Cai, Tianxiang Jiang, Junhong Han, and Bo Zhang. 2022. "Histone Chaperones and Digestive Cancer: A Review of the Literature" Cancers 14, no. 22: 5584. https://doi.org/10.3390/cancers14225584
APA StyleZhao, Z., Cai, Z., Jiang, T., Han, J., & Zhang, B. (2022). Histone Chaperones and Digestive Cancer: A Review of the Literature. Cancers, 14(22), 5584. https://doi.org/10.3390/cancers14225584







