The Potential of MicroRNAs as Non-Invasive Prostate Cancer Biomarkers: A Systematic Literature Review Based on a Machine Learning Approach
Abstract
Simple Summary
Abstract
1. Introduction
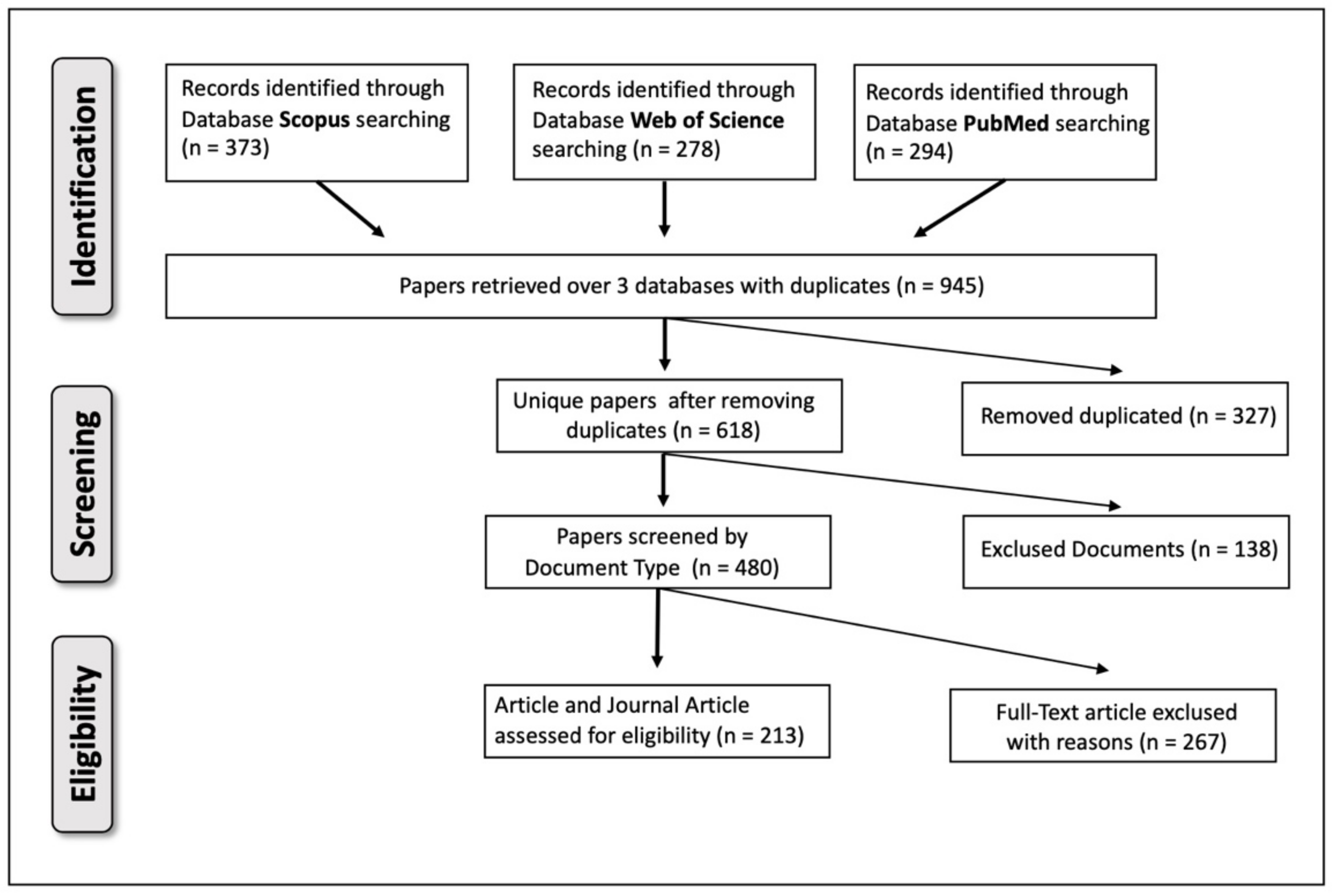
2. Methods
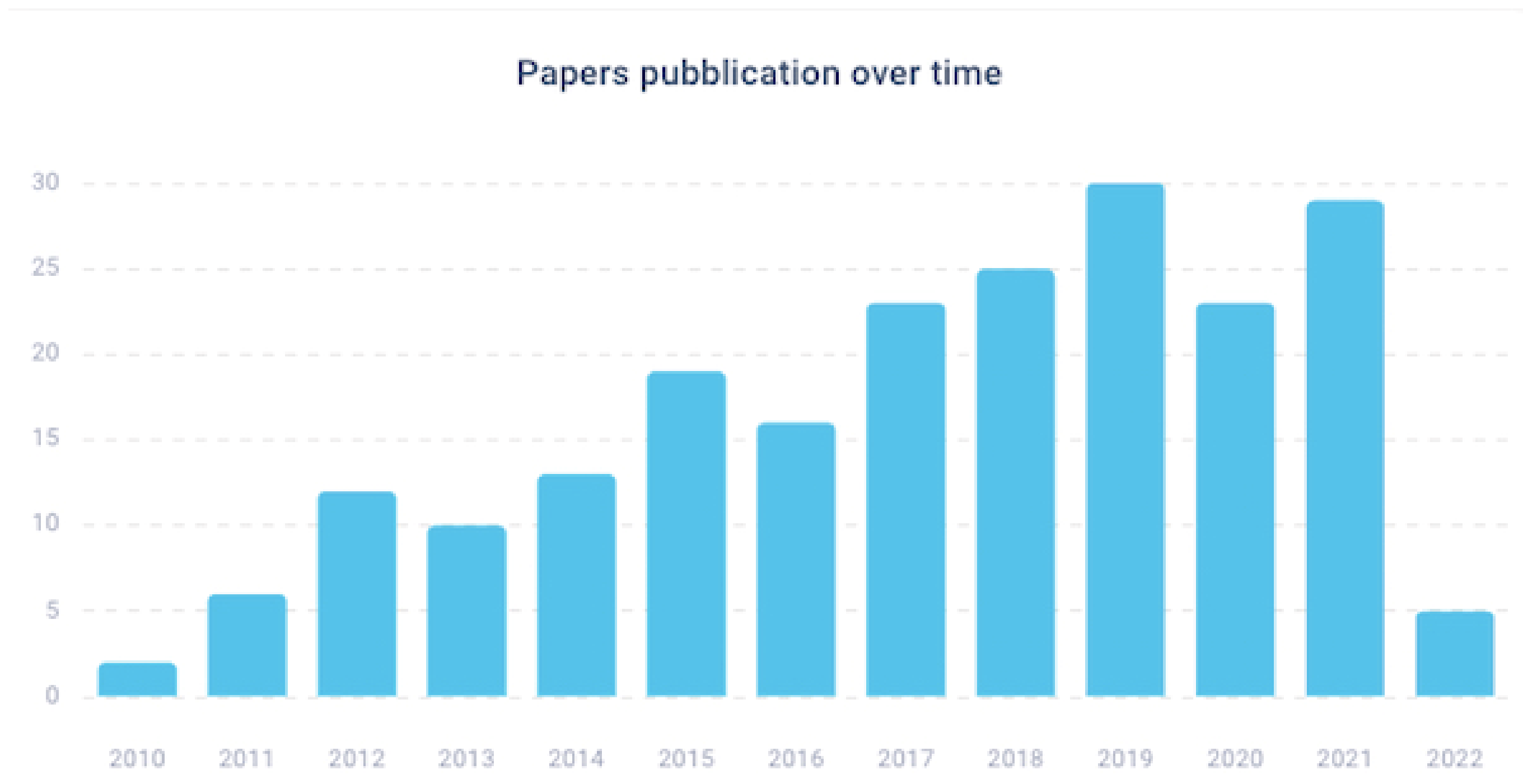
2.1. Paper Location and Selection
2.2. Paper Analysis
- The study was conducted on human cells, tissue or patients with PCa (not xenografts or other animal models).
- The study measured the expression of miRs in serum/plasma/urine or cells/tissues.
- The study investigated the association between prognosis outcomes and miRs expression.
- The study tested the prognostic role of target genes instead of the miR itself.
- The study involved other non-coding RNAs with as yet unknown functions, such as circular RNAs, long non-coding RNAs and small nucleolar RNAs.
- The clinical study lacked key information such as hazard ratio (HR), 95% confidence intervals (CI), p value, and survival curves.
- The study was a review, an editorial article, a meta-analysis, a letter to editors, a short communication, a conference paper, an erratum, a chapter book, a note, a personal opinion and commentary, or a retracted publication.
- k sets of relevant keywords (where each set represents a topic).
- The document-term matrix, i.e., a matrix describing how much each paper is statistically related to a specific topic (namely, the topic proportion).
2.3. Results Presentation
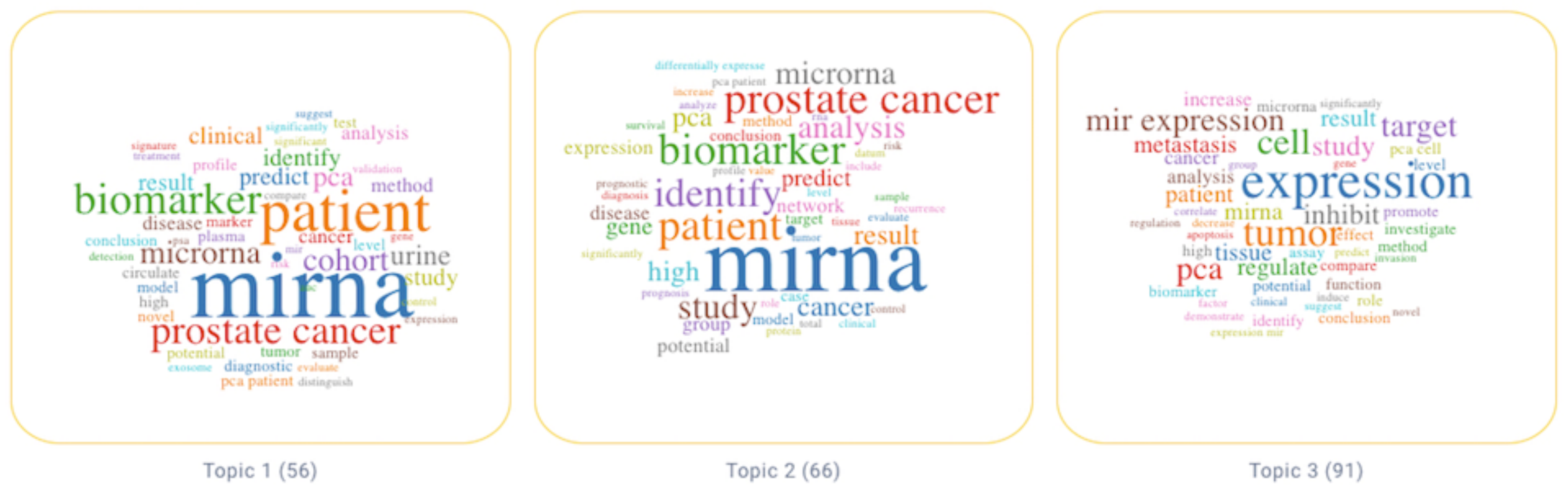
3. Results
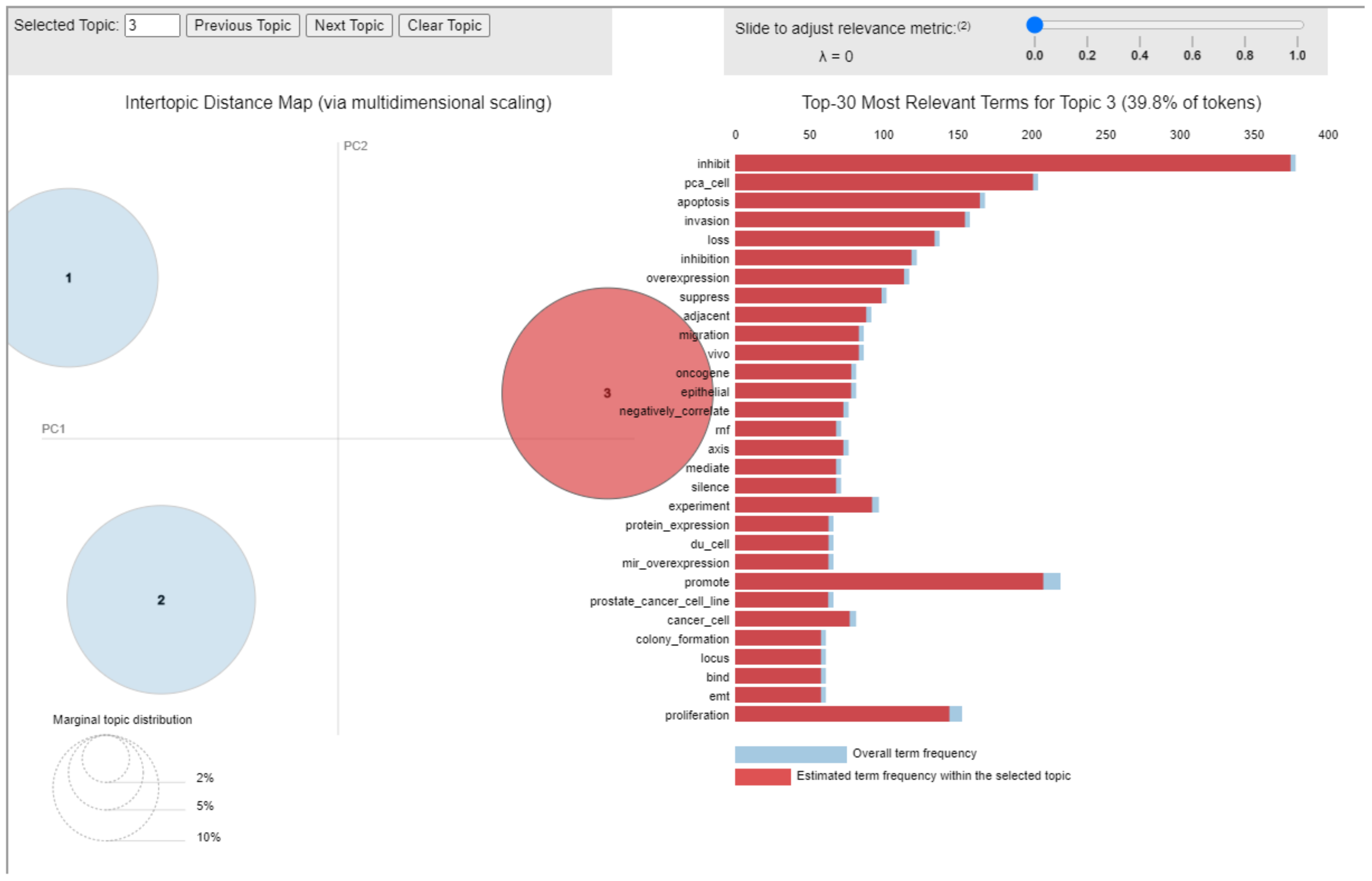
3.1. Topic 3—Study of miRs in Human PCa
3.2. Topic 2—Potential of miRs as Biomarkers in Translational Research of PCa
3.3. Topic 1—Use of miRs as Biomarkers for PCa in the Clinical Setting
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Coronado, G.D.; Argenbright, K.; Brenner, A.T.; Castaneda, S.F.; Dominitz, J.A.; Green, B.; Issaka, R.B.; Levin, T.R.; Reuland, D.S.; et al. Mailed fecal immunochemical test outreach for colorectal cancer screening: Summary of a Centers for Disease Control and Prevention-sponsored Summit. CA Cancer J. Clin. 2020, 70, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Tucci, P.; Agostini, M.; Grespi, F.; Markert, E.K.; Terrinoni, A.; Vousden, K.H.; Muller, P.A.; Dotsch, V.; Kehrloesser, S.; Sayan, B.S.; et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 15312–15317. [Google Scholar] [CrossRef] [PubMed]
- Aghdam, A.M.; Amiri, A.; Salarinia, R.; Masoudifar, A.; Ghasemi, F.; Mirzaei, H. MicroRNAs as Diagnostic, Prognostic, and Therapeutic Biomarkers in Prostate Cancer. Crit. Rev. Eukaryot. Gene Expr. 2019, 29, 127–139. [Google Scholar] [CrossRef]
- Casanova-Salas, I.; Rubio-Briones, J.; Fernandez-Serra, A.; Lopez-Guerrero, J.A. miRNAs as biomarkers in prostate cancer. Clin. Transl. Oncol. 2012, 14, 803–811. [Google Scholar] [CrossRef]
- Moher, D.; Shamseer, L.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A.; Group, P.-P. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 2015, 4, 1. [Google Scholar] [CrossRef]
- Gurbuz, V.; Sozen, S.; Bilen, C.Y.; Konac, E.J.O.L. miR-148a, miR-152 and miR-200b promote prostate cancer metastasis by targeting DNMT1 and PTEN expression. Oncol. Lett. 2021, 22, 1–14. [Google Scholar] [CrossRef]
- Williams, L.V.; Veliceasa, D.; Vinokour, E.; Volpert, O.V.J.P.O. miR-200b inhibits prostate cancer EMT, growth and metastasis. PLoS ONE 2013, 8, e83991. [Google Scholar] [CrossRef]
- Basu, S.; Chaudhary, A.; Chowdhury, P.; Karmakar, D.; Basu, K.; Karmakar, D.; Chatterjee, J.; Sengupta, S. Evaluating the role of hsa-miR-200c in reversing the epithelial to mesenchymal transition in prostate cancer. Gene 2020, 730, 144264. [Google Scholar] [CrossRef]
- Vulli, A.; Srinivasu, P.N.; Sashank, M.S.K.; Shafi, J.; Choi, J.; Ijaz, M.F.J.S. Fine-Tuned DenseNet-169 for Breast Cancer Metastasis Prediction Using FastAI and 1-Cycle Policy. Sensors 2022, 22, 2988. [Google Scholar] [CrossRef]
- Singh, J.; Thakur, D.; Gera, T.; Shah, B.; Abuhmed, T.; Ali, F.J.I.A. Classification and analysis of android malware images using feature fusion technique. IEEE Access 2021, 9, 90102–90117. [Google Scholar] [CrossRef]
- Denyer, D.; Tranfield, D. Producing a systematic review. In The Sage Handbook of Organizational Research Methods; Sage Publications Ltd.: Thousand Oaks, CA, USA, 2009; pp. 671–689. [Google Scholar]
- Petticrew, M. Systematic reviews from astronomy to zoology: Myths and misconceptions. BMJ 2001, 322, 98–101. [Google Scholar] [CrossRef]
- Cumpston, M.; Li, T.; Page, M.J.; Chandler, J.; Welch, V.A.; Higgins, J.P.; Thomas, J. Updated guidance for trusted systematic reviews: A new edition of the Cochrane Handbook for Systematic Reviews of Interventions. Cochrane Database Syst. Rev. 2019, 10, ED000142. [Google Scholar] [CrossRef]
- Ammirato, S.; Felicetti, A.; Rogano, D.; Linzalone, R.; Corvello, V. Digitalising the Systematic Literature Review process: The MySLR platform. Knowl. Manag. Res. Pract. 2022, 1–18. [Google Scholar] [CrossRef]
- Blei, D.M.; Ng, A.Y.; Jordan, M.I. Latent dirichlet allocation. J. Mach. Learn. Res. 2003, 3, 993–1022. [Google Scholar]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, B. Topic Modeling using Topics from Many Domains, Lifelong Learning and Big Data. In Proceedings of the 31st International Conference on Machine Learning, Proceedings of Machine Learning Research, Bejing, China, 22–24 June 2014; pp. 703–711. [Google Scholar]
- Chang, Y.; Deng, Q.; Guan, Z.; Cheng, Y.; Sun, Y. MiR-1273 g-3p Promotes Malignant Progression and has Prognostic Implications in Prostate Cancer. Mol. Biotechnol. 2022, 64, 17–24. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhao, S.; Luo, L.; Xiang, Q.; Zhu, Z.; Wang, J.; Liu, Y.; Luo, J. miR-199b-5p-DDR1-ERK signalling axis suppresses prostate cancer metastasis via inhibiting epithelial-mesenchymal transition. Br. J. Cancer 2021, 124, 982–994. [Google Scholar] [CrossRef]
- Yang, B.; Diao, H.; Wang, P.; Guan, F.; Liu, H. microRNA-877-5p exerts tumor-suppressive functions in prostate cancer through repressing transcription of forkhead box M1. Bioengineered 2021, 12, 9094–9102. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Cheng, B.; Jia, R.; Tan, B.; Liu, W. Altered expression of microRNA-92b-3p predicts survival outcomes of patients with prostate cancer and functions as an oncogene in tumor progression. Oncol. Lett. 2021, 21, 4. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.Q.; He, Y.H.; Wang, S.B.; Yang, S.; Wang, Y.J.; Nan, C.J.; Bao, Y.F.; Xie, Q.P.; Chen, Y.H. MiR-130b/TNF-alpha/NF-kappaB/VEGFA loop inhibits prostate cancer angiogenesis. Clin. Transl. Oncol. 2020, 22, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Lv, C.; Feng, Y.; Yu, S. Activation of the KDM5A/miRNA-495/YTHDF2/m6A-MOB3B axis facilitates prostate cancer progression. J. Exp. Clin. Cancer Res. 2020, 39, 223. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Luo, L.; Xiang, Q.; Wang, J.; Liu, Y.; Deng, Y.; Zhao, Z. MiRNA-671-5p Promotes prostate cancer development and metastasis by targeting NFIA/CRYAB axis. Cell Death Dis. 2020, 11, 949. [Google Scholar] [CrossRef]
- Zang, Y.; Zhu, J.; Li, Q.; Tu, J.; Li, X.; Hu, R.; Yang, D. miR-137-3p Modulates the Progression of Prostate Cancer by Regulating the JNK3/EZH2 Axis. Onco. Targets Ther. 2020, 13, 7921–7932. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, X.; Zhang, Q.; Chen, X. miR-138-5p inhibits the malignant progression of prostate cancer by targeting FOXC1. Cancer Cell Int. 2020, 20, 297. [Google Scholar] [CrossRef]
- Zhao, S.; Jie, C.; Xu, P.; Diao, Y. MicroRNA-140 inhibit prostate cancer cell invasion and migration by targeting YES proto-oncogene 1. J. Cell Biochem. 2020, 121, 482–488. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, J.; Xue, D.; Li, Z.; Liu, Y.; Dong, L. MiR-515-5p acts as a tumor suppressor via targeting TRIP13 in prostate cancer. Int. J. Biol. Macromol. 2019, 129, 227–232. [Google Scholar] [CrossRef]
- Lo, U.G.; Pong, R.C.; Yang, D.; Gandee, L.; Hernandez, E.; Dang, A.; Lin, C.J.; Santoyo, J.; Ma, S.; Sonavane, R.; et al. IFNgamma-Induced IFIT5 Promotes Epithelial-to-Mesenchymal Transition in Prostate Cancer via miRNA Processing. Cancer Res. 2019, 79, 1098–1112. [Google Scholar] [CrossRef]
- Feng, F.; Liu, H.; Chen, A.; Xia, Q.; Zhao, Y.; Jin, X.; Huang, J. miR-148-3p and miR-152-3p synergistically regulate prostate cancer progression via repressing KLF4. J. Cell Biochem. 2019, 120, 17228–17239. [Google Scholar] [CrossRef]
- Zheng, C.; Guo, K.; Chen, B.; Wen, Y.; Xu, Y. miR-214-5p inhibits human prostate cancer proliferation and migration through regulating CRMP5. Cancer Biomark. 2019, 26, 193–202. [Google Scholar] [CrossRef]
- Xing, Q.; Xie, H.; Zhu, B.; Sun, Z.; Huang, Y. MiR-455-5p Suppresses the Progression of Prostate Cancer by Targeting CCR5. Biomed. Res. Int. 2019, 2019, 6394784. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Su, X.P.; Li, Y.N.; Guo, Y.H. MicroRNA-425-5p promotes the development of prostate cancer via targeting forkhead box J3. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 547–554. [Google Scholar] [CrossRef]
- Ray, J.; Hoey, C.; Huang, X.; Jeon, J.; Taeb, S.; Downes, M.R.; Boutros, P.C.; Liu, S.K. MicroRNA198 suppresses prostate tumorigenesis by targeting MIB1. Oncol. Rep. 2019, 42, 1047–1056. [Google Scholar] [CrossRef]
- Ling, X.H.; Fu, H.; Chen, Z.Y.; Lu, J.M.; Zhuo, Y.J.; Chen, J.H.; Zhong, W.D.; Jia, Z. miR505 suppresses prostate cancer progression by targeting NRCAM. Oncol. Rep. 2019, 42, 991–1004. [Google Scholar] [CrossRef]
- Qu, H.W.; Jin, Y.; Cui, Z.L.; Jin, X.B. MicroRNA-373-3p inhibits prostate cancer progression by targeting AKT1. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6252–6259. [Google Scholar] [CrossRef]
- Bhagirath, D.; Yang, T.L.; Bucay, N.; Sekhon, K.; Majid, S.; Shahryari, V.; Dahiya, R.; Tanaka, Y.; Saini, S. microRNA-1246 Is an Exosomal Biomarker for Aggressive Prostate Cancer. Cancer Res. 2018, 78, 1833–1844. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, D.; Lv, J.; Wang, S.; Zhang, Q. miR-410-3p promotes prostate cancer progression via regulating PTEN/AKT/mTOR signaling pathway. Biochem. Biophys Res. Commun. 2018, 503, 2459–2465. [Google Scholar] [CrossRef]
- Xu, S.; Ge, J.; Zhang, Z.; Zhou, W. miR-141 inhibits prostatic cancer cell proliferation and migration, and induces cell apoptosis via targeting of RUNX1. Oncol. Rep. 2018, 39, 1454–1460. [Google Scholar] [CrossRef]
- Li, J.; Fu, F.; Wan, X.; Huang, S.; Wu, D.; Li, Y. Up-regulated miR-29c inhibits cell proliferation and glycolysis by inhibiting SLC2A3 expression in prostate cancer. Gene 2018, 665, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.Y.; Chen, G.; Zhang, Y.Q.; He, H.C.; Liang, Y.X.; Ye, J.H.; Liang, Y.K.; Mo, R.J.; Lu, J.M.; Zhuo, Y.J.; et al. MicroRNA-30d promotes angiogenesis and tumor growth via MYPT1/c-JUN/VEGFA pathway and predicts aggressive outcome in prostate cancer. Mol. Cancer 2017, 16, 48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, X.; Wang, Y.; Peng, R.; Lin, Z.; Wang, Y.; Hu, B.; Wang, J.; Shi, G. Low expression of microRNA-30c promotes prostate cancer cells invasion involved in downregulation of KRAS protein. Oncol. Lett. 2017, 14, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Ayub, S.G.; Kaul, D.; Ayub, T. An androgen-regulated miR-2909 modulates TGFbeta signalling through AR/miR-2909 axis in prostate cancer. Gene 2017, 631, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.M.; McKenna, M.M.; Walsh, C.P.; McKenna, D.J. miR-24 regulates CDKN1B/p27 expression in prostate cancer. Prostate 2016, 76, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tao, T.; Liu, C.; Guan, H.; Huang, Y.; Xu, B.; Chen, M. Downregulation of miR-195 promotes prostate cancer progression by targeting HMGA1. Oncol. Rep. 2016, 36, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Liu, X.; Wang, M. miR-503 suppresses tumor cell proliferation and metastasis by directly targeting RNF31 in prostate cancer. Biochem. Biophys Res. Commun. 2015, 464, 1302–1308. [Google Scholar] [CrossRef]
- Lin, Z.Y.; Huang, Y.Q.; Zhang, Y.Q.; Han, Z.D.; He, H.C.; Ling, X.H.; Fu, X.; Dai, Q.S.; Cai, C.; Chen, J.H.; et al. MicroRNA-224 inhibits progression of human prostate cancer by downregulating TRIB1. Int. J. Cancer 2014, 135, 541–550. [Google Scholar] [CrossRef]
- Arora, S.; Saini, S.; Fukuhara, S.; Majid, S.; Shahryari, V.; Yamamura, S.; Chiyomaru, T.; Deng, G.; Tanaka, Y.; Dahiya, R. MicroRNA-4723 Inhibits Prostate Cancer Growth through Inactivation of the Abelson Family of Nonreceptor Protein Tyrosine Kinases. PLoS ONE 2013, 8, e78023. [Google Scholar] [CrossRef]
- Majid, S.; Dar, A.A.; Saini, S.; Arora, S.; Shahryari, V.; Zaman, M.S.; Chang, I.; Yamamura, S.; Tanaka, Y.; Deng, G.; et al. miR-23b represses proto-oncogene Src kinase and functions as methylation-silenced tumor suppressor with diagnostic and prognostic significance in prostate cancer. Cancer Res. 2012, 72, 6435–6446. [Google Scholar] [CrossRef]
- Saini, S.; Majid, S.; Shahryari, V.; Arora, S.; Yamamura, S.; Chang, I.; Zaman, M.S.; Deng, G.; Tanaka, Y.; Dahiya, R. miRNA-708 control of CD44(+) prostate cancer-initiating cells. Cancer Res. 2012, 72, 3618–3630. [Google Scholar] [CrossRef]
- Santo, G.D.; Frasca, M.; Bertoli, G.; Castiglioni, I.; Cava, C. Identification of key miRNAs in prostate cancer progression based on miRNA-mRNA network construction. Comput. Struct. Biotechnol. J. 2022, 20, 864–873. [Google Scholar] [CrossRef]
- Camargo, J.A.; Lopes, R.E.; Ferreira, G.F.D.; Viana, N.I.; Guimaraes, V.; Leite, K.R.M.; Nahas, W.C.; Srougi, M.; Antunes, A.A.; Reis, S.T. The role of single nucleotide polymorphisms of miRNAs 100 and 146a as prognostic factors for prostate cancer. Int. J. Biol. Markers 2021, 36, 50–56. [Google Scholar] [CrossRef]
- Wang, T.H.; Lee, C.Y.; Lee, T.Y.; Huang, H.D.; Hsu, J.B.; Chang, T.H. Biomarker Identification through Multiomics Data Analysis of Prostate Cancer Prognostication Using a Deep Learning Model and Similarity Network Fusion. Cancers 2021, 13, 2528. [Google Scholar] [CrossRef]
- Liu, H.; Li, L.; Fan, Y.; Lu, Y.; Zhu, C.; Xia, W. Construction of Potential Gene Expression and Regulation Networks in Prostate Cancer Using Bioinformatics Tools. Oxid. Med. Cell Longev. 2021, 2021, 8846951. [Google Scholar] [CrossRef]
- Wei, J.; Yin, Y.; Deng, Q.; Zhou, J.; Wang, Y.; Yin, G.; Yang, J.; Tang, Y. Integrative Analysis of MicroRNA and Gene Interactions for Revealing Candidate Signatures in Prostate Cancer. Front. Genet. 2020, 11, 176. [Google Scholar] [CrossRef]
- Parra-Medina, R.; Lopez-Kleine, L.; Ramirez-Clavijo, S.; Payan-Gomez, C. Identification of candidate miRNAs in early-onset and late-onset prostate cancer by network analysis. Sci. Rep. 2020, 10, 12345. [Google Scholar] [CrossRef]
- Yang, Y.; Jia, B.; Zhao, X.; Wang, Y.; Ye, W. miR-93-5p may be an important oncogene in prostate cancer by bioinformatics analysis. J. Cell Biochem. 2019, 120, 10463–10483. [Google Scholar] [CrossRef]
- Baumann, B.; Acosta, A.M.; Richards, Z.; Deaton, R.; Sapatynska, A.; Murphy, A.; Kajdacsy-Balla, A.; Gann, P.H.; Nonn, L. Association of High miR-182 Levels with Low-Risk Prostate Cancer. Am. J. Pathol. 2019, 189, 911–923. [Google Scholar] [CrossRef]
- Barcelo, M.; Castells, M.; Bassas, L.; Vigues, F.; Larriba, S. Semen miRNAs Contained in Exosomes as Non-Invasive Biomarkers for Prostate Cancer Diagnosis. Sci. Rep. 2019, 9, 13772. [Google Scholar] [CrossRef]
- Ibrahim, N.H.; Abdellateif, M.S.; Thabet, G.; Kassem, S.H.; El-Salam, M.A.; El-Leithy, A.A.; Selim, M.M. Combining PHI and miRNAs as Biomarkers in Prostate Cancer Diagnosis and Prognosis. Clin. Lab. 2019, 65, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Marrone, M.T.; Joshu, C.E.; Peskoe, S.B.; De Marzo, A.M.; Heaphy, C.M.; Lupold, S.E.; Meeker, A.K.; Platz, E.A. Adding the Team into T1 Translational Research: A Case Study of Multidisciplinary Team Science in the Evaluation of Biomarkers of Prostate Cancer Risk and Prognosis. Clin. Chem. 2019, 65, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Xi, Q.; Liu, H.; Guo, X.; Zhang, J.; Zhang, Z.; Li, Y.; Yang, G.; Zhou, D.; Yang, H.; et al. miR-21 promotes NLRP3 inflammasome activation to mediate pyroptosis and endotoxic shock. Cell Death Dis. 2019, 10, 461. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Wu, Y.P.; Yin, H.B.; Xue, X.Y.; Gou, X. Molecular network-based identification of competing endogenous RNAs and mRNA signatures that predict survival in prostate cancer. J. Transl. Med. 2018, 16, 274. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Chen, F.; Shen, L.; Tang, X.; Du, C.; Sun, Z.; Ding, H.; Chen, J.; Shen, B. Biomarker microRNAs for prostate cancer metastasis: Screened with a network vulnerability analysis model. J. Transl. Med. 2018, 16, 134. [Google Scholar] [CrossRef]
- Schmidt, L.; Fredsoe, J.; Kristensen, H.; Strand, S.H.; Rasmussen, A.; Hoyer, S.; Borre, M.; Mouritzen, P.; Orntoft, T.; Sorensen, K.D. Training and validation of a novel 4-miRNA ratio model (MiCaP) for prediction of postoperative outcome in prostate cancer patients. Ann. Oncol. 2018, 29, 2003–2009. [Google Scholar] [CrossRef]
- Zidan, H.E.; Abdul-Maksoud, R.S.; Elsayed, W.S.H.; Desoky, E.A.M. Diagnostic and prognostic value of serum miR-15a and miR-16-1 expression among egyptian patients with prostate cancer. IUBMB Life 2018, 70, 437–444. [Google Scholar] [CrossRef]
- Chen, Z.; Zhan, Y.; Chi, J.; Guo, S.; Zhong, X.; He, A.; Zheng, J.; Gong, Y.; Li, X.; Zhou, L. Using microRNAs as Novel Predictors of Urologic Cancer Survival: An Integrated Analysis. EBioMedicine 2018, 34, 94–107. [Google Scholar] [CrossRef]
- Yan, H.B.; Zhang, Y.; Cen, J.M.; Wang, X.; Gan, B.L.; Huang, J.C.; Li, J.Y.; Song, Q.H.; Li, S.H.; Chen, G. Expression of microRNA-99a-3p in Prostate Cancer Based on Bioinformatics Data and Meta-Analysis of a Literature Review of 965 Cases. Med. Sci. Monit. 2018, 24, 4807–4822. [Google Scholar] [CrossRef]
- Ali, R.; El Tabbakh, S.; El Delgawy, W.; Kotb, A.; Desouky, M.N. microRNA-141 as a diagnostic and prognostic biomarker for prostate cancer in Egyptian population: Pilot study. Afr. J. Urol. 2018, 24, 347–352. [Google Scholar] [CrossRef]
- Souza, M.F.; Kuasne, H.; Barros-Filho, M.C.; Ciliao, H.L.; Marchi, F.A.; Fuganti, P.E.; Paschoal, A.R.; Rogatto, S.R.; Colus, I.M.S. Circulating mRNAs and miRNAs as candidate markers for the diagnosis and prognosis of prostate cancer. PLoS ONE 2017, 12, e0184094. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.M.; Mahon, K.L.; Spielman, C.; Gurney, H.; Mallesara, G.; Stockler, M.R.; Bastick, P.; Briscoe, K.; Marx, G.; Swarbrick, A.; et al. Phase 2 study of circulating microRNA biomarkers in castration-resistant prostate cancer. Br. J. Cancer 2017, 116, 1002–1011. [Google Scholar] [CrossRef]
- Wei, W.; Leng, J.; Shao, H.; Wang, W. MiR-1, a Potential Predictive Biomarker for Recurrence in Prostate Cancer After Radical Prostatectomy. Am. J. Med. Sci. 2017, 353, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Zedan, A.H.; Blavnsfeldt, S.G.; Hansen, T.F.; Nielsen, B.S.; Marcussen, N.; Pleckaitis, M.; Osther, P.J.S.; Sorensen, F.B. Heterogeneity of miRNA expression in localized prostate cancer with clinicopathological correlations. PLoS ONE 2017, 12, e0179113. [Google Scholar] [CrossRef] [PubMed]
- Waseem, M.; Ahmad, M.K.; Srivatava, V.K.; Rastogi, N.; Serajuddin, M.; Kumar, S.; Mishra, D.P.; Sankhwar, S.N.; Mahdi, A.A. Evaluation of miR-711 as Novel Biomarker in Prostate Cancer Progression. Asian Pac. J. Cancer Prev. 2017, 18, 2185–2191. [Google Scholar] [CrossRef]
- Cochetti, G.; Poli, G.; Guelfi, G.; Boni, A.; Egidi, M.G.; Mearini, E. Different levels of serum microRNAs in prostate cancer and benign prostatic hyperplasia: Evaluation of potential diagnostic and prognostic role. Onco Targets Ther. 2016, 9, 7545–7553. [Google Scholar] [CrossRef]
- Yang, B.; Liu, Z.; Ning, H.; Zhang, K.; Pan, D.; Ding, K.; Huang, W.; Kang, X.L.; Wang, Y.; Chen, X. MicroRNA-21 in peripheral blood mononuclear cells as a novel biomarker in the diagnosis and prognosis of prostate cancer. Cancer Biomark. 2016, 17, 223–230. [Google Scholar] [CrossRef]
- Sun, Y.; Jia, X.; Hou, L.; Liu, X. Screening of Differently Expressed miRNA and mRNA in Prostate Cancer by Integrated Analysis of Transcription Data. Urology 2016, 94, 313.e1–313.e6. [Google Scholar] [CrossRef]
- Huang, Z.; Zhang, L.; Yi, X.; Yu, X. Diagnostic and prognostic values of tissue hsa-miR-30c and hsa-miR-203 in prostate carcinoma. Tumour Biol. 2016, 37, 4359–4365. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Liang, M.; Du, M.; Xia, S.; Dittmar, R.; Wang, D.; See, W.; Costello, B.A.; Quevedo, F.; et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 2015, 67, 33–41. [Google Scholar] [CrossRef]
- Xiaoli, Z.; Yawei, W.; Lianna, L.; Haifeng, L.; Hui, Z. Screening of Target Genes and Regulatory Function of miRNAs as Prognostic Indicators for Prostate Cancer. Med. Sci. Monit. 2015, 21, 3748–3759. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, W.; Kang, X.L.; Cen, S.; Wang, Y.; Chen, X. High-Level Expression of microRNA-21 in Peripheral Blood Mononuclear Cells Is a Diagnostic and Prognostic Marker in Prostate Cancer. Genet. Test Mol. Biomark. 2015, 19, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Peskoe, S.B.; Ribas, J.; Rafiqi, F.; Kudrolli, T.; Meeker, A.K.; De Marzo, A.M.; Platz, E.A.; Lupold, S.E. Investigation of miR-21, miR-141, and miR-221 expression levels in prostate adenocarcinoma for associated risk of recurrence after radical prostatectomy. Prostate 2014, 74, 1655–1662. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Goldberger, H.; Dimtchev, A.; Marian, C.; Soldin, O.; Li, X.; Collins, S.P.; Suy, S.; Kumar, D. Circulatory miR-628-5p is downregulated in prostate cancer patients. Tumour Biol. 2014, 35, 4867–4873. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.P.; Levesque, E.; Guillemette, C.; Yu, C.C.; Huang, C.Y.; Lin, V.C.; Chung, I.C.; Chen, L.C.; Laverdiere, I.; Lacombe, L.; et al. Genetic variants in microRNAs and microRNA target sites predict biochemical recurrence after radical prostatectomy in localized prostate cancer. Int. J. Cancer 2014, 135, 2661–2667. [Google Scholar] [CrossRef] [PubMed]
- Melbo-Jorgensen, C.; Ness, N.; Andersen, S.; Valkov, A.; Donnem, T.; Al-Saad, S.; Kiselev, Y.; Berg, T.; Nordby, Y.; Bremnes, R.M.; et al. Stromal expression of MiR-21 predicts biochemical failure in prostate cancer patients with Gleason score 6. PLoS ONE 2014, 9, e113039. [Google Scholar] [CrossRef]
- Santos, J.I.; Teixeira, A.L.; Dias, F.; Maurício, J.; Lobo, F.; Morais, A.; Medeiros, R. Influence of peripheral whole-blood microRNA-7 and microRNA-221 high expression levels on the acquisition of castration-resistant prostate cancer: Evidences from in vitro and in vivo studies. Tumor Biol. 2014, 35, 7105–7113. [Google Scholar] [CrossRef]
- Selth, L.A.; Townley, S.L.; Bert, A.G.; Stricker, P.D.; Sutherland, P.D.; Horvath, L.G.; Goodall, G.J.; Butler, L.M.; Tilley, W.D. Circulating microRNAs predict biochemical recurrence in prostate cancer patients. Br. J. Cancer 2013, 109, 641–650. [Google Scholar] [CrossRef]
- Mavridis, K.; Stravodimos, K.; Scorilas, A. Downregulation and prognostic performance of microRNA 224 expression in prostate cancer. Clin. Chem. 2013, 59, 261–269. [Google Scholar] [CrossRef]
- Chen, Z.H.; Zhang, G.L.; Li, H.R.; Luo, J.D.; Li, Z.X.; Chen, G.M.; Yang, J. A panel of five circulating microRNAs as potential biomarkers for prostate cancer. Prostate 2012, 72, 1443–1452. [Google Scholar] [CrossRef]
- Alshalalfa, M.; Bader, G.D.; Goldenberg, A.; Morris, Q.; Alhajj, R. Detecting microRNAs of high influence on protein functional interaction networks: A prostate cancer case study. BMC Syst. Biol. 2012, 6, 112. [Google Scholar] [CrossRef]
- Watahiki, A.; Wang, Y.; Morris, J.; Dennis, K.; O’Dwyer, H.M.; Gleave, M.; Gout, P.W.; Wang, Y. MicroRNAs associated with metastatic prostate cancer. PLoS ONE 2011, 6, e24950. [Google Scholar] [CrossRef]
- Shin, S.; Park, Y.H.; Jung, S.H.; Jang, S.H.; Kim, M.Y.; Lee, J.Y.; Chung, Y.J. Urinary exosome microRNA signatures as a noninvasive prognostic biomarker for prostate cancer. NPJ Genom. Med. 2021, 6, 45. [Google Scholar] [CrossRef]
- Jeon, J.; Olkhov-Mitsel, E.; Xie, H.; Yao, C.Q.; Zhao, F.; Jahangiri, S.; Cuizon, C.; Scarcello, S.; Jeyapala, R.; Watson, J.D.; et al. Temporal Stability and Prognostic Biomarker Potential of the Prostate Cancer Urine miRNA Transcriptome. J. Natl. Cancer Inst. 2020, 112, 247–255. [Google Scholar] [CrossRef]
- Mello-Grand, M.; Bruno, A.; Sacchetto, L.; Cristoni, S.; Gregnanin, I.; Dematteis, A.; Zitella, A.; Gontero, P.; Peraldo-Neia, C.; Ricotta, R.; et al. Two Novel Ceramide-Like Molecules and miR-5100 Levels as Biomarkers Improve Prediction of Prostate Cancer in Gray-Zone PSA. Front. Oncol. 2021, 11, 769158. [Google Scholar] [CrossRef]
- Rajendiran, S.; Maji, S.; Haddad, A.; Lotan, Y.; Nandy, R.R.; Vishwanatha, J.K.; Chaudhary, P. MicroRNA-940 as a Potential Serum Biomarker for Prostate Cancer. Front. Oncol. 2021, 11, 628094. [Google Scholar] [CrossRef]
- Giglio, S.; De Nunzio, C.; Cirombella, R.; Stoppacciaro, A.; Faruq, O.; Volinia, S.; Baldassarre, G.; Tubaro, A.; Ishii, H.; Croce, C.M.; et al. A preliminary study of micro-RNAs as minimally invasive biomarkers for the diagnosis of prostate cancer patients. J. Exp. Clin. Cancer Res. 2021, 40, 79. [Google Scholar] [CrossRef]
- Kim, S.; Park, S.; Cho, Y.S.; Kim, Y.; Tae, J.H.; No, T.I.; Shim, J.S.; Jeong, Y.; Kang, S.H.; Lee, K.H. Electrical Cartridge Sensor Enables Reliable and Direct Identification of MicroRNAs in Urine of Patients. ACS Sens. 2021, 6, 833–841. [Google Scholar] [CrossRef]
- Nayak, B.; Khan, N.; Garg, H.; Rustagi, Y.; Singh, P.; Seth, A.; Dinda, A.K.; Kaushal, S. Role of miRNA-182 and miRNA-187 as potential biomarkers in prostate cancer and its correlation with the staging of prostate cancer. Int. Braz. J. Urol. 2020, 46, 614–623. [Google Scholar] [CrossRef]
- Mercadal, M.; Herrero, C.; Lopez-Rodrigo, O.; Castells, M.; de la Fuente, A.; Vigues, F.; Bassas, L.; Larriba, S. Impact of Extracellular Vesicle Isolation Methods on Downstream Mirna Analysis in Semen: A Comparative Study. Int. J. Mol. Sci. 2020, 21, 5949. [Google Scholar] [CrossRef]
- Fredsoe, J.; Rasmussen, A.K.I.; Mouritzen, P.; Borre, M.; Orntoft, T.; Sorensen, K.D. A five-microRNA model (pCaP) for predicting prostate cancer aggressiveness using cell-free urine. Int. J. Cancer 2019, 145, 2558–2567. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Peng, J.H. Increased Expression of miR-494 in Serum of Patients with Prostate Cancer and its Potential Diagnostic Value. Clin. Lab. 2019, 65. [Google Scholar] [CrossRef] [PubMed]
- Fredsoe, J.; Rasmussen, A.K.I.; Thomsen, A.R.; Mouritzen, P.; Hoyer, S.; Borre, M.; Orntoft, T.F.; Sorensen, K.D. Diagnostic and Prognostic MicroRNA Biomarkers for Prostate Cancer in Cell-free Urine. Eur. Urol. Focus 2018, 4, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Endzelins, E.; Berger, A.; Melne, V.; Bajo-Santos, C.; Sobolevska, K.; Abols, A.; Rodriguez, M.; Santare, D.; Rudnickiha, A.; Lietuvietis, V.; et al. Detection of circulating miRNAs: Comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer 2017, 17, 730. [Google Scholar] [CrossRef] [PubMed]
- Foj, L.; Ferrer, F.; Serra, M.; Arevalo, A.; Gavagnach, M.; Gimenez, N.; Filella, X. Exosomal and Non-Exosomal Urinary miRNAs in Prostate Cancer Detection and Prognosis. Prostate 2017, 77, 573–583. [Google Scholar] [CrossRef]
- Torres-Ferreira, J.; Ramalho-Carvalho, J.; Gomez, A.; Menezes, F.D.; Freitas, R.; Oliveira, J.; Antunes, L.; Bento, M.J.; Esteller, M.; Henrique, R.; et al. MiR-193b promoter methylation accurately detects prostate cancer in urine sediments and miR-34b/c or miR-129-2 promoter methylation define subsets of clinically aggressive tumors. Mol. Cancer 2017, 16, 26. [Google Scholar] [CrossRef]
- Daniunaite, K.; Dubikaityte, M.; Gibas, P.; Bakavicius, A.; Rimantas Lazutka, J.; Ulys, A.; Jankevicius, F.; Jarmalaite, S. Clinical significance of miRNA host gene promoter methylation in prostate cancer. Hum. Mol. Genet. 2017, 26, 2451–2461. [Google Scholar] [CrossRef]
- Rodríguez-Báez, A.; Comoto-Santacruz, D.A.; Huerta-Núñez, L.F.E.; Campos-Saucedo, J.G.; Estrada-Carrasco, C.E.J. Circulating miRNA expression as a clinical option for the detection of prostate cancer. Rev. Mex. Urol. 2017, 77, 199–206. [Google Scholar]
- Alhasan, A.H.; Scott, A.W.; Wu, J.J.; Feng, G.; Meeks, J.J.; Thaxton, C.S.; Mirkin, C.A. Circulating microRNA signature for the diagnosis of very high-risk prostate cancer. Proc. Natl. Acad. Sci. USA 2016, 113, 10655–10660. [Google Scholar] [CrossRef]
- Stuopelyte, K.; Daniunaite, K.; Jankevicius, F.; Jarmalaite, S. Detection of miRNAs in urine of prostate cancer patients. Medicina 2016, 52, 116–124. [Google Scholar] [CrossRef]
- Wang, J.; Ye, H.; Zhang, D.; Hu, Y.; Yu, X.; Wang, L.; Zuo, C.; Yu, Y.; Xu, G.; Liu, S. MicroRNA-410-5p as a potential serum biomarker for the diagnosis of prostate cancer. Cancer Cell Int. 2016, 16, 12. [Google Scholar] [CrossRef]
- Salido-Guadarrama, A.I.; Morales-Montor, J.G.; Rangel-Escareno, C.; Langley, E.; Peralta-Zaragoza, O.; Cruz Colin, J.L.; Rodriguez-Dorantes, M. Urinary microRNA-based signature improves accuracy of detection of clinically relevant prostate cancer within the prostate-specific antigen grey zone. Mol. Med. Rep. 2016, 13, 4549–4560. [Google Scholar] [CrossRef]
- Li, Z.; Ma, Y.Y.; Wang, J.; Zeng, X.F.; Li, R.; Kang, W.; Hao, X.K. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther. 2016, 9, 139–148. [Google Scholar] [CrossRef]
- Kachakova, D.; Mitkova, A.; Popov, E.; Popov, I.; Vlahova, A.; Dikov, T.; Christova, S.; Mitev, V.; Slavov, C.; Kaneva, R. Combinations of serum prostate-specific antigen and plasma expression levels of let-7c, miR-30c, miR-141, and miR-375 as potential better diagnostic biomarkers for prostate cancer. DNA Cell Biol. 2015, 34, 189–200. [Google Scholar] [CrossRef]
- Wach, S.; Al-Janabi, O.; Weigelt, K.; Fischer, K.; Greither, T.; Marcou, M.; Theil, G.; Nolte, E.; Holzhausen, H.J.; Stohr, R.; et al. The combined serum levels of miR-375 and urokinase plasminogen activator receptor are suggested as diagnostic and prognostic biomarkers in prostate cancer. Int. J. Cancer 2015, 137, 1406–1416. [Google Scholar] [CrossRef]
- Guzel, E.; Karatas, O.F.; Semercioz, A.; Ekici, S.; Aykan, S.; Yentur, S.; Creighton, C.J.; Ittmann, M.; Ozen, M. Identification of microRNAs differentially expressed in prostatic secretions of patients with prostate cancer. Int. J. Cancer 2015, 136, 875–879. [Google Scholar] [CrossRef]
- Casanova-Salas, I.; Rubio-Briones, J.; Calatrava, A.; Mancarella, C.; Masia, E.; Casanova, J.; Fernandez-Serra, A.; Rubio, L.; Ramirez-Backhaus, M.; Arminan, A.; et al. Identification of miR-187 and miR-182 as biomarkers of early diagnosis and prognosis in patients with prostate cancer treated with radical prostatectomy. J. Urol. 2014, 192, 252–259. [Google Scholar] [CrossRef]
- Bryant, R.J.; Pawlowski, T.; Catto, J.W.; Marsden, G.; Vessella, R.L.; Rhees, B.; Kuslich, C.; Visakorpi, T.; Hamdy, F.C. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 2012, 106, 768–774. [Google Scholar] [CrossRef]
- Brase, J.C.; Johannes, M.; Schlomm, T.; Falth, M.; Haese, A.; Steuber, T.; Beissbarth, T.; Kuner, R.; Sultmann, H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int. J. Cancer 2011, 128, 608–616. [Google Scholar] [CrossRef]
- Moltzahn, F.; Olshen, A.B.; Baehner, L.; Peek, A.; Fong, L.; Stoppler, H.; Simko, J.; Hilton, J.F.; Carroll, P.; Blelloch, R. Microfluidic-based multiplex qRT-PCR identifies diagnostic and prognostic microRNA signatures in the sera of prostate cancer patients. Cancer Res. 2011, 71, 550–560. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.J. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]



| Year | Ref. | miRNAs | Main Regulatory Effect | Clinicopathological Data | Category | Number of Patients | p Value |
|---|---|---|---|---|---|---|---|
| 2022 | [21] | miR-1273 g-3p | Promotes tumor progression by increasing cell proliferation, migration and invasion | Age | ≤65; >65 | 54; 51 | 0.378 |
| PSA (ng/mL) | <10; ≥10 | 51; 54 | 0.284 | ||||
| Differentiation | Poor; well moderate | 48; 57 | 0.139 | ||||
| Gleason score | ≤7; >7 | 55; 50 | 0.024 | ||||
| TNM stage | I-II; III-IV | 57; 48 | 0.008 | ||||
| Clinical stage | T1- T2 | 54; 51 | 0.015 | ||||
| Lymph node metastasis | No; Yes | 56; 49 | 0.014 | ||||
| 2021 | [22] | miR-199-5p | Suppresses PCa metastasis by inhibiting EMT pathway | Age | ≤65; >65 | 58; 36 | 0.424 |
| PSA (ng/mL) | <10; 10–20; >20 | 27; 30; 37 | 0.061 | ||||
| Gleason score | <7; =7; >7 | 38;28; 28 | <0.001 | ||||
| Clinical stage | ≤T2; ≥T3 | 65; 29 | 0.278 | ||||
| Lymph node involvement | Neg; Pos | 89; 5 | 0.636 | ||||
| Distal metastases | Yes; No | 38; 56 | 0.003 | ||||
| [23] | miR-877-5p | Suppresses PCa via forkhead box M1 | Age | <60; ≥65 | 53; 48 | 0.940 | |
| Tumor size | ≤3; >3 | 62; 39 | 0.849 | ||||
| PSA (ng/mL) | ≤10; >10 | 65; 36 | 0.196 | ||||
| Surgical margin | Neg; Pos | 72; 29 | 0.433 | ||||
| Prostate volume (ml) | ≤50, >50 | 66; 35 | 0.878 | ||||
| TNM stage | I–II; III | 54; 47 | 0.011 | ||||
| Gleason score | ≤7; >7 | 68; 33 | 0.047 | ||||
| [24] | miR-92b-3p | Suppresses PCa by inhibiting cell proliferation, migration, and invasion | Age | <60; ≥60 | 41; 67 | 0.685 | |
| PSA (ng/mL) | <10; ≥10 | 34; 74 | 0.009 | ||||
| Bone metastasis | Neg; Pos | 55; 53 | 0.033 | ||||
| Gleason score | ≤7; >7 | 59; 49 | 0.001 | ||||
| 2020 | [25] | miR-130b | Inhibits PCa angiogenesis via TNF-α/NF-kB/VEGFA axis | N/A | N/A | N/A | N/A |
| [26] | miR-495 | Promotes cancer progression via KDM5A/miRNA-495/YTHDF2/m6A-MOB3B axis | N/A | N/A | N/A | N/A | |
| [27] | miR-671-5p | Promotes PCa development and metastasis via NFIA/CRYAB axis | Age | ≤72; >72 | 23; 17 | 0.31 | |
| Clinical stage | T2; T3–T4 | 26; 14 | 0.75 | ||||
| Gleason score | <7; ≥7 | 5; 35 | 0.70 | ||||
| Lymph node metastasis | N0; N1 | 28; 12 | 0.42 | ||||
| Distant metastasis | M0; M1 | 25; 15 | 0.004 | ||||
| [28] | miR-137- 3p | Inhibits PCa progression via JNK3/EZH2 axis | Gleason score | 6; 7; 8 | 4; 10; 6 | N/A | |
| Grading System | 1, 2, 3, 4 | 6; 4; 4; 6 | |||||
| Tumor Stage | I; II; III, IV | 4; 9; 6; 1 | |||||
| [29] | miR-138-5p | Inhibits PCa progression via FOXC1 | Age | <60; ≥60 | 24; 36 | 0.830 | |
| Tumor size (cm) | <4; ≥4 | 28; 32 | 0.526 | ||||
| Gleason score | ≤7; >7 | 40; 20 | 0.025 | ||||
| Lymph node metastasis | No; Yes | 37; 23 | 0.009 | ||||
| Bone metastasis | No; Yes | 35; 25 | 0.109 | ||||
| [30] | miR-140 | Inhibits PCa cell invasion and migration via YES proto-oncogene 1 | N/A | N/A | N/A | N/A | |
| 2019 | [31] | miR-515-5p | Inhibits PCa progression via TRIP13 | Gleason score | <7; =7; >7 | 45; 15; 36 | <1 |
| Clinical stage | T1–T2; T3–T4 | 41; 55 | 0.007 | ||||
| [32] | miR-106a-363 cluster | Inhibits PCa progression by inhibiting IFNϒ pathway | N/A | N/A | N/A | N/A | |
| [33] | miR-148—3p miR-152-3p | Inhibits PCa progression by repressing KLF4 | Age | ≤65; >65 | 26; 16 | N/A | |
| PSA (ng/mL) | ≤10; >10 | 23; 19 | |||||
| Tumor size (mm) | ≤20; >20 | 20; 22 | |||||
| pT-stage | pT2; pT3a; pT3b | 19; 16; 7 | |||||
| Gleason grade | 1; 2; 3; 4; 5 | 12; 16; 3; 5; 6 | |||||
| [34] | miR-214-5p | Inhibits PCa proliferation and migration by increasing levels of CRMP5 | N/A | N/A | N/A | N/A | |
| [35] | miR-455-5p | Suppresses PCa progression by targeting CCR5 | Gleason score | <7; =7; >7 | 32; 55; 19 | <0.001 | |
| PSA (ng/mL) | ≤10; >10 | 44; 63 | 0.006 | ||||
| [36] | miR-425-5p | Promotes PCa development by targeting FOXJ3 | N/A | N/A | N/A | N/A | |
| [37] | miR-198 | Suppresses PCa by targeting MIB1 | Gleason score | <7; >7 | 149; 13 | 0.02 | |
| [38] | miR-505 | Suppresses PCa progression by targeting NRCAM | Tissue | Cancer; non cancer | 50; 23 | 0.432 | |
| Age | ≤60; >60 | 12; 68 | 0.331 | ||||
| Gleason score | ≤7; >7 | 31; 19 | 0.032 | ||||
| Pathological grade | ≤2; >2 | 4; 46 | 0.010 | ||||
| Tumor stage | T1; T2-T4 | 29; 21 | 0.351 | ||||
| Lymph node metastasis | N0; N1 | 43; 7 | 15; 2 | ||||
| Distant metastasis | M0; M1 | 44; 6 | 0.093 | ||||
| 2018 | [39] | miR-373-3p | Inhibits PCa progression by targeting AKT1 | N/A | N/A | N/A | N/A |
| [40] | miR-1246 | Inhibits PCa cell proliferation, invasiveness, and migration via EMT pathway | Pathological stage | pT2a; pT2b; pT3; pT4 | 3; 17; 11; 36 | 0.002 | |
| Gleason score | ≤7; >7 | 31; 34 | 0.7263 | ||||
| Lymph node metastasis | Yes; No | 25; 43 | 0.0436 | ||||
| Age | 40–59; 60–79 | 18; 49 | 0.9576 | ||||
| Serum PSA | ≤6.78; >6.78 | 32; 33 | 0.1778 | ||||
| Race | White; Black | 61; 7 | 0.3528 | ||||
| [41] | miR-410-3p | Promotes PCa progression by regulating PTEN/AKT/mTOR signaling pathway | Age | <70; ≥70 | 16; 26 | 0.596 | |
| Metastasis | No; Yes | 9; 33 | 0.001 | ||||
| Gleason score | <7; ≥7 | 19; 23 | 0.006 | ||||
| Clinical stage | T1; T2-T3 | 17; 25 | 0.003 | ||||
| PSA levels (ng/mL) | <10; ≥10 | 20; 22 | 0.370 | ||||
| [42] | miR-141 | Inhibits PCa cell proliferation, migration, and induces cell apoptosis by targeting RUNX1 | N/A | N/A | N/A | N/A | |
| [43] | miR-29c | Inhibits PCa cell proliferation and glycolysis by inhibiting SLC2A3 expression | Metastasis | No; Yes | 127; 14 | <0.001 | |
| Gleason score | <7; 8; 9 | 227; 49; 76 | <0.001 | ||||
| Pathological stage | II; IIIa; IIIb; IV | 148; 31; 81; 4 | <0.01 | ||||
| 2017 | [44] | miR-30d | Promotes angiogenesis and tumor growth via MYPT1/c-JUN/VEGFA pathway | Age | <66; ≥66 | 93; 20 | 0.613 |
| PSA levels (ng/mL) | <4; ≥4 | 22; 89 | 0.003 | ||||
| Gleason score | <8; ≥8 | 87; 19 | 0.010 | ||||
| Clinical stage | <T2a; ≥T2a | 65; 44 | 0.007 | ||||
| Pathological stage | T2a–T2c; T3a–T4 | 71; 37 | 0.004 | ||||
| Metastasis | No; Yes | 94; 19 | 0.001 | ||||
| Overall survival | Alive; Died | 99; 14 | 0.097 | ||||
| [45] | miR-30c | Promotes PCa cells invasion by downregulating KRAS protein | N/A | N/A | N/A | N/A | |
| [46] | miR-2909 | Promotes oncogenic functions by attenuating TGFβ signaling | N/A | N/A | N/A | N/A | |
| 2016 | [47] | miR-24 | Inhibits PCa by regulating CDKN1B/p27 | Age | ≥41; ≤67 | N/A | N/A |
| PSA levels (ng/mL) | ≥4.5; ≤17.7 | ||||||
| Gleason score | ≥5; ≤8 | ||||||
| TMN | pT2; pT3a | ||||||
| [48] | miR-195 | Promotes PCa progression by targeting HMGA1 | N/A | N/A | N/A | N/A | |
| 2015 | [49] | miR-503 | Suppresses PCa cell proliferation and metastasis by targeting RNF31 | Age | <70; ≥70 | 77; 63 | 0.886 |
| Lymph node metastasis | No; yes | 124; 16 | 0.051 | ||||
| Clinical stage | T1; T2–T3 | 85; 55 | 0.004 | ||||
| Gleason score | <7; =7; >7 | 65; 34; 41 | <0.001 | ||||
| PSA levels | <4; 4–10; >10 | 6; 45; 79 | <0.001 | ||||
| 2014 | [50] | miR-224 | Inhibits PCa progression by targeting TRIB1 | Age | <66; ≥66 | 89; 25 | 0.08 |
| PSA levels (ng/mL) | <4; ≥4 | 24; 90 | 0.02 | ||||
| Gleason score | <8; ≥8; | 91; 23 | 0.09 | ||||
| Clinical stage | <T2a; ≥T2a | 66; 48 | 0.04 | ||||
| Pathological stage | T2a; T2c; T3a–T4 | 76; 38 | 0.08 | ||||
| Metastasis | No; Yes | 91; 23 | <0.001 | ||||
| 2013 | [51] | miR-4723 | Inhibits PCa growth by targeting AbI kinase | Gleason grade | 4–6; 7; 8–10 | 51; 33; 14 | N/A |
| Pathological stage | pT2; pT3 | 65; 19 | |||||
| Biochemical recurrence | Yes; no | 47; 42 | |||||
| 2012 | [52] | miR-23b | Suppresses PCa by repressing proto-oncogene Src kinase | Pathological stage | pT2; pT3–pT4 | 89; 61 | <1 × 10−4 |
| Gleason score | 4–6; 7; 8–10 | 46; 52; 34 | <0.001 | ||||
| Biochemical recurrence | yes | 36 | <1 × 10−4 | ||||
| [53] | miR-708 | Promotes PCa progression by regulating CD44+ and AKT2 | Age | 40–59; 60–89 | 33; 68 | 0.6557 | |
| Gleason score | 4–6; 7; 8–10 | 36; 41; 23 | 8 × 10−4 | ||||
| Pathological stage | pT2; pT3; pT4 | 57; 42; 2 | 0.1095 | ||||
| Biochemical recurrence | Yes; No | 24; 76 | 0.0138 | ||||
| [5] | miR-205 | Inhibits PCa cell migration and metastasis via the EMT pathway | Metastasis | N/A | N/A | <1 × 10−5 | |
| Lymph node involvement | <0.01 | ||||||
| Biochemical recurrence | 0.00191 | ||||||
| Gleason score | |||||||
| PSA levels |
| Year | Ref. | miRNAs | Bioinformatic Analysis | Clinicopathological Data | Category | Number of Patients | p Value |
|---|---|---|---|---|---|---|---|
| 2022 | [54] | miR-25-3p, miR-93-3p, miR-122-5p, miR-183-5p, miR-615-3p, miR-7-5p, miR-375 and miR-92a-3p | N/A | Gleason score | 6–9 | 493 | <0.01 |
| 2021 | [55] | miR-146a | N/A | Gleason score | <7; ≥7 | 100 | 0.43 |
| Clinical stage | pT2; pT3 | 0.004 | |||||
| mi-100 | Biochemical recurrence | Yes; No | 0.011 | ||||
| PSA levels (ng/mL) | <10; ≥10 | 0.003 | |||||
| [56] | miR-143, miR-378a | N/A | Overall | 494 188; 290; 9 46; 245; 64; 136; 3 | 5.33 × 10−9 <0.001 | ||
| TNM stage | 2; 3; 4 | ||||||
| Gleason score | 6; 7; 8; 9; 10 | ||||||
| C-index | 0.684 | ||||||
| [57] | 70 miRs | PANTHER | N/A | N/A | N/A | N/A | |
| 2020 | [58] | miR-17-5p, miR-20a-5p, miR-92a-3p, miR-93-5p | Cancer Genome Atlas | N/A | N/A | N/A | N/A |
| [59] | 69 miRNAs | CytoHubba | Clinical stage Gleason score PSA levels Race | T2 7 ≤10 | 49 | N/A | |
| 2019 | [60] | miR-93-5p | Cancer Genome Atlas and Gene Ontology | N/A | N/A | N/A | N/A |
| [61] | miR-182 | N/A | Age | 60 | 133 | ||
| Race/ethnicity | White, black | 0.97 | |||||
| Gleason score | 5–9 | 0.22 | |||||
| Clinical stage | T2–T3 | 0.16 | |||||
| [62] | miR-142-3p, miR-142-5p, miR-223-3p, miR-342-3p, miR-374b-5p | N/A | PSA levels | 4–10 ng/mL | 24 | 0.02 | |
| Gleason score | 6–8 | ||||||
| Clinical stage | T1–T3 | ||||||
| [63] | miR-21, miR-221 | N/A | Gleason score | 6–10 | 100 | <0.01 | |
| Clinical stage | T1–T4 | ||||||
| [64] | miR-21, miR-141, miR-221 | N/A | N/A | N/A | N/A | N/A | |
| [65] | miR-21 | Gene Expression Omnibus | N/A | N/A | N/A | N/A | |
| 2018 | [66] | 13 miRNAs | miRcode, Gene Ontology | N/A | N/A | 499 | <0.05 |
| [67] | miR-101-3p, miR-145-5p, miR-204-5p, miR-198, miR-152 | Gene expression omnibus | N/A | N/A | 142 | <0.05 | |
| [68] | miR-23a, miR-10b-5p, miR-133a, miR-374-5p | N/A | Age | 65 | 123 | ||
| Clinical stage | T2–T3 | ||||||
| Gleason score | I–V | ||||||
| [69] | miR-15a, miR-16-1 | N/A | Age | 65 | 70 | 0.02, 0.007 0.001 | |
| Gleason score | ≤7, >7 | ||||||
| Clinical stage | ≤T2, >T2 | ||||||
| [70] | miR-21 | Cancer genome Atlas; Gene Expression Omnibus | Clinical stage | T2–T4 | N/A | <0.001, | |
| Tumor stage | III–IV | ||||||
| PSA levels | ≥10 | ||||||
| Gleason score | ≥7 | ||||||
| [71] | miR-99a-3p | Cancer genome Atlas; Gene Expression Omnibus, Kyoto Encyclopedia of Genes and Genomes | Age | <60, ≥60 | 201, 203 | 0.917 | |
| Race | White, black, Asian | 146, 7, 2 | 0.759 | ||||
| Stage | I–II, III–IV | 187, 300 | 0.347 | ||||
| [72] | miR-141 | N/A | Age | 67 | 30 | 0.389 | |
| PSA levels (ng/mL) | 30 | <0.001, | |||||
| Prostate volume (g) | 89 | <0.001, | |||||
| 2017 | [73] | miR-200c, miR-200b | N/A | Age | <65, ≥65 | 30; 72 | 0.007 |
| Ancestry | Caucasian; African | 79; 23 | 0.94 | ||||
| Smoking habit | Yes; No | 31; 71 | 0.96 | ||||
| Alcohol consumption | Yes; No | 58; 44 | 0.55 | ||||
| Family history of cancer | Yes; Yes, prostate; No | 62; 16; 40 | 0.09 | ||||
| [74] | 14 miRNAs | N/A | Age | N/A | 89 | N/A | |
| PSA levels | |||||||
| Metastasis | |||||||
| [75] | miR-1 | N/A | N/A | N/A | 78 | <0.001 | |
| [76] | miR-21, miR-34a, miR-125b, miR-126, miR-143, miR-145 | N/A | Age | 52–71 | 49 | 0.016 | |
| PSA level | <10; 10–20; >20 | 28; 17; 4 | |||||
| Gleason score | ≤6; 7; ≥8 | 19; 28; 2 | |||||
| Clinical stage | T2a; T2b; T2c; T3a; T3b | 4; 5; 32; 3; 5 | |||||
| [77] | miR-711 | GO, KEGG | Age | <60, ≥60 | 13; 61 | <0.05 | |
| Smoking habits | Yes; No | 49; 25 | |||||
| PSA levels | Median; High | 23; 51 | |||||
| Gleason score | 7; ≥7 | 31; 43 | |||||
| 2016 | [78] | Let-7c, let-7e, let-7i, miR-26a-5p, miR-26b-5p, miR-24-3p, miR-23b-3p, miR-27-b-3p, miR-106a-5p, miR-20b-5p, miR-18b-5p, miR-19b-2-5p, miR-363-3p, miR-497, miR-195, miR-25-3p, miR-30c-5p, miR-622, miR-874-3p, miR-346, miR-940 | N/A | Age | >75 | 64 | N/A |
| PSA levels (ng/mL) | >3, <10 | ||||||
| Gleason score | 6; 7; 8 | 35; 15; 4 | |||||
| Clinical stage | cT1c; cT2a; T2b; T2c | 40; 4; 8; 12 | |||||
| [79] | miR-21 | N/A | N/A | N/A | 92 | <0.05 | |
| [80] | miR-301a | GEO | 197 | <0.01 | |||
| [81] | miR-30c; miR-2013 | N/A | Age | <50; >50 | 21; 23 | 0.091 | |
| TNM stage | I + II; III + IV | 19; 25 | 0.039 | ||||
| Metastatic status | Yes; No | 18; 26 | <0.001 | ||||
| Clinical stage | T1 + T2; T3; T4 | 17; 14; 13 | 0.0167 | ||||
| 2015 | [82] | miR-1290 miR-375 | N/A | Overall | 23 | N/A | |
| Gleason score | 7; 8; 9 | 10; 4; 9 | |||||
| Clinical stage | T1-T4 | 23 | |||||
| [83] | miR-29a, miR-10a, miR-221 | TCGA, GO, KEGG | 551 | 1 × 10−5 | |||
| [84] | miR-21 | N/A | Clinical stage | N/A | 75 | <0.001 | |
| Lymph node metastasis | |||||||
| Tumor differentiation | |||||||
| 2014 | [85] | miR-21; miR-141; miR-221 | N/A | Age | 58.5 ± 7 | 59 | 0.0149; <1 × 10−4; 2 × 10−4 |
| Race | White | ||||||
| Pathologic stage | T2–T3 | ||||||
| Gleason score | 6–8 | ||||||
| [86] | miR-628-5p | N/A | N/A | N/A | 36 | <1 × 10−4 | |
| [87] | miR-605 | N/A | PSA levels (ng/mL) | 2–7; 8–10 | 846 | <0.001 | |
| Gleason score | <0.001 | ||||||
| Pathologic stage | <0.001 | ||||||
| Surgical margin | <0.001 | ||||||
| [88] | miR-21 | N/A | Age | ≤65; <65 | 357; 176 | ||
| Clinical stage | T2; T3a; T3b | 324; 114; 47 | <0.001 | ||||
| PSA levels (ng/mL) | <10; >10 | 308; 221 | <0.001 | ||||
| Gleason score | 6; 7; 8; >8 | 183; 300; 19; 33 | <0.001 | ||||
| Tumor size (mm) | 0–20; >20 | 250; 285 | <0.001 | ||||
| [89] | miR-7 | N/A | N/A | N/A | N/A | 0.012; | |
| miR-221 | 0.002 | ||||||
| miR-222 | 0.002 | ||||||
| 2013 | [90] | miR-141, miR-146b-3p, mir-194 | N/A | Age | 60 | 16 | 0.0857 |
| PSA levels | <10; ≥10 | 11; 5 | 0.0282 | ||||
| Gleason score | 7; 9 | 12; 4 | 0.001 | ||||
| Clinical stage | T2-T3 | 6; 10 | 0.001 | ||||
| [91] | miR-224 | N/A | Overall | 9.23 ± 0.69 64.8 ± 0.74 | 73 | ||
| PSA levels (ng/mL) | |||||||
| Age | |||||||
| Gleason score | <0.001 | ||||||
| Clinical stage | 0.005 | ||||||
| 2012 | [92] | Let-7e, let-7c, miR-346; miR-622, miR-940, miR-1285 | N/A | Age | 73 ± 8 | 105 | 0.18 <0.001 |
| PSA levels (ng/mL) | 0–4; 4.1–20; >20 | 42; 28; 31 | |||||
| Gleason score | 6–7; 8–9 | 62; 37 | |||||
| [93] | miR-96, miR-182, miR-143 | GEO | N/A | N/A | N/A | N/A | |
| 2011 | [94] | miR-16, miR-34a, miR-126, miR-145, miR-205 | MicroCosm, KEGG | N/A | N/A | N/A | 0.001 |
| Year | Ref. | miRNAs | Biological Fluid | Number of Patients | Interval Time of miRNA Processed after Sample Collection |
|---|---|---|---|---|---|
| 2021 | [95] | miR-21, miR-16, miR-142-3p, miR-451, miR-636 | Urine | 149 | Exosomes isolation from samples and incubation overnight at 4 °C |
| [96] | miR-3195, let-7b-5p, miR-144-3p, miR-451, miR-148a-3p, miR-512-5p, miR-431-5p | Urine | 149 | Overnight at −80 °C | |
| [97] | miR-5100 | Plasma | 102 | After 1 h | |
| [98] | miR-940 | Serum and urine | 32 | N/A | |
| [99] | miR4732-3p, mir-98-5p, miR-let-7a-5p, miR-26b-5p, miR-21-5p | Plasma | 290 | N/A | |
| [100] | miR-21, miR-1246, miR-let-7b | Urine | 10 | After 20 min | |
| 2020 | [101] | miR-182, miR-187 | Urine | 63 | N/A |
| [102] | miR-142-3p, miR-142-5p, miR-223-3p | Semen | 7 | After 30 min at 37 °C | |
| 2019 | [103] | miR-151 a-5p, miR-204-5p, miR-222-3p, miR-23b-3p, miR-331-3p | Urine | 215 | Fresh urine samples |
| [104] | miR-494 | Serum | 90 | N/A | |
| 2018 | [105] | miR-222-3p miR-24-3p/miR-30c-5p | Urine | 215 | N/A |
| 2017 | [106] | miR-375, miR-200c-3p, miR-21-5p, let-7a-5p | Plasma | 50 | Within 2 h |
| [107] | miR-21, miR-141, miR-214, miR-375, let-7c | Urine | 60 | Stored at 4 °C and processed within 4 h | |
| [108] | miR-193b | Tissue and urine | 180 | Fresh samples | |
| [109] | miR-155, miR-152, miR-137 and miR-31 | Tissue | 129 | N/A | |
| [110] | miR-32-5p, miR-455-4p, miR-184, miR-31-5p, miR-200b-3p, miR-19b-3p, miR-34a-5p, miR-32-5p, miR-143-5p, miR-200b-3p and miR-375 | Blood | 34 | Fresh samples | |
| 2016 | [111] | miR-200c, miR-605, miR-135a, miR-433 and miR-106a | Serum | 16 | N/A |
| [112] | miR-21, miR-19a and miR-19b | Urine | 143 | within half an hour | |
| [113] | miR-410-5p | Serum | 149 | within 1 h | |
| [114] | miR-100, miR-200b | Urine | Samples were stored at −80 °C for one week and further processed | ||
| 2015 | [115] | miR-141 | Serum | 11 | 1 h at room temperature |
| [116] | let-7c, miR-30c, miR-141 and miR-375 | Plasma | 11 | 1 h | |
| [117] | miR-375 | Serum | 146 | Samples stored at −80 °C until RNA isolation | |
| [118] | miR-133b, miR-221, miR-361-3p | Prostate secretion samples | 23 | Samples stored at −80 °C until RNA isolation | |
| 2014 | [119] | miR-187 and miR-182 | Tissue and urine | 92 | Fresh samples |
| 2012 | [120] | miR-107 and miR-574-3p | Plasma and urine | 78 and 135 | 10 min plasma samples; stored at 4 °C for up 4 h urine samples |
| 2011 | [121] | miR-375 and miR-141 | Serum | 71 | 30 min at room temperature |
| [122] | 384 human miRNAs | Serum | 36 | N/A | |
| 2008 | [123] | miR-100, miR-125b, miR-141, miR-143, miR-205, miR296 | Plasma and serum | 25 | Within 2 h |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bevacqua, E.; Ammirato, S.; Cione, E.; Curcio, R.; Dolce, V.; Tucci, P. The Potential of MicroRNAs as Non-Invasive Prostate Cancer Biomarkers: A Systematic Literature Review Based on a Machine Learning Approach. Cancers 2022, 14, 5418. https://doi.org/10.3390/cancers14215418
Bevacqua E, Ammirato S, Cione E, Curcio R, Dolce V, Tucci P. The Potential of MicroRNAs as Non-Invasive Prostate Cancer Biomarkers: A Systematic Literature Review Based on a Machine Learning Approach. Cancers. 2022; 14(21):5418. https://doi.org/10.3390/cancers14215418
Chicago/Turabian StyleBevacqua, Emilia, Salvatore Ammirato, Erika Cione, Rosita Curcio, Vincenza Dolce, and Paola Tucci. 2022. "The Potential of MicroRNAs as Non-Invasive Prostate Cancer Biomarkers: A Systematic Literature Review Based on a Machine Learning Approach" Cancers 14, no. 21: 5418. https://doi.org/10.3390/cancers14215418
APA StyleBevacqua, E., Ammirato, S., Cione, E., Curcio, R., Dolce, V., & Tucci, P. (2022). The Potential of MicroRNAs as Non-Invasive Prostate Cancer Biomarkers: A Systematic Literature Review Based on a Machine Learning Approach. Cancers, 14(21), 5418. https://doi.org/10.3390/cancers14215418








