The Inflammasomes Adaptor Protein PYCARD Is a Potential Pyroptosis Biomarker Related to Immune Response and Prognosis in Clear Cell Renal Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Processing and Sample Collection
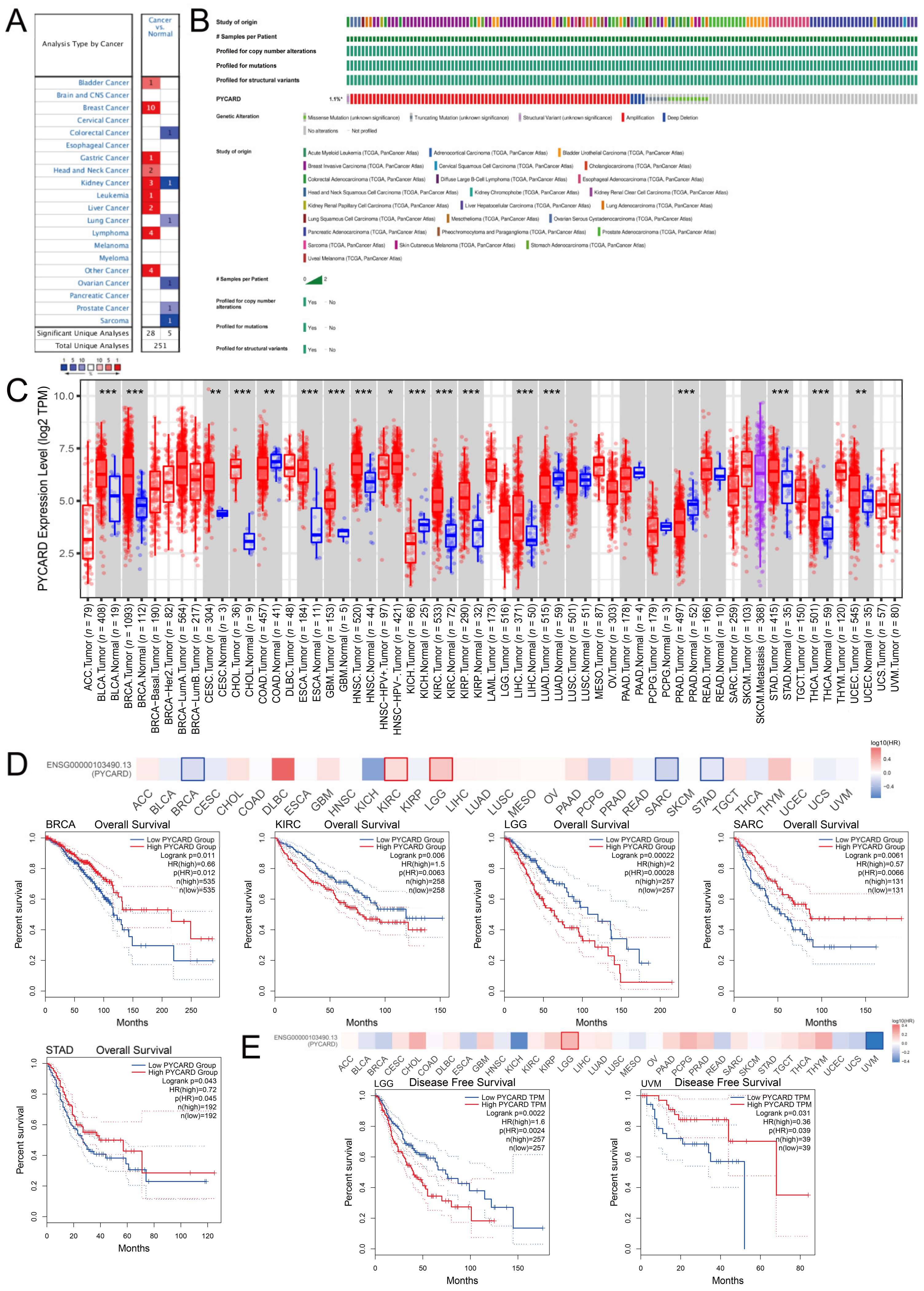
2.2. PYCARD Expression, Genomic Alterations and Prognostic Value in Human Cancers
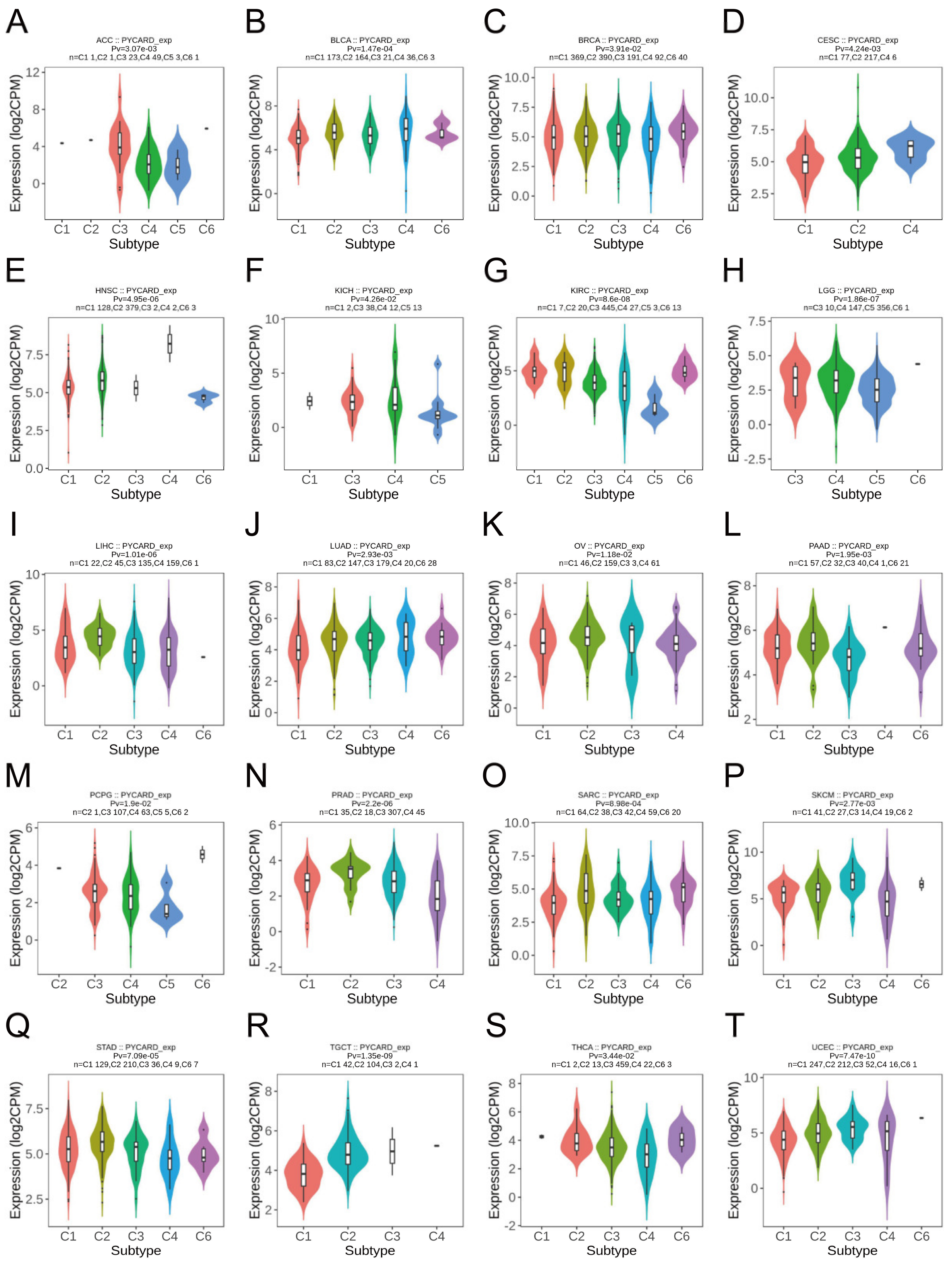
2.3. PYCARD Expression Varied in Different Immune Subtypes
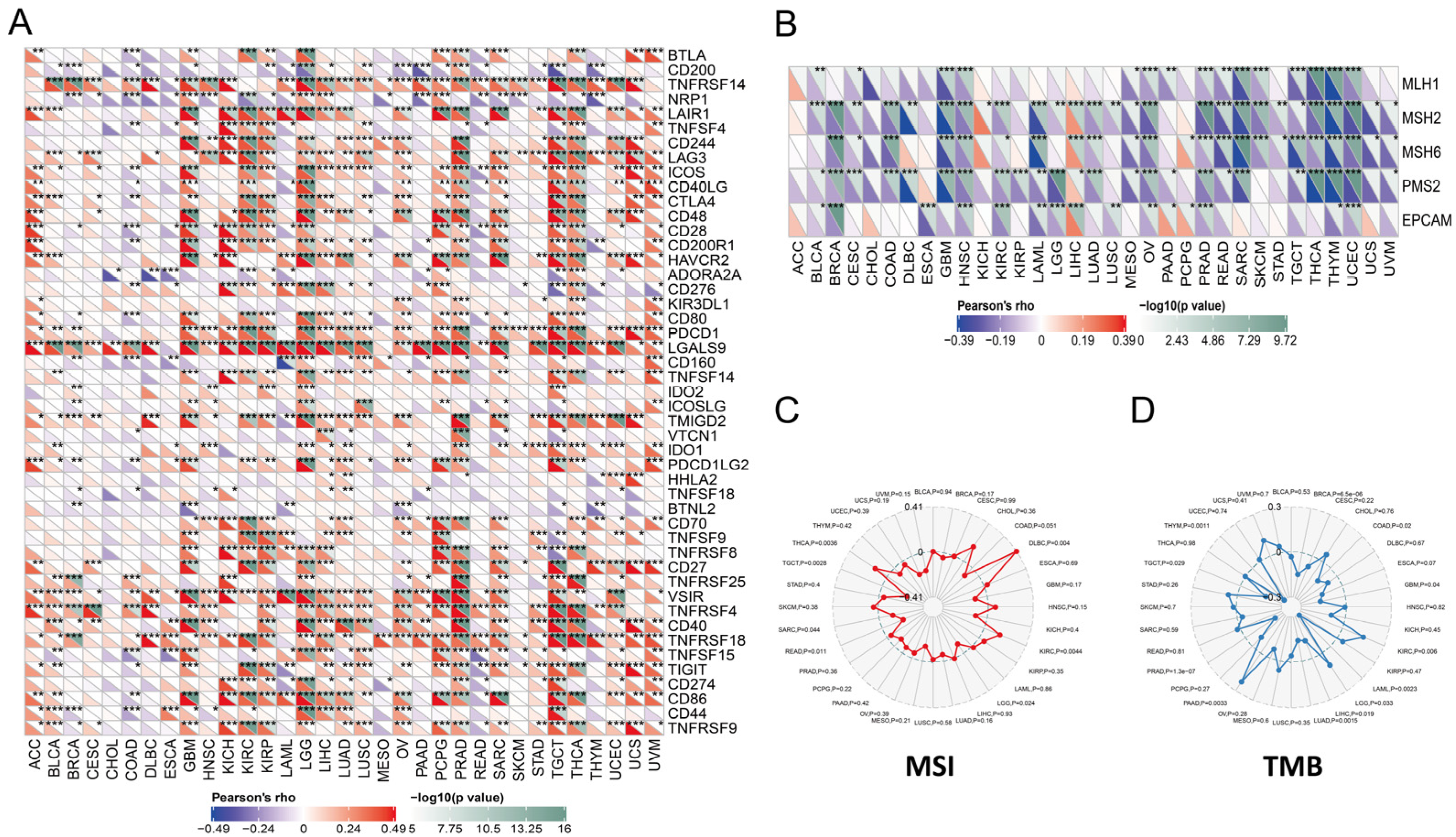
2.4. Relationships between PYCARD Expression and Immune Checkpoint (ICP) Genes, Mis-Match Repair (MMR) Genes, Microsatellite Instability (MSI), Tumor Mutational Burden (TMB), and ESTIMATE Scores
2.5. PYCARD Expression and Survival Analysis in ccRCC
2.6. Immunohistochemical (IHC) Staining Analysis
2.7. Real-Time Quantitative PCR (RT-qPCR) Analysis
2.8. Immunotherapy Response and Single-Cell Analysis of PYCARD
2.9. Analysis of Co-Expression and Protein–Protein Interaction (PPI) Networks
2.10. PYCARD Subgroups Analysis
2.11. PYCARD Expression and Drug Response
2.12. Statistical Analysis
3. Results
3.1. PYCARD Expression, Genomic Alterations and Prognostic Ability in Human Cancers
3.2. PYCARD Expression Varied in Different Immune Subtypes
3.3. PYCARD Expression Correlated to Immune Checkpoint (ICP) Genes, Mis-Match Repair (MMR) Genes, Microsatellite Instability (MSI), Tumor Mutational Burden (TMB), and ESTIMATE Scores
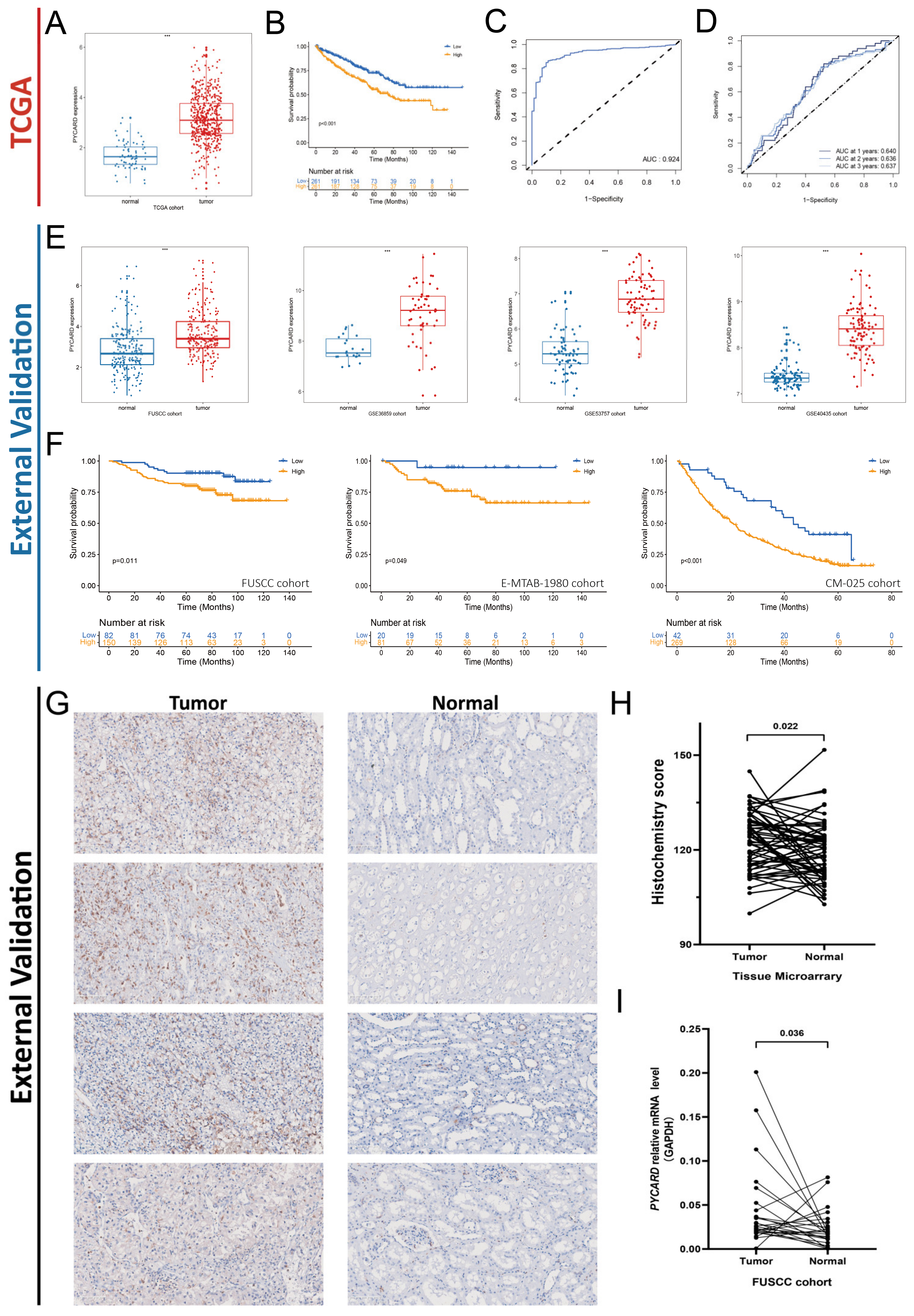
3.4. PYCARD Expression in ccRCC External Validation Cohorts
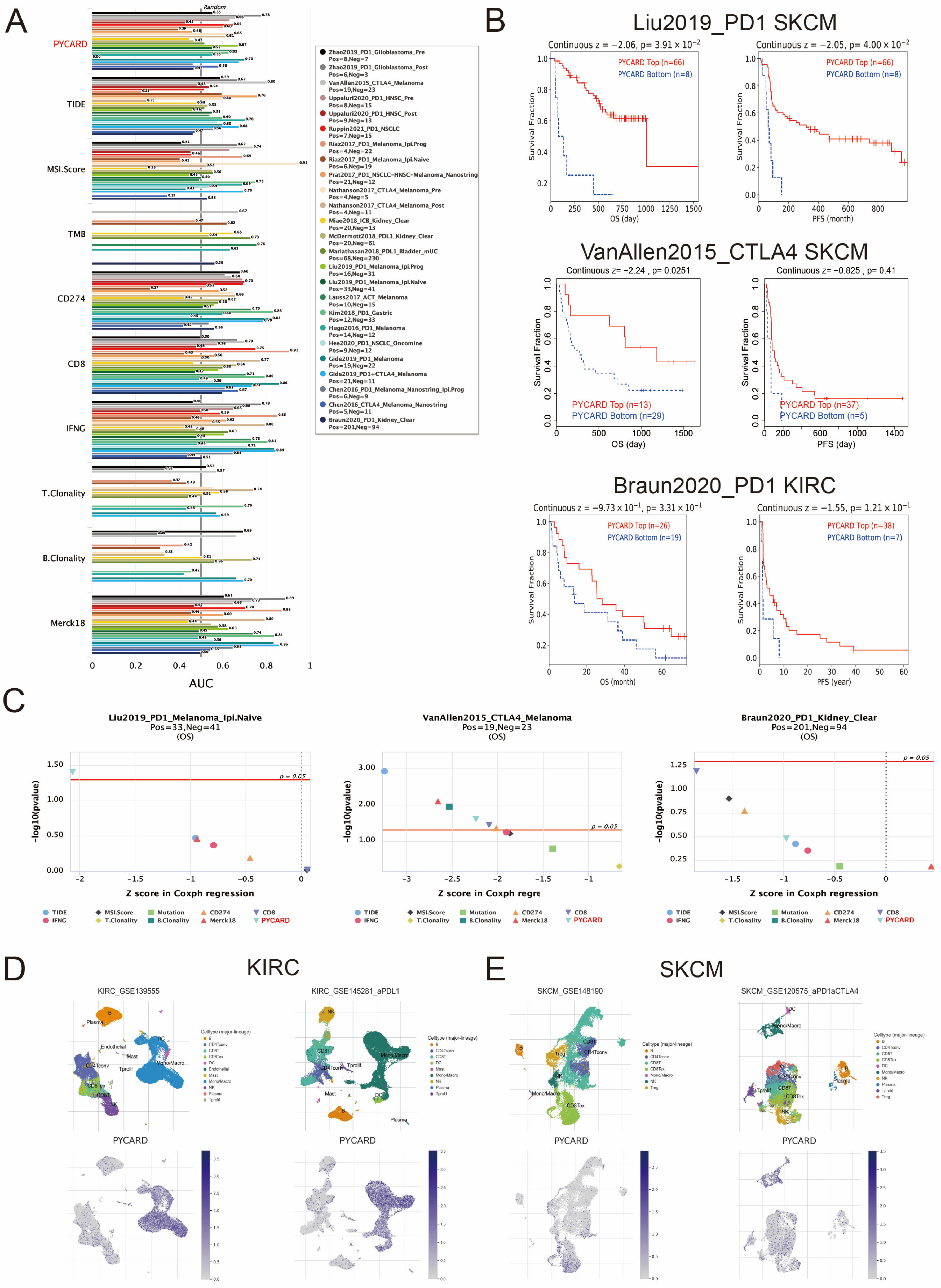
3.5. PYCARD Expression Correlated to Immune Response
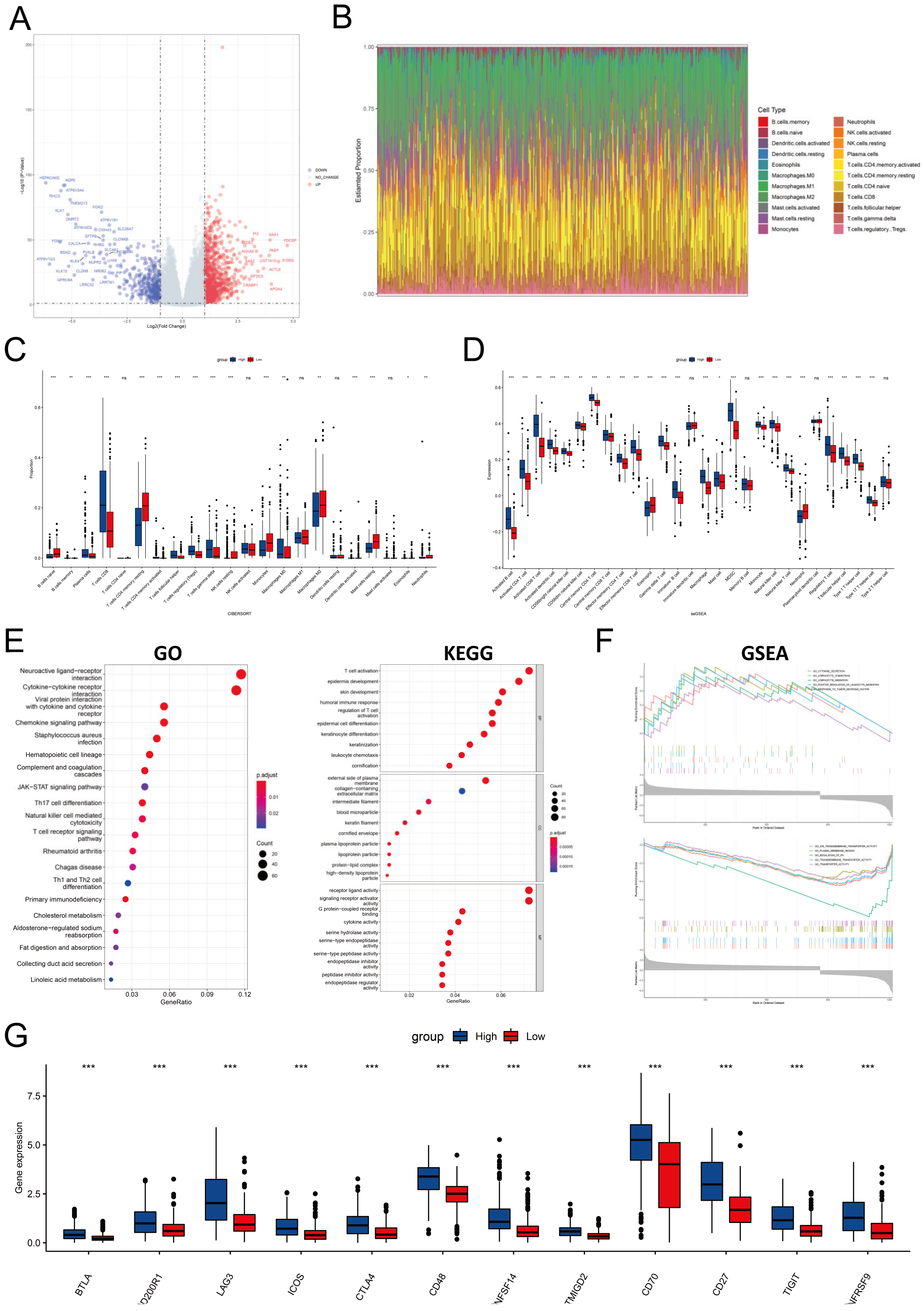
3.6. PYCARD Enriched Process of Immune Response, Inflammation and Apoptosis
3.7. PYCARD Expression Affected Immune Cell Infiltrations and Immune Regulation
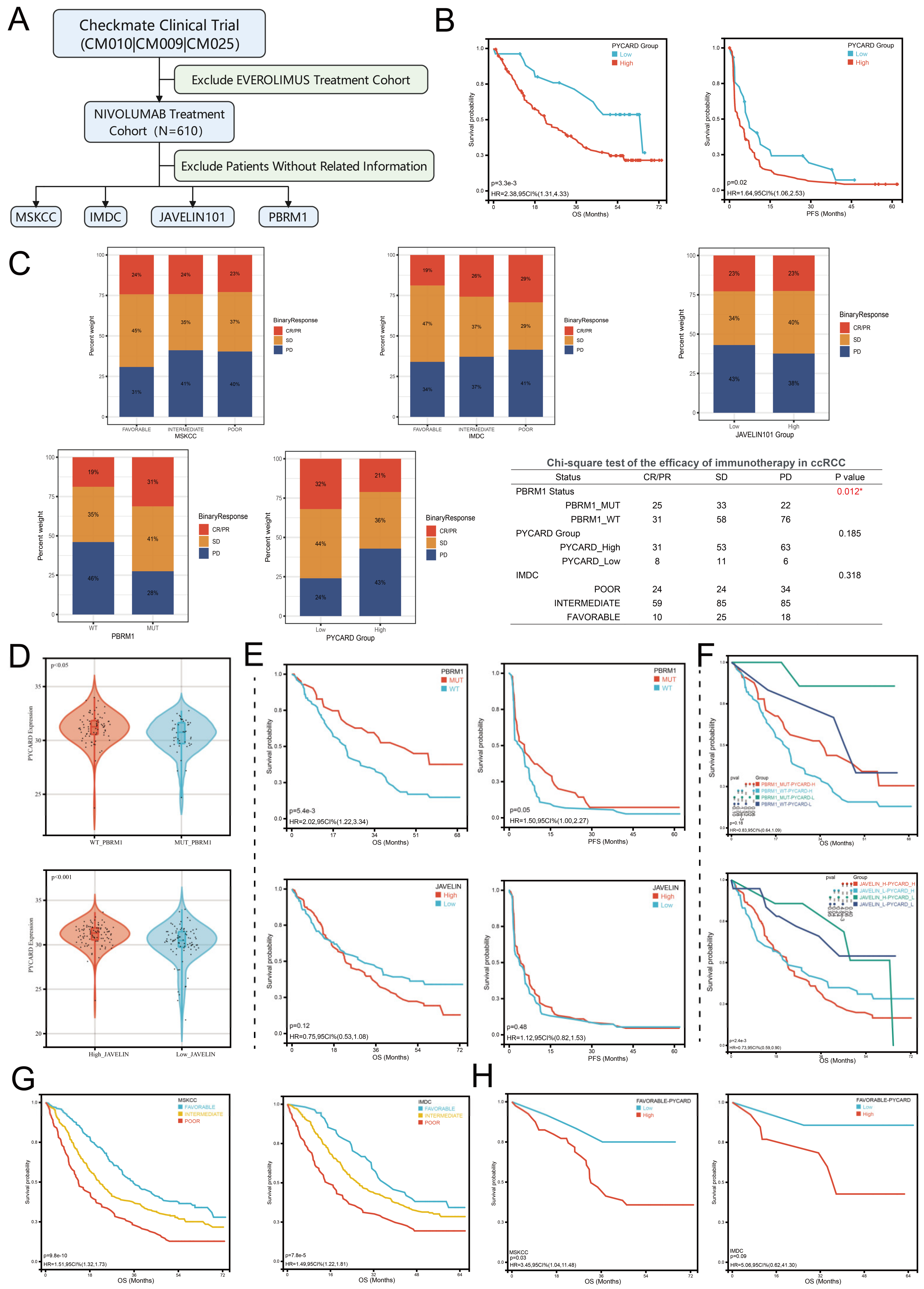
3.8. PYCARD Expression Correlated with Drug Response
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Thornberry, N.A.; Bull, H.G.; Calaycay, J.R.; Chapman, K.T.; Howard, A.D.; Kostura, M.J.; Miller, D.K.; Molineaux, S.M.; Weidner, J.R.; Aunins, J.; et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 1992, 356, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Ghayur, T.; Banerjee, S.; Hugunin, M.; Butler, D.; Herzog, L.; Carter, A.; Quintal, L.; Sekut, L.; Talanian, R.; Paskind, M.; et al. Caspase-1 processes IFN-gamma-inducing factor and regulates LPS-induced IFN-gamma production. Nature 1997, 386, 619–623. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Kuida, K.; Tsutsui, H.; Ku, G.; Hsiao, K.; Fleming, M.A.; Hayashi, N.; Higashino, K.; Okamura, H.; Nakanishi, K.; et al. Activation of interferon-gamma inducing factor mediated by interleukin-1beta converting enzyme. Science 1997, 275, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Fink, S.L.; Cookson, B.T. Apoptosis, pyroptosis, and necrosis: Mechanistic description of dead and dying eukaryotic cells. Infect. Immun. 2005, 73, 1907–1916. [Google Scholar] [CrossRef]
- Lin, G.; Feng, Q.; Zhan, F.; Yang, F.; Niu, Y.; Li, G. Generation and Analysis of Pyroptosis-Based and Immune-Based Signatures for Kidney Renal Clear Cell Carcinoma Patients, and Cell Experiment. Front. Genet. 2022, 13, 809794. [Google Scholar] [CrossRef]
- Sun, Z.; Jing, C.; Guo, X.; Zhang, M.; Kong, F.; Wang, Z.; Jiang, S.; Wang, H. Comprehensive Analysis of the Immune Infiltrates of Pyroptosis in Kidney Renal Clear Cell Carcinoma. Front. Oncol. 2021, 11, 716854. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Masumoto, J.; Taniguchi, S.; Ayukawa, K.; Sarvotham, H.; Kishino, T.; Niikawa, N.; Hidaka, E.; Katsuyama, T.; Higuchi, T.; Sagara, J. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J. Biol. Chem. 1999, 274, 33835–33838. [Google Scholar] [CrossRef]
- Zhou, R.; Yazdi, A.S.; Menu, P.; Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011, 469, 221–225. [Google Scholar] [CrossRef]
- Broz, P.; Pelegrín, P.; Shao, F. The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 2020, 20, 143–157. [Google Scholar] [CrossRef]
- Machida, E.O.; Brock, M.V.; Hooker, C.M.; Nakayama, J.; Ishida, A.; Amano, J.; Picchi, M.A.; Belinsky, S.A.; Herman, J.G.; Taniguchi, S.; et al. Hypermethylation of ASC/TMS1 is a sputum marker for late-stage lung cancer. Cancer Res. 2006, 66, 6210–6218. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, C.; Wang, X.; Ding, X.; Deng, J.; Liang, H. Methylation of ASC/TMS1 promoter is associated with poor prognosis of patients with gastric cancer. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2016, 18, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Brunetto, E.; De Monte, L.; Balzano, G.; Camisa, B.; Laino, V.; Riba, M.; Heltai, S.; Bianchi, M.; Bordignon, C.; Falconi, M.; et al. The IL-1/IL-1 receptor axis and tumor cell released inflammasome adaptor ASC are key regulators of TSLP secretion by cancer associated fibroblasts in pancreatic cancer. J. Immunother. Cancer 2019, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Creagh, E.M.; Conroy, H.; Martin, S.J. Caspase-activation pathways in apoptosis and immunity. Immunol. Rev. 2003, 193, 10–21. [Google Scholar] [CrossRef]
- Ohtsuka, T.; Ryu, H.; Minamishima, Y.A.; Macip, S.; Sagara, J.; Nakayama, K.I.; Aaronson, S.A.; Lee, S.W. ASC is a Bax adaptor and regulates the p53-Bax mitochondrial apoptosis pathway. Nat. Cell Biol. 2004, 6, 121–128. [Google Scholar] [CrossRef]
- Singh, R.; Letai, A.; Sarosiek, K. Regulation of apoptosis in health and disease: The balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 2019, 20, 175–193. [Google Scholar] [CrossRef]
- Sattler, M.; Liang, H.; Nettesheim, D.; Meadows, R.P.; Harlan, J.E.; Eberstadt, M.; Yoon, H.S.; Shuker, S.B.; Chang, B.S.; Minn, A.J.; et al. Structure of Bcl-xL-Bak peptide complex: Recognition between regulators of apoptosis. Science 1997, 275, 983–986. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef]
- Escudier, B.; Motzer, R.J.; Sharma, P.; Wagstaff, J.; Plimack, E.R.; Hammers, H.J.; Donskov, F.; Gurney, H.; Sosman, J.A.; Zalewski, P.G.; et al. Treatment Beyond Progression in Patients with Advanced Renal Cell Carcinoma Treated with Nivolumab in CheckMate 025. Eur. Urol. 2017, 72, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Feng, J.; Wu, X.; Bai, L.; Xu, W.; Zhu, L.; Liu, Y.; Xu, F.; Zhang, X.; Yang, G.; et al. A proteogenomic analysis of clear cell renal cell carcinoma in a Chinese population. Nat. Commun. 2022, 13, 2052. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Yoshizato, T.; Shiraishi, Y.; Maekawa, S.; Okuno, Y.; Kamura, T.; Shimamura, T.; Sato-Otsubo, A.; Nagae, G.; Suzuki, H.; et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013, 45, 860–867. [Google Scholar] [CrossRef]
- Braun, D.A.; Hou, Y.; Bakouny, Z.; Ficial, M.; Sant’ Angelo, M.; Forman, J.; Ross-Macdonald, P.; Berger, A.C.; Jegede, O.A.; Elagina, L.; et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat. Med. 2020, 26, 909–918. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Kalyana-Sundaram, S.; Mahavisno, V.; Varambally, R.; Yu, J.; Briggs, B.B.; Barrette, T.R.; Anstet, M.J.; Kincead-Beal, C.; Kulkarni, P.; et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007, 9, 166–180. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Mizuno, H.; Kitada, K.; Nakai, K.; Sarai, A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009, 2, 18. [Google Scholar] [CrossRef] [PubMed]
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.S.W.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 2019, 35, 4200–4202. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautès-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, F.; Dündar, F.; Diehl, S.; Grüning, B.A.; Manke, T. deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014, 42, W187–W191. [Google Scholar] [CrossRef] [PubMed]
- Blanche, P.; Dartigues, J.F.; Jacqmin-Gadda, H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat. Med. 2013, 32, 5381–5397. [Google Scholar] [CrossRef]
- Xu, W.; Anwaier, A.; Ma, C.; Liu, W.; Tian, X.; Palihati, M.; Hu, X.; Qu, Y.; Zhang, H.; Ye, D.; et al. Multi-omics reveals novel prognostic implication of SRC protein expression in bladder cancer and its correlation with immunotherapy response. Ann. Med. 2021, 53, 596–610. [Google Scholar] [CrossRef]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Sun, D.; Wang, J.; Han, Y.; Dong, X.; Ge, J.; Zheng, R.; Shi, X.; Wang, B.; Li, Z.; Ren, P.; et al. TISCH: A comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021, 49, D1420–D1430. [Google Scholar] [CrossRef]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Franz, M.; Rodriguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, W.C.; Sunshine, M.; Liu, H.; Varma, S.; Kohn, K.W.; Morris, J.; Doroshow, J.; Pommier, Y. CellMiner: A web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012, 72, 3499–3511. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef]
- Topalian, S.L.; Drake, C.G.; Pardoll, D.M. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 2015, 27, 450–461. [Google Scholar] [CrossRef]
- Hause, R.J.; Pritchard, C.C.; Shendure, J.; Salipante, S.J. Classification and characterization of microsatellite instability across 18 cancer types. Nat. Med. 2016, 22, 1342–1350. [Google Scholar] [CrossRef]
- Vilar, E.; Gruber, S.B. Microsatellite instability in colorectal cancer-the stable evidence. Nat. Rev. Clin. Oncol. 2010, 7, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014, 371, 2189–2199, Erratum in N. Engl. J. Med. 2015, 373, 1984. [Google Scholar] [CrossRef]
- Carbone, D.P.; Reck, M.; Paz-Ares, L.; Creelan, B.; Horn, L.; Steins, M.; Felip, E.; van den Heuvel, M.M.; Ciuleanu, T.E.; Badin, F.; et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 376, 2415–2426. [Google Scholar] [CrossRef]
- Miao, D.; Margolis, C.A.; Gao, W.; Voss, M.H.; Li, W.; Martini, D.J.; Norton, C.; Bossé, D.; Wankowicz, S.M.; Cullen, D.; et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018, 359, 801–806. [Google Scholar] [CrossRef]
- Liang, A.; Zhong, S.; Xi, B.; Zhou, C.; Jiang, X.; Zhu, R.; Yang, Y.; Zhong, L.; Wan, D. High expression of PYCARD is an independent predictor of unfavorable prognosis and chemotherapy resistance in glioma. Ann. Transl. Med. 2021, 9, 986. [Google Scholar] [CrossRef]
- Walker, L.S.; Sansom, D.M. The emerging role of CTLA4 as a cell-extrinsic regulator of T cell responses. Nat. Rev. Immunol. 2011, 11, 852–863. [Google Scholar] [CrossRef]
- Ruffo, E.; Wu, R.C.; Bruno, T.C.; Workman, C.J.; Vignali, D.A.A. Lymphocyte-activation gene 3 (LAG3): The next immune checkpoint receptor. Semin. Immunol. 2019, 42, 101305. [Google Scholar] [CrossRef]
- Koizumi, M.; Watanabe, T.; Masumoto, J.; Sunago, K.; Imamura, Y.; Kanemitsu, K.; Kumagi, T.; Hiasa, Y. Apoptosis-associated speck-like protein containing a CARD regulates the growth of pancreatic ductal adenocarcinoma. Sci. Rep. 2021, 11, 22351. [Google Scholar] [CrossRef]
- Liu, Q.; Jin, J.; Ying, J.; Cui, Y.; Sun, M.; Zhang, L.; Fan, Y.; Xu, B.; Zhang, Q. Epigenetic inactivation of the candidate tumor suppressor gene ASC/TMS1 in human renal cell carcinoma and its role as a potential therapeutic target. Oncotarget 2015, 6, 22706–22723. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [CrossRef] [PubMed]
- McDermott, D.F.; Huseni, M.A.; Atkins, M.B.; Motzer, R.J.; Rini, B.I.; Escudier, B.; Fong, L.; Joseph, R.W.; Pal, S.K.; Reeves, J.A.; et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat. Med. 2018, 24, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Robbins, P.B.; Powles, T.; Albiges, L.; Haanen, J.B.; Larkin, J.; Mu, X.J.; Ching, K.A.; Uemura, M.; Pal, S.K.; et al. Avelumab plus axitinib versus sunitinib in advanced renal cell carcinoma: Biomarker analysis of the phase 3 JAVELIN Renal 101 trial. Nat. Med. 2020, 26, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Reinherz, E.L. CD2BP1 modulates CD2-dependent T cell activation via linkage to protein tyrosine phosphatase (PTP)-PEST. J. Immunol. 2006, 176, 5898–5907. [Google Scholar] [CrossRef]
- Chou, W.C.; Guo, Z.; Guo, H.; Chen, L.; Zhang, G.; Liang, K.; Xie, L.; Tan, X.; Gibson, S.A.; Rampanelli, E.; et al. AIM2 in regulatory T cells restrains autoimmune diseases. Nature 2021, 591, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Zappasodi, R.; Serganova, I.; Cohen, I.J.; Maeda, M.; Shindo, M.; Senbabaoglu, Y.; Watson, M.J.; Leftin, A.; Maniyar, R.; Verma, S.; et al. CTLA-4 blockade drives loss of T(reg) stability in glycolysis-low tumours. Nature 2021, 591, 652–658. [Google Scholar] [CrossRef] [PubMed]

| Immune Infiltrating Cell Markers | KIRC | SKCM | |||||||
|---|---|---|---|---|---|---|---|---|---|
| None | Purity adj. | None | Purity adj. | ||||||
| Cell Type | Marker | Cor | p | Cor | p | Cor | p | Cor | p |
| CD8+ T cell | CD8A | 0.491 | *** | 0.422 | *** | 0.224 | *** | 0.142 | *** |
| CD8B | 0.532 | *** | 0.472 | *** | 0.242 | *** | 0.161 | ** | |
| T cell (general) | CD2 | 0.557 | *** | 0.490 | *** | 0.222 | *** | 0.133 | *** |
| CD3D | 0.616 | *** | 0.559 | *** | 0.247 | *** | 0.166 | *** | |
| CD3E | 0.564 | *** | 0.500 | *** | 0.259 | *** | 0.181 | 0.446 | |
| B cell | CD19 | 0.468 | *** | 0.406 | *** | 0.129 | ** | 0.036 | 0.060 |
| CD79A | 0.493 | *** | 0.426 | *** | 0.184 | *** | 0.088 | 0.318 | |
| Monocyte | CD86 | 0.449 | *** | 0.397 | *** | 0.068 | 0.140 | −0.047 | 0.197 |
| CD115 (CSF1R) | 0.444 | *** | 0.401 | *** | 0.047 | 0.308 | −0.060 | 0.159 | |
| TAM | CCL2 | 0.014 | 0.741 | -0.068 | 0.144 | 0.034 | 0.467 | −0.066 | 0.315 |
| CD68 | 0.356 | *** | 0.332 | *** | 0.138 | ** | 0.047 | 0.118 | |
| IL10 | 0.287 | *** | 0.206 | *** | 0.018 | 0.691 | −0.073 | 0.173 | |
| M1 Macrophage | INOS (NOS2) | −0.101 | * | −0.179 | *** | −0.063 | 0.173 | −0.064 | ** |
| IRF5 | 0.383 | *** | 0.366 | *** | 0.198 | *** | 0.126 | 0.250 | |
| CD80 | 0.337 | *** | 0.297 | *** | 0.049 | 0.286 | −0.054 | *** | |
| COX2 (PTGS2) | −0.133 | ** | −0.197 | *** | −0.143 | ** | -0.169 | *** | |
| M2 Macrophage | CD163 | 0.203 | *** | 0.164 | *** | −0.052 | 0.261 | −0.169 | * |
| VSIG4 | 0.403 | *** | 0.366 | *** | −0.009 | 0.846 | −0.094 | ** | |
| MS4A4 | 0.283 | *** | 0.219 | *** | −0.020 | 0.661 | −0.124 | 0.103 | |
| Neutrophils | CD66b (CEACAM8) | −0.095 | * | -0.096 | * | −0.086 | 0.062 | −0.076 | * |
| ITGAM | 0.426 | *** | 0.392 | *** | 0.165 | *** | 0.103 | ** | |
| CCR7 | 0.399 | *** | 0.321 | *** | 0.234 | *** | 0.137 | 0.056 | |
| Natural killer cell | KIR2DL1 | 0.003 | 0.937 | −0.029 | 0.530 | 0.130 | ** | 0.090 | 0.081 |
| KIR2DL3 | 0.067 | 0.122 | 0.057 | 0.220 | 0.145 | ** | 0.082 | *** | |
| KIR2DL4 | 0.260 | *** | 0.223 | *** | 0.245 | *** | 0.183 | * | |
| KIR3DL1 | −0.001 | 0.982 | −0.022 | 0.631 | 0.169 | *** | 0.118 | * | |
| KIR3DL2 | 0.200 | *** | 0.166 | *** | 0.178 | *** | 0.103 | 0.052 | |
| KIR3DL3 | 0.066 | 0.130 | 0.030 | 0.521 | 0.111 | * | 0.091 | 0.233 | |
| KIR2DS4 | 0.061 | 0.162 | 0.043 | 0.355 | 0.109 | * | 0.056 | 0.233 | |
| Dentritic cell | HLA-DPB1 | 0.502 | *** | 0.462 | *** | 0.248 | *** | 0.056 | ** |
| HLA-DQB1 | 0.313 | *** | 0.252 | *** | 0.235 | *** | 0.153 | * | |
| HLA-DRA | 0.410 | *** | 0.361 | *** | 0.194 | *** | 0.103 | ** | |
| HLA-DPA1 | 0.392 | *** | 0.323 | *** | 0.216 | *** | 0.135 | 0.152 | |
| HDAC1 (CD1C) | 0.226 | *** | 0.142 | ** | 0.156 | *** | 0.067 | *** | |
| BDCA4 (NRP1) | −0.280 | *** | −0.381 | *** | −0.278 | *** | −0.348 | *** | |
| CD11c (ITGAX) | 0.422 | *** | 0.408 | *** | 0.249 | *** | 0.180 | ** | |
| Th1 | T-bet (TBX21) | 0.277 | *** | 0.209 | *** | 0.225 | *** | 0.145 | 0.783 |
| STAT4 | 0.310 | *** | 0.225 | *** | 0.111 | * | 0.013 | 0.097 | |
| STAT1 | 0.300 | *** | 0.230 | *** | 0.137 | ** | 0.078 | 0.097 | |
| IFN-γ (IFNG) | 0.496 | *** | 0.433 | *** | 0.193 | *** | 0.078 | * | |
| TNF-α (TNF) | 0.295 | *** | 0.250 | *** | 0.182 | *** | 0.098 | * | |
| Th2 | GATA3 | 0.241 | *** | 0.222 | *** | 0.206 | *** | 0.098 | *** |
| STAT6 | −0.111 | * | −0.109 | * | 0.155 | *** | 0.164 | *** | |
| STAT5A | 0.424 | *** | 0.360 | *** | 0.250 | *** | 0.245 | *** | |
| IL13 | 0.033 | 0.442 | −0.020 | 0.662 | 0.039 | 0.397 | 0.245 | *** | |
| Tfh | BCL6 | −0.113 | ** | −0.139 | ** | −0.149 | ** | −0.183 | 0.569 |
| IL21 | 0.142 | *** | 0.120 | ** | 0.101 | * | 0.027 | 0.102 | |
| Th17 | STAT3 | −0.133 | ** | −0.206 | *** | −0.061 | 0.185 | −0.076 | 0.994 |
| IL17A | 0.064 | 0.137 | 0.019 | 0.686 | 0.013 | 0.771 | 0.000 | ** | |
| Treg | FOXP3 | 0.544 | *** | 0.498 | *** | 0.225 | *** | 0.143 | 0.668 |
| CCR8 | 0.370 | *** | 0.301 | *** | 0.076 | 0.099 | −0.020 | 0.068 | |
| STAT5B | −0.306 | *** | −0.350 | *** | 0.083 | 0.072 | 0.085 | 0.448 | |
| TGFB1 | 0.140 | ** | 0.088 | 0.058 | 0.044 | 0.343 | −0.036 | *** | |
| T cell exhaustion | PDCD1 | 0.597 | *** | 0.552 | *** | 0.295 | *** | 0.232 | 0.194 |
| CTLA4 | 0.446 | *** | 0.391 | *** | 0.134 | ** | 0.061 | *** | |
| LAG3 | 0.559 | *** | 0.512 | *** | 0.250 | *** | 0.177 | 0.902 | |
| TIM-3 (HAVCR2) | 0.131 | ** | 0.075 | 0.107 | 0.104 | * | -0.006 | *** | |
| GZMB | 0.336 | *** | 0.271 | *** | 0.274 | *** | 0.209 | *** | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, J.-Q.; Tian, X.; Xu, W.-H.; Anwaier, A.; Ye, S.-Q.; Zhu, S.-X.; Wang, Y.; Gu, J.; Shi, G.-H.; Qu, Y.-Y.; et al. The Inflammasomes Adaptor Protein PYCARD Is a Potential Pyroptosis Biomarker Related to Immune Response and Prognosis in Clear Cell Renal Cell Carcinoma. Cancers 2022, 14, 4992. https://doi.org/10.3390/cancers14204992
Su J-Q, Tian X, Xu W-H, Anwaier A, Ye S-Q, Zhu S-X, Wang Y, Gu J, Shi G-H, Qu Y-Y, et al. The Inflammasomes Adaptor Protein PYCARD Is a Potential Pyroptosis Biomarker Related to Immune Response and Prognosis in Clear Cell Renal Cell Carcinoma. Cancers. 2022; 14(20):4992. https://doi.org/10.3390/cancers14204992
Chicago/Turabian StyleSu, Jia-Qi, Xi Tian, Wen-Hao Xu, Aihetaimujiang Anwaier, Shi-Qi Ye, Shu-Xuan Zhu, Yue Wang, Jun Gu, Guo-Hai Shi, Yuan-Yuan Qu, and et al. 2022. "The Inflammasomes Adaptor Protein PYCARD Is a Potential Pyroptosis Biomarker Related to Immune Response and Prognosis in Clear Cell Renal Cell Carcinoma" Cancers 14, no. 20: 4992. https://doi.org/10.3390/cancers14204992
APA StyleSu, J.-Q., Tian, X., Xu, W.-H., Anwaier, A., Ye, S.-Q., Zhu, S.-X., Wang, Y., Gu, J., Shi, G.-H., Qu, Y.-Y., Zhang, H.-L., & Ye, D.-W. (2022). The Inflammasomes Adaptor Protein PYCARD Is a Potential Pyroptosis Biomarker Related to Immune Response and Prognosis in Clear Cell Renal Cell Carcinoma. Cancers, 14(20), 4992. https://doi.org/10.3390/cancers14204992






