RNA-seq and ChIP-seq Identification of Unique and Overlapping Targets of GLI Transcription Factors in Melanoma Cell Lines
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Plasmids and Cell Transfection
2.3. MTT Assay
2.4. Western Blot
2.5. RNA-Sequencing
2.6. Chromatin Immunoprecipitation Sequencing (ChIP-seq)
2.7. Bioinformatic Analysis of RNA-seq Data
2.8. Bioinformatic Analysis of ChIP-seq Data
2.9. Quantitative PCR Analysis
2.10. Statistical Analysis
3. Results
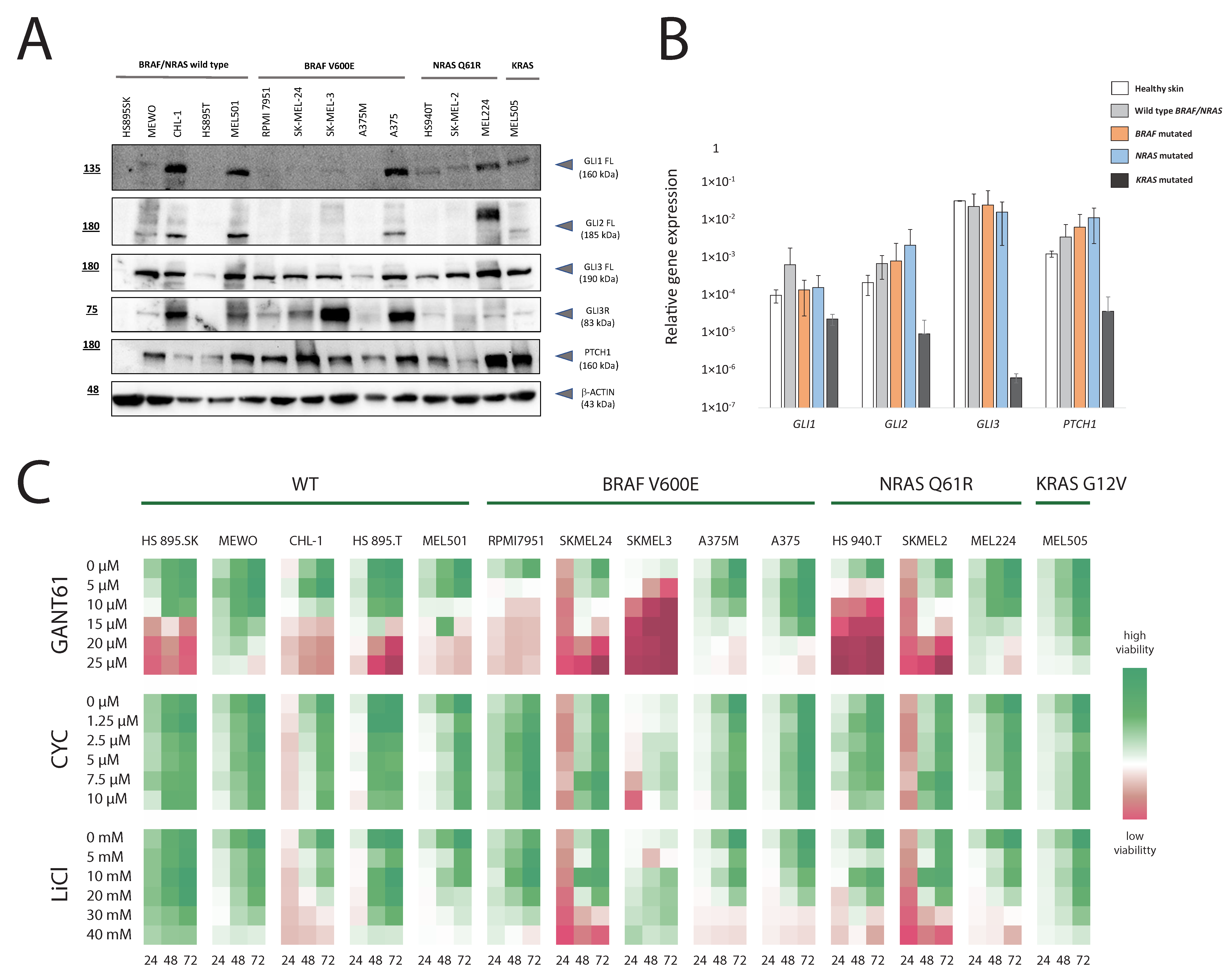
3.1. HH-GLI Signaling Pathway in Melanoma Cell Lines with Different Genetic Background and Their Response to Pathway Inhibition
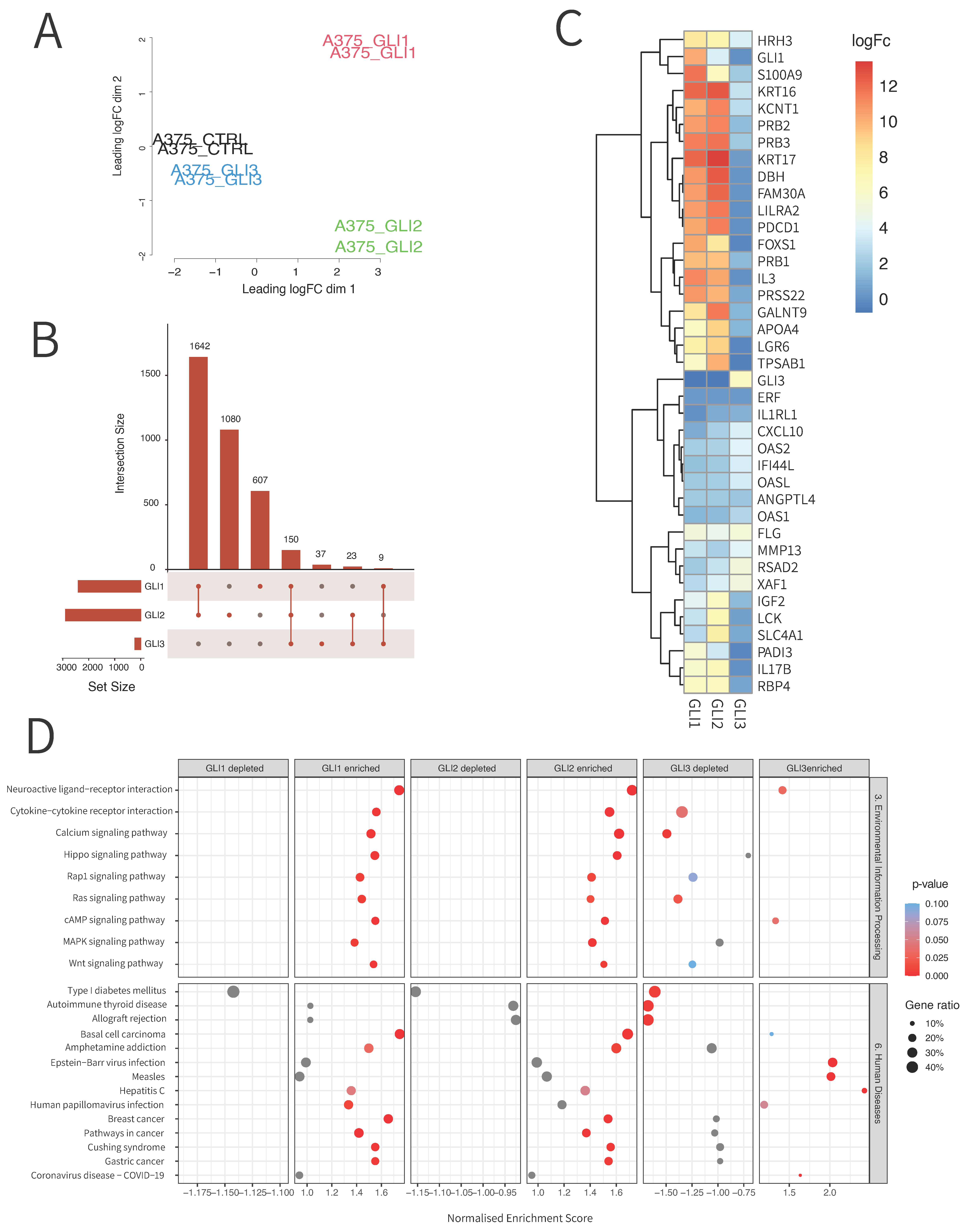
3.2. RNA Sequencing Reveals Unique and Overlapping Targets of GLI Transcription Factors
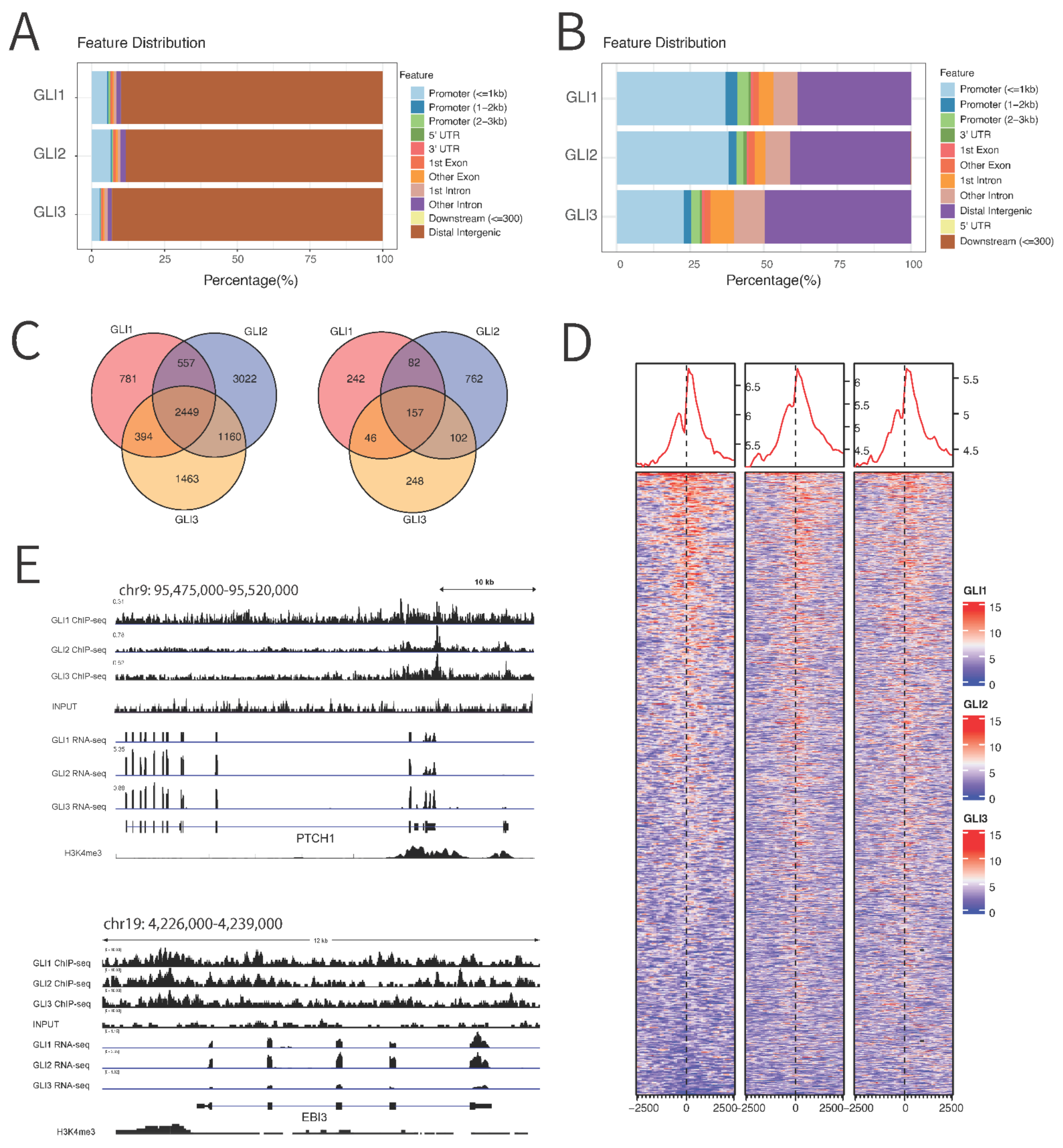
3.3. Chromatin Immunoprecipitation (ChIP) Sequencing Reveals Novel Binding Targets of GLI Proteins
3.4. qPCR Validation of RNA Sequencing and ChIP Sequencing Data Confirms 15 Novel GLI Target Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davis, L.E.; Shalin, S.C.; Tackett, A.J. Current State of Melanoma Diagnosis and Treatment. Cancer Biol. Ther. 2019, 20, 1366–1379. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF Gene in Human Cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Rovida, E.; Stecca, B. Mitogen-Activated Protein Kinases and Hedgehog-GLI Signaling in Cancer: A Crosstalk Providing Therapeutic Opportunities? Semin. Cancer Biol. 2015, 35, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Raducu, M.; Fung, E.; Serres, S.; Infante, P.; Barberis, A.; Fischer, R.; Bristow, C.; Thézénas, M.-L.; Finta, C.; Christianson, J.C.; et al. SCF (Fbxl17) Ubiquitylation of Sufu Regulates Hedgehog Signaling and Medulloblastoma Development. EMBO J. 2016, 35, 1400–1416. [Google Scholar] [CrossRef]
- Berman, D.M.; Karhadkar, S.S.; Maitra, A.; Montes De Oca, R.; Gerstenblith, M.R.; Briggs, K.; Parker, A.R.; Shimada, Y.; Eshleman, J.R.; Watkins, D.N.; et al. Widespread Requirement for Hedgehog Ligand Stimulation in Growth of Digestive Tract Tumours. Nature 2003, 425, 846–851. [Google Scholar] [CrossRef]
- Sheng, T.; Li, C.; Zhang, X.; Chi, S.; He, N.; Chen, K.; McCormick, F.; Gatalica, Z.; Xie, J. Activation of the Hedgehog Pathway in Advanced Prostate Cancer. Mol. Cancer 2004, 3, 29. [Google Scholar] [CrossRef]
- Varnat, F.; Duquet, A.; Malerba, M.; Zbinden, M.; Mas, C.; Gervaz, P.; Ruiz i Altaba, A. Human Colon Cancer Epithelial Cells Harbour Active HEDGEHOG-GLI Signalling That Is Essential for Tumour Growth, Recurrence, Metastasis and Stem Cell Survival and Expansion. EMBO Mol. Med. 2009, 1, 338–351. [Google Scholar] [CrossRef]
- Marini, K.D.; Payne, B.J.; Watkins, D.N.; Martelotto, L.G. Mechanisms of Hedgehog Signalling in Cancer. Growth Factors 2011, 29, 221–234. [Google Scholar] [CrossRef]
- Ozretić, P.; Trnski, D.; Musani, V.; Maurac, I.; Kalafatić, D.; Orešković, S.; Levanat, S.; Sabol, M. Non-Canonical Hedgehog Signaling Activation in Ovarian Borderline Tumors and Ovarian Carcinomas. Int. J. Oncol. 2017, 51, 1869–1877. [Google Scholar] [CrossRef]
- Giroux-Leprieur, E.; Costantini, A.; Ding, V.W.; He, B. Hedgehog Signaling in Lung Cancer: From Oncogenesis to Cancer Treatment Resistance. Int. J. Mol. Sci. 2018, 19, E2835. [Google Scholar] [CrossRef]
- O’Reilly, K.E.; Vega-Saenz de Miera, E.; Segura, M.F.; Friedman, E.; Poliseno, L.; Han, S.W.; Zhong, J.; Zavadil, J.; Pavlick, A.; Hernando, E.; et al. Hedgehog Pathway Blockade Inhibits Melanoma Cell Growth in Vitro and in Vivo. Pharmaceuticals 2013, 6, 1429–1450. [Google Scholar] [CrossRef]
- Jalili, A.; Mertz, K.D.; Romanov, J.; Wagner, C.; Kalthoff, F.; Stuetz, A.; Pathria, G.; Gschaider, M.; Stingl, G.; Wagner, S.N. NVP-LDE225, a Potent and Selective SMOOTHENED Antagonist Reduces Melanoma Growth In Vitro and In Vivo. PLoS ONE 2013, 8, e69064. [Google Scholar] [CrossRef]
- Gunarta, I.K.; Li, R.; Nakazato, R.; Suzuki, R.; Boldbaatar, J.; Suzuki, T.; Yoshioka, K. Critical Role of Glioma-associated Oncogene Homolog 1 in Maintaining Invasive and Mesenchymal-like Properties of Melanoma Cells. Cancer Sci. 2017, 108, 1602–1611. [Google Scholar] [CrossRef]
- Faião-Flores, F.; Alves-Fernandes, D.K.; Pennacchi, P.C.; Sandri, S.; Vicente, A.L.S.A.; Scapulatempo-Neto, C.; Vazquez, V.L.; Reis, R.M.; Chauhan, J.; Goding, C.R.; et al. Targeting the Hedgehog Transcription Factors GLI1 and GLI2 Restores Sensitivity to Vemurafenib-Resistant Human Melanoma Cells. Oncogene 2017, 36, 1849–1861. [Google Scholar] [CrossRef]
- Sabol, M.; Trnski, D.; Musani, V.; Ozretić, P.; Levanat, S. Role of GLI Transcription Factors in Pathogenesis and Their Potential as New Therapeutic Targets. Int. J. Mol. Sci. 2018, 19, 2562. [Google Scholar] [CrossRef]
- Lo Re, A.E.; Fernandez-Barrena, M.G.; Almada, L.L.; Mills, L.D.; Elsawa, S.F.; Lund, G.; Ropolo, A.; Molejon, M.I.; Vaccaro, M.I.; Fernandez-Zapico, M.E. Novel AKT1-GLI3-VMP1 Pathway Mediates KRAS Oncogene-Induced Autophagy in Cancer Cells. J. Biol. Chem. 2012, 287, 25325–25334. [Google Scholar] [CrossRef]
- Riobo, N.A.; Saucy, B.; DiLizio, C.; Manning, D.R. Activation of Heterotrimeric G Proteins by Smoothened. Proc. Natl. Acad. Sci. USA 2006, 103, 12607. [Google Scholar] [CrossRef]
- Pietrobono, S.; Gagliardi, S.; Stecca, B. Non-Canonical Hedgehog Signaling Pathway in Cancer: Activation of GLI Transcription Factors Beyond Smoothened. Front. Genet. 2019, 10, 556. [Google Scholar] [CrossRef]
- Antonucci, L.; Di Magno, L.; D’Amico, D.; Manni, S.; Serrao, S.M.; Di Pastena, F.; Bordone, R.; Yurtsever, Z.N.; Caimano, M.; Petroni, M.; et al. Mitogen-Activated Kinase Kinase Kinase 1 Inhibits Hedgehog Signaling and Medulloblastoma Growth through GLI1 Phosphorylation. Int. J. Oncol. 2018, 54, 505–514. [Google Scholar] [CrossRef]
- Zubčić, V.; Rinčić, N.; Kurtović, M.; Trnski, D.; Musani, V.; Ozretić, P.; Levanat, S.; Leović, D.; Sabol, M. GANT61 and Lithium Chloride Inhibit the Growth of Head and Neck Cancer Cell Lines Through the Regulation of GLI3 Processing by GSK3β. Int. J. Mol. Sci. 2020, 21, E6410. [Google Scholar] [CrossRef]
- Mazumdar, T.; DeVecchio, J.; Shi, T.; Jones, J.; Agyeman, A.; Houghton, J.A. Hedgehog Signaling Drives Cellular Survival in Human Colon Carcinoma Cells. Cancer Res. 2011, 71, 1092–1102. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fang, Z.; Wang, A.; Luo, C.; Cheng, X.; Lu, M. Lithium Suppresses Hedgehog Signaling via Promoting ITCH E3 Ligase Activity and Gli1–SUFU Interaction in PDA Cells. Front. Pharmacol. 2017, 8, 820. [Google Scholar] [CrossRef] [PubMed]
- Plaisant, M.; Giorgetti-Peraldi, S.; Gabrielson, M.; Loubat, A.; Dani, C.; Peraldi, P. Inhibition of Hedgehog Signaling Decreases Proliferation and Clonogenicity of Human Mesenchymal Stem Cells. PLoS ONE 2011, 6, e16798. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.A.; Niu, B.; Cheah, K.S.E.; Alman, B. Unique and Overlapping GLI1 and GLI2 Transcriptional Targets in Neoplastic Chondrocytes. PLoS ONE 2019, 14, e0211333. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. 2007. Available online: Https://Bioinfogp.Cnb.Csic.Es/Tools/Venny/Index.Html (accessed on 10 August 2022).
- Ben-Ari Fuchs, S.; Lieder, I.; Stelzer, G.; Mazor, Y.; Buzhor, E.; Kaplan, S.; Bogoch, Y.; Plaschkes, I.; Shitrit, A.; Rappaport, N.; et al. GeneAnalytics: An Integrative Gene Set Analysis Tool for Next Generation Sequencing, RNAseq and Microarray Data. OMICS 2016, 20, 139–151. [Google Scholar] [CrossRef]
- Réda, J.; Vachtenheim, J.; Vlčková, K.; Horák, P.; Vachtenheim, J.; Ondrušová, L. Widespread Expression of Hedgehog Pathway Components in a Large Panel of Human Tumor Cells and Inhibition of Tumor Growth by GANT61: Implications for Cancer Therapy. Int. J. Mol. Sci. 2018, 19, 2682. [Google Scholar] [CrossRef]
- Lex, R.K.; Ji, Z.; Falkenstein, K.N.; Zhou, W.; Henry, J.L.; Ji, H.; Vokes, S.A. GLI Transcriptional Repression Regulates Tissue-Specific Enhancer Activity in Response to Hedgehog Signaling. eLife 2020, 9, e50670. [Google Scholar] [CrossRef]
- Yin, W.-C.; Satkunendran, T.; Mo, R.; Morrissy, S.; Zhang, X.; Huang, E.S.; Uusküla-Reimand, L.; Hou, H.; Son, J.E.; Liu, W.; et al. Dual Regulatory Functions of SUFU and Targetome of GLI2 in SHH Subgroup Medulloblastoma. Dev. Cell 2020, 52, 132. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Wang, L.-X.; Li, Y.; Chen, G.-Z. Network-Based Co-Expression Analysis for Exploring the Potential Diagnostic Biomarkers of Metastatic Melanoma. PLoS ONE 2018, 13, e0190447. [Google Scholar] [CrossRef]
- Metri, R.; Mohan, A.; Nsengimana, J.; Pozniak, J.; Molina-Paris, C.; Newton-Bishop, J.; Bishop, D.; Chandra, N. Identification of a Gene Signature for Discriminating Metastatic from Primary Melanoma Using a Molecular Interaction Network Approach. Sci. Rep. 2017, 7, 17314. [Google Scholar] [CrossRef]
- Huang, W.-C.; Jang, T.-H.; Tung, S.-L.; Yen, T.-C.; Chan, S.-H.; Wang, L.-H. A Novel MiR-365-3p/EHF/Keratin 16 Axis Promotes Oral Squamous Cell Carcinoma Metastasis, Cancer Stemness and Drug Resistance via Enhancing Β5-Integrin/c-Met Signaling Pathway. J. Exp. Clin. Cancer Res. 2019, 38, 89. [Google Scholar] [CrossRef]
- Elazezy, M.; Schwentesius, S.; Stegat, L.; Wikman, H.; Werner, S.; Mansour, W.Y.; Failla, A.V.; Peine, S.; Müller, V.; Thiery, J.P.; et al. Emerging Insights into Keratin 16 Expression during Metastatic Progression of Breast Cancer. Cancers 2021, 13, 3869. [Google Scholar] [CrossRef]
- Ujiie, D.; Okayama, H.; Saito, K.; Ashizawa, M.; Thar Min, A.K.; Endo, E.; Kase, K.; Yamada, L.; Kikuchi, T.; Hanayama, H.; et al. KRT17 as a Prognostic Biomarker for Stage II Colorectal Cancer. Carcinogenesis 2020, 41, 591–599. [Google Scholar] [CrossRef]
- Li, J.; Chen, Q.; Deng, Z.; Chen, X.; Liu, H.; Tao, Y.; Wang, X.; Lin, S.; Liu, N. KRT17 Confers Paclitaxel-Induced Resistance and Migration to Cervical Cancer Cells. Life Sci. 2019, 224, 255–262. [Google Scholar] [CrossRef]
- Li, D.; Ni, X.-F.; Tang, H.; Zhang, J.; Zheng, C.; Lin, J.; Wang, C.; Sun, L.; Chen, B. KRT17 Functions as a Tumor Promoter and Regulates Proliferation, Migration and Invasion in Pancreatic Cancer via MTOR/S6k1 Pathway. Cancer Manag. Res. 2020, 12, 2087–2095. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, M.-Q.; Lei, L.; Fei, L.-R.; Zheng, Y.-W.; Huang, W.-J.; Li, Z.-H.; Liu, C.-C.; Xu, H.-T. Overexpression of KRT17 Promotes Proliferation and Invasion of Non-Small Cell Lung Cancer and Indicates Poor Prognosis. Cancer Manag. Res. 2019, 11, 7485–7497. [Google Scholar] [CrossRef]
- Xiong, T.-F.; Pan, F.-Q.; Li, D. Expression and Clinical Significance of S100 Family Genes in Patients with Melanoma. Melanoma Res. 2019, 29, 23–29. [Google Scholar] [CrossRef]
- Sakurai, M.; Miki, Y.; Takagi, K.; Suzuki, T.; Ishida, T.; Ohuchi, N.; Sasano, H. Interaction with Adipocyte Stromal Cells Induces Breast Cancer Malignancy via S100A7 Upregulation in Breast Cancer Microenvironment. Breast Cancer Res. 2017, 19, 70. [Google Scholar] [CrossRef]
- Muoio, M.G.; Talia, M.; Lappano, R.; Sims, A.H.; Vella, V.; Cirillo, F.; Manzella, L.; Giuliano, M.; Maggiolini, M.; Belfiore, A.; et al. Activation of the S100A7/RAGE Pathway by IGF-1 Contributes to Angiogenesis in Breast Cancer. Cancers 2021, 13, 621. [Google Scholar] [CrossRef]
- Tian, T.; Li, X.; Hua, Z.; Ma, J.; Wu, X.; Liu, Z.; Chen, H.; Cui, Z. S100A7 Promotes the Migration, Invasion and Metastasis of Human Cervical Cancer Cells through Epithelial-Mesenchymal Transition. Oncotarget 2017, 8, 24964–24977. [Google Scholar] [CrossRef]
- Lin, M.; Xia, B.; Qin, L.; Chen, H.; Lou, G. S100A7 Regulates Ovarian Cancer Cell Metastasis and Chemoresistance Through MAPK Signaling and Is Targeted by MiR-330-5p. DNA Cell Biol. 2018, 37, 491–500. [Google Scholar] [CrossRef]
- Hibino, T.; Sakaguchi, M.; Miyamoto, S.; Yamamoto, M.; Motoyama, A.; Hosoi, J.; Shimokata, T.; Ito, T.; Tsuboi, R.; Huh, N.-H. S100A9 Is a Novel Ligand of EMMPRIN That Promotes Melanoma Metastasis. Cancer Res. 2013, 73, 172–183. [Google Scholar] [CrossRef]
- Hwang, S.-H.; Ahn, J.-H.; Lee, M. Upregulation of S100A9 Contributes to the Acquired Resistance to BRAF Inhibitors. Genes Genom. 2019, 41, 1273–1280. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, Q.; Xu, Q.; Zhang, S.; Cao, L. Predictive Value of S100A9 for Lymph Node Metastasis in Cervical Cancer. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2020, 45, 701–708. [Google Scholar] [CrossRef]
- Lv, Z.; Li, W.; Wei, X. S100A9 Promotes Prostate Cancer Cell Invasion by Activating TLR4/NF-ΚB/Integrin Β1/FAK Signaling. OncoTargets Ther. 2020, 13, 6443–6452. [Google Scholar] [CrossRef]
- Zhou, C.; Shen, S.; Moran, R.; Deng, N.; Marbán, E.; Melmed, S. Pituitary Somatotroph Adenoma-Derived Exosomes: Characterization of Nonhormonal Actions. J. Clin. Endocrinol. Metab. 2022, 107, 379–397. [Google Scholar] [CrossRef]
- Yang, X.; Liang, R.; Liu, C.; Liu, J.A.; Cheung, M.P.L.; Liu, X.; Man, O.Y.; Guan, X.-Y.; Lung, H.L.; Cheung, M. SOX9 Is a Dose-Dependent Metastatic Fate Determinant in Melanoma. J. Exp. Clin. Cancer Res. 2019, 38, 17. [Google Scholar] [CrossRef]
- Cheng, P.F.; Shakhova, O.; Widmer, D.S.; Eichhoff, O.M.; Zingg, D.; Frommel, S.C.; Belloni, B.; Raaijmakers, M.I.; Goldinger, S.M.; Santoro, R.; et al. Methylation-Dependent SOX9 Expression Mediates Invasion in Human Melanoma Cells and Is a Negative Prognostic Factor in Advanced Melanoma. Genome Biol. 2015, 16, 42. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Liu, Z.; Zhang, R.; Ma, S.; Lin, T.; Li, Y.; Yang, S.; Zhang, W.; Wang, Y. Long Non-Coding RNA LEF1-AS1 Promotes Migration, Invasion and Metastasis of Colon Cancer Cells Through MiR-30-5p/SOX9 Axis. Onco Targets Ther. 2020, 13, 2957–2972. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Wu, L.; Ma, N.; Tang, F.; Yu, Z.; Jiang, Z.; Li, Y.; Zong, Z.; Hu, K. SOX9-Activated FARSA-AS1 Predetermines Cell Growth, Stemness, and Metastasis in Colorectal Cancer through Upregulating FARSA and SOX9. Cell Death Dis. 2020, 11, 1071. [Google Scholar] [CrossRef] [PubMed]
- Yasumoto, M.; Sakamoto, E.; Ogasawara, S.; Isobe, T.; Kizaki, J.; Sumi, A.; Kusano, H.; Akiba, J.; Torimura, T.; Akagi, Y.; et al. Muscle RAS Oncogene Homolog (MRAS) Recurrent Mutation in Borrmann Type IV Gastric Cancer. Cancer Med. 2017, 6, 235–244. [Google Scholar] [CrossRef]
- Young, L.C.; Hartig, N.; Muñoz-Alegre, M.; Oses-Prieto, J.A.; Durdu, S.; Bender, S.; Vijayakumar, V.; Vietri Rudan, M.; Gewinner, C.; Henderson, S.; et al. An MRAS, SHOC2, and SCRIB Complex Coordinates ERK Pathway Activation with Polarity and Tumorigenic Growth. Mol. Cell 2013, 52, 679–692. [Google Scholar] [CrossRef]
- Sun, Y.; Jia, X.; Hou, L.; Liu, X. Screening of Differently Expressed MiRNA and MRNA in Prostate Cancer by Integrated Analysis of Transcription Data. Urology 2016, 94, 313.e1–313.e6. [Google Scholar] [CrossRef]
- Faruk, M.; Ibrahim, S.; Aminu, S.M.; Adamu, A.; Abdullahi, A.; Suleiman, A.M.; Rafindadi, A.H.; Mohammed, A.; Iliyasu, Y.; Idoko, J.; et al. Prognostic Significance of BIRC7/Livin, Bcl-2, P53, Annexin V, PD-L1, DARC, MSH2 and PMS2 in Colorectal Cancer Treated with FOLFOX Chemotherapy with or without Aspirin. PLoS ONE 2021, 16, e0245581. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, P.; Xu, W.; Xia, W. High BIRC7 Expression Might Be an Independent Prognostic Indicator of Poor Recurrence-Free Survival in Patients with Prostate Cancer. Technol. Cancer Res. Treat. 2018, 17, 1533033818809694. [Google Scholar] [CrossRef]
- Liu, K.; Yu, Q.; Li, H.; Xie, C.; Wu, Y.; Ma, D.; Sheng, P.; Dai, W.; Jiang, H. BIRC7 Promotes Epithelial-Mesenchymal Transition and Metastasis in Papillary Thyroid Carcinoma through Restraining Autophagy. Am. J. Cancer Res. 2020, 10, 78–94. [Google Scholar]
- Li, J.; Yang, Z.; Huang, S.; Li, D. BIRC7 and STC2 Expression Are Associated With Tumorigenesis and Poor Outcome in Extrahepatic Cholangiocarcinoma. Technol. Cancer Res. Treat. 2020, 19, 1533033820971676. [Google Scholar] [CrossRef]
- Sun, K.; Liao, Q.; Chen, Z.; Chen, T.; Zhang, J. Expression of Livin and PlGF in Human Osteosarcoma Is Associated with Tumor Progression and Clinical Outcome. Oncol. Lett. 2018, 16, 4953–4960. [Google Scholar] [CrossRef]
- Hsieh, C.-H.; Lin, Y.-J.; Wu, C.-P.; Lee, H.-T.; Shyu, W.-C.; Wang, C.-C. Livin Contributes to Tumor Hypoxia-Induced Resistance to Cytotoxic Therapies in Glioblastoma Multiforme. Clin. Cancer Res. 2015, 21, 460–470. [Google Scholar] [CrossRef]
- Lazar, I.; Perlman, R.; Lotem, M.; Peretz, T.; Ben-Yehuda, D.; Kadouri, L. The Clinical Effect of the Inhibitor of Apopotosis Protein Livin in Melanoma. Oncology 2012, 82, 197–204. [Google Scholar] [CrossRef]
- Yuan, B.; Ran, B.; Wang, S.; Liu, Z.; Zheng, Z.; Chen, H. SiRNA Directed against Livin Inhibits Tumor Growth and Induces Apoptosis in Human Glioma Cells. J. Neurooncol. 2012, 107, 81–87. [Google Scholar] [CrossRef]
- Zhuang, L.; Shen, L.-D.; Li, K.; Yang, R.-X.; Zhang, Q.-Y.; Chen, Y.; Gao, C.-L.; Dong, C.; Bi, Q.; Tao, J.-N.; et al. Inhibition of Livin Expression Suppresses Cell Proliferation and Enhances Chemosensitivity to Cisplatin in Human Lung Adenocarcinoma Cells. Mol. Med. Rep. 2015, 12, 547–552. [Google Scholar] [CrossRef]
- Liu, J.; Yang, Y.; Li, H.; Liu, Y.; Sun, Y.; Wu, J.; Xiong, Z.; Jin, T. IL1R2 Polymorphisms Are Associated with Increased Risk of Esophageal Cancer. Curr. Mol. Med. 2020, 20, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qiang, J.; Yang, X.; Wang, D.; Rehman, A.U.; He, X.; Chen, W.; Sheng, D.; Zhou, L.; Jiang, Y.-Z.; et al. IL1R2 Blockade Suppresses Breast Tumorigenesis and Progression by Impairing USP15-Dependent BMI1 Stability. Adv. Sci. 2020, 7, 1901728. [Google Scholar] [CrossRef]
- Torricelli, C.; Carron, J.; Carvalho, B.F.; Macedo, L.T.; Rinck-Junior, J.A.; Lima, C.S.P.; Lourenço, G.J. Influence of IL1B (Rs16944) and IL1R2 (Rs4141134) Polymorphisms on Aggressiveness and Prognosis of Cutaneous Melanoma. Melanoma Res. 2021, 31, 476–481. [Google Scholar] [CrossRef]
- Yuan, M.; Wang, L.; Huang, H.; Li, Y.; Zheng, X.; Shao, Q.; Jiang, J. IL-1R2 Expression in Human Gastric Cancer and Its Clinical Significance. Biosci. Rep. 2021, 41, BSR20204425. [Google Scholar] [CrossRef]
- Mar, A.-C.; Chu, C.-H.; Lee, H.-J.; Chien, C.-W.; Cheng, J.-J.; Yang, S.-H.; Jiang, J.-K.; Lee, T.-C. Interleukin-1 Receptor Type 2 Acts with c-Fos to Enhance the Expression of Interleukin-6 and Vascular Endothelial Growth Factor A in Colon Cancer Cells and Induce Angiogenesis. J. Biol. Chem. 2015, 290, 22212–22224. [Google Scholar] [CrossRef]
- Ballerini, P.; Struski, S.; Cresson, C.; Prade, N.; Toujani, S.; Deswarte, C.; Dobbelstein, S.; Petit, A.; Lapillonne, H.; Gautier, E.-F.; et al. RET Fusion Genes Are Associated with Chronic Myelomonocytic Leukemia and Enhance Monocytic Differentiation. Leukemia 2012, 26, 2384–2389. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Zhang, Y.; Wang, S.; Liu, Y.; Zheng, L.; Yang, J.; Huang, W.; Ye, Y.; Luo, W.; Xiao, D. Hes1 Is Involved in the Self-Renewal and Tumourigenicity of Stem-like Cancer Cells in Colon Cancer. Sci. Rep. 2014, 4, 3963. [Google Scholar] [CrossRef]
- Gao, F.; Huang, W.; Zhang, Y.; Tang, S.; Zheng, L.; Ma, F.; Wang, Y.; Tang, H.; Li, X. Hes1 Promotes Cell Proliferation and Migration by Activating Bmi-1 and PTEN/Akt/GSK3β Pathway in Human Colon Cancer. Oncotarget 2015, 6, 38667–38680. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Liu, X.; Wang, M.; Wang, P.; Yang, J.; Zhang, S. Lutein Inhibits Proliferation, Invasion and Migration of Hypoxic Breast Cancer Cells via Downregulation of HES1. Int. J. Oncol. 2018, 52, 2119–2129. [Google Scholar] [CrossRef]
- Cenciarelli, C.; Marei, H.E.; Zonfrillo, M.; Casalbore, P.; Felsani, A.; Giannetti, S.; Trevisi, G.; Althani, A.; Mangiola, A. The Interference of Notch1 Target Hes1 Affects Cell Growth, Differentiation and Invasiveness of Glioblastoma Stem Cells through Modulation of Multiple Oncogenic Targets. Oncotarget 2017, 8, 17873–17886. [Google Scholar] [CrossRef]
- Jiang, J.; Liu, X. Upregulated EBI3 Correlates with Poor Outcome and Tumor Progression in Breast Cancer. Oncol. Res. Treat. 2018, 41, 111–115. [Google Scholar] [CrossRef]
- Hou, Y.-M.; Dong, J.; Liu, M.-Y.; Yu, S. Expression of Epstein-Barr Virus-Induced Gene 3 in Cervical Cancer: Association with Clinicopathological Parameters and Prognosis. Oncol. Lett. 2016, 11, 330–334. [Google Scholar] [CrossRef]
- Yonekura, S. Epstein-Barr Virus-Induced Gene 3 as a Novel Biomarker in Metastatic Melanoma With Infiltrating CD8+ T Cells: A Study Based on The Cancer Genome Atlas (TCGA). Anticancer Res. 2022, 42, 511–517. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, J.-Q.; Shi, Y.; Zhu, X.; Liu, Z.; Li, M.-S.; Yu, J.; Wu, L.-C.; He, Y.; Zhang, G.; et al. Epstein-Barr Virus-Induced Gene 3-Deficiency Leads to Impaired Antitumor T-Cell Responses and Accelerated Tumor Growth. Oncoimmunology 2015, 4, e989137. [Google Scholar] [CrossRef]
- Han, Y.; Li, X.; Yan, J.; Ma, C.; Wang, X.; Pan, H.; Zheng, X.; Zhang, Z.; Gao, B.; Ji, X.-Y. Bioinformatic Analysis Identifies Potential Key Genes in the Pathogenesis of Melanoma. Front. Oncol. 2020, 10, 581985. [Google Scholar] [CrossRef]
- Leick, K.M.; Rodriguez, A.B.; Melssen, M.M.; Benamar, M.; Lindsay, R.S.; Eki, R.; Du, K.-P.; Parlak, M.; Abbas, T.; Engelhard, V.H.; et al. The Barrier Molecules Junction Plakoglobin, Filaggrin, and Dystonin Play Roles in Melanoma Growth and Angiogenesis. Ann. Surg. 2019, 270, 712–722. [Google Scholar] [CrossRef]
- Wu, J.; Lu, Y.; Qin, A.; Qiao, Z.; Jiang, X. Overexpression of RAB34 Correlates with Poor Prognosis and Tumor Progression in Hepatocellular Carcinoma. Oncol. Rep. 2017, 38, 2967–2974. [Google Scholar] [CrossRef]
- Sun, L.; Xu, X.; Chen, Y.; Zhou, Y.; Tan, R.; Qiu, H.; Jin, L.; Zhang, W.; Fan, R.; Hong, W.; et al. Rab34 Regulates Adhesion, Migration, and Invasion of Breast Cancer Cells. Oncogene 2018, 37, 3698–3714. [Google Scholar] [CrossRef]
- Wang, H.; Gao, Y.; Chen, L.; Li, Y.; Jiang, C. RAB34 Was a Progression- and Prognosis-Associated Biomarker in Gliomas. Tumour Biol. 2015, 36, 1573–1578. [Google Scholar] [CrossRef]
- Leucci, E.; Vendramin, R.; Spinazzi, M.; Laurette, P.; Fiers, M.; Wouters, J.; Radaelli, E.; Eyckerman, S.; Leonelli, C.; Vanderheyden, K.; et al. Melanoma Addiction to the Long Non-Coding RNA SAMMSON. Nature 2016, 531, 518–522. [Google Scholar] [CrossRef]
- Han, S.; Yan, Y.; Ren, Y.; Hu, Y.; Wang, Y.; Chen, L.; Zhi, Z.; Zheng, Y.; Shao, Y.; Liu, J. LncRNA SAMMSON Mediates Adaptive Resistance to RAF Inhibition in BRAF-Mutant Melanoma Cells. Cancer Res. 2021, 81, 2918–2929. [Google Scholar] [CrossRef]
- Somura, H.; Iizuka, N.; Tamesa, T.; Sakamoto, K.; Hamaguchi, T.; Tsunedomi, R.; Yamada-Okabe, H.; Sawamura, M.; Eramoto, M.; Miyamoto, T.; et al. A Three-Gene Predictor for Early Intrahepatic Recurrence of Hepatocellular Carcinoma after Curative Hepatectomy. Oncol. Rep. 2008, 19, 489–495. [Google Scholar] [CrossRef]
- Berberich, A.; Bartels, F.; Tang, Z.; Knoll, M.; Pusch, S.; Hucke, N.; Kessler, T.; Dong, Z.; Wiestler, B.; Winkler, F.; et al. LAPTM5-CD40 Crosstalk in Glioblastoma Invasion and Temozolomide Resistance. Front. Oncol. 2020, 10, 747. [Google Scholar] [CrossRef]
- Li, X.; Su, Y.; Zhang, J.; Zhu, Y.; Xu, Y.; Wu, G. LAPTM5 Plays a Key Role in the Diagnosis and Prognosis of Testicular Germ Cell Tumors. Int. J. Genom. 2021, 2021, 8816456. [Google Scholar] [CrossRef]
- Yao, C.D.; Haensel, D.; Gaddam, S.; Patel, T.; Atwood, S.X.; Sarin, K.Y.; Whitson, R.J.; McKellar, S.; Shankar, G.; Aasi, S.; et al. AP-1 and TGFß Cooperativity Drives Non-Canonical Hedgehog Signaling in Resistant Basal Cell Carcinoma. Nat. Commun. 2020, 11, 5079. [Google Scholar] [CrossRef]
- Barros-Silva, J.D.; Linn, D.E.; Steiner, I.; Guo, G.; Ali, A.; Pakula, H.; Ashton, G.; Peset, I.; Brown, M.; Clarke, N.W.; et al. Single-Cell Analysis Identifies LY6D as a Marker Linking Castration-Resistant Prostate Luminal Cells to Prostate Progenitors and Cancer. Cell Rep. 2018, 25, 3504–3518.e6. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Fan, J.; Gao, W.; Wu, Y.; Zhao, Q.; Chen, B.; Ding, Y.; Wen, S.; Nan, X.; Wang, B. LY6D as a Chemoresistance Marker Gene and Therapeutic Target for Laryngeal Squamous Cell Carcinoma. Stem Cells Dev. 2020, 29, 774–785. [Google Scholar] [CrossRef] [PubMed]
- Mayama, A.; Takagi, K.; Suzuki, H.; Sato, A.; Onodera, Y.; Miki, Y.; Sakurai, M.; Watanabe, T.; Sakamoto, K.; Yoshida, R.; et al. OLFM4, LY6D and S100A7 as Potent Markers for Distant Metastasis in Estrogen Receptor-Positive Breast Carcinoma. Cancer Sci. 2018, 109, 3350–3359. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Lemon, W.; Liu, P.-Y.; Yi, Y.; Morrison, C.; Yang, P.; Sun, Z.; Szoke, J.; Gerald, W.L.; Watson, M.; et al. A Gene Expression Signature Predicts Survival of Patients with Stage I Non-Small Cell Lung Cancer. PLoS Med. 2006, 3, e467. [Google Scholar] [CrossRef]
- Kang, X.; Kong, F.; Huang, K.; Li, L.; Li, Z.; Wang, X.; Zhang, W.; Wu, X. LncRNA MIR210HG Promotes Proliferation and Invasion of Non-Small Cell Lung Cancer by Upregulating Methylation of CACNA2D2 Promoter via Binding to DNMT1. Onco Targets Ther. 2019, 12, 3779–3790. [Google Scholar] [CrossRef]
- Warnier, M.; Roudbaraki, M.; Derouiche, S.; Delcourt, P.; Bokhobza, A.; Prevarskaya, N.; Mariot, P. CACNA2D2 Promotes Tumorigenesis by Stimulating Cell Proliferation and Angiogenesis. Oncogene 2015, 34, 5383–5394. [Google Scholar] [CrossRef]
- Rossi, E.; Picozzi, P.; Bodega, B.; Lavazza, C.; Carlo-Stella, C.; Marozzi, A.; Ginelli, E. Forced Expression of RDH10 Gene Retards Growth of HepG2 Cells. Cancer Biol. Ther. 2007, 6, 238–244. [Google Scholar] [CrossRef]
- Sun, E.; Liu, K.; Zhao, K.; Wang, L. Serine/Threonine Kinase 32C Is Overexpressed in Bladder Cancer and Contributes to Tumor Progression. Cancer Biol. Ther. 2019, 20, 307–320. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, X.; Cao, W.; Hua, Z.-C. Tumor-Specifically Hypoxia-Induced Therapy of SPRY1/2 Displayed Differential Therapeutic Efficacy for Melanoma. Am. J. Cancer Res. 2015, 5, 792–801. [Google Scholar]
- Ahn, J.-H.; Han, B.-I.; Lee, M. Induction of Resistance to BRAF Inhibitor Is Associated with the Inability of Spry2 to Inhibit BRAF-V600E Activity in BRAF Mutant Cells. Biomol. Ther. 2015, 23, 320–326. [Google Scholar] [CrossRef]
- Fong, C.W.; Chua, M.-S.; McKie, A.B.; Ling, S.H.M.; Mason, V.; Li, R.; Yusoff, P.; Lo, T.L.; Leung, H.Y.; So, S.K.S.; et al. Sprouty 2, an Inhibitor of Mitogen-Activated Protein Kinase Signaling, Is down-Regulated in Hepatocellular Carcinoma. Cancer Res. 2006, 66, 2048–2058. [Google Scholar] [CrossRef]
- Holderfield, M.; Deuker, M.M.; McCormick, F.; McMahon, M. Targeting RAF Kinases for Cancer Therapy: BRAF Mutated Melanoma and Beyond. Nat. Rev. Cancer 2014, 14, 455–467. [Google Scholar] [CrossRef]
- Stecca, B.; Mas, C.; Clement, V.; Zbinden, M.; Correa, R.; Piguet, V.; Beermann, F.; Ruiz, I.; Altaba, A. Melanomas Require HEDGEHOG-GLI Signaling Regulated by Interactions between GLI1 and the RAS-MEK/AKT Pathways. Proc. Natl. Acad. Sci. USA 2007, 104, 5895–5900. [Google Scholar] [CrossRef]
- Vlčková, K.; Réda, J.; Ondrušová, L.; Krayem, M.; Ghanem, G.; Vachtenheim, J. GLI Inhibitor GANT61 Kills Melanoma Cells and Acts in Synergy with Obatoclax. Int. J. Oncol. 2016, 49, 953–960. [Google Scholar] [CrossRef]
- Liang, G.; Liu, M.; Wang, Q.; Shen, Y.; Mei, H.; Li, D.; Liu, W. Itraconazole Exerts Its Anti-Melanoma Effect by Suppressing Hedgehog, Wnt, and PI3K/MTOR Signaling Pathways. Oncotarget 2017, 8, 28510–28525. [Google Scholar] [CrossRef]
- Pietrobono, S.; Gaudio, E.; Gagliardi, S.; Zitani, M.; Carrassa, L.; Migliorini, F.; Petricci, E.; Manetti, F.; Makukhin, N.; Bond, A.G.; et al. Targeting Non-Canonical Activation of GLI1 by the SOX2-BRD4 Transcriptional Complex Improves the Efficacy of HEDGEHOG Pathway Inhibition in Melanoma. Oncogene 2021, 40, 3799–3814. [Google Scholar] [CrossRef]
- Chen, J.K.; Taipale, J.; Cooper, M.K.; Beachy, P.A. Inhibition of Hedgehog Signaling by Direct Binding of Cyclopamine to Smoothened. Genes Dev. 2002, 16, 2743–2748. [Google Scholar] [CrossRef]
- Trnski, D.; Sabol, M.; Gojević, A.; Martinić, M.; Ozretić, P.; Musani, V.; Ramić, S.; Levanat, S. GSK3β and Gli3 Play a Role in Activation of Hedgehog-Gli Pathway in Human Colon Cancer-Targeting GSK3β Downregulates the Signaling Pathway and Reduces Cell Proliferation. Biochim. Biophys. Acta 2015, 1852, 2574–2584. [Google Scholar] [CrossRef]
- Lauth, M.; Bergström, A.; Shimokawa, T.; Toftgård, R. Inhibition of GLI-Mediated Transcription and Tumor Cell Growth by Small-Molecule Antagonists. Proc. Natl. Acad. Sci. USA 2007, 104, 8455–8460. [Google Scholar] [CrossRef]
- Han, J.; Sang, F.; Chang, J.; Hua, Y.; Shi, W.; Tang, L.; Liu, L. Arsenic Trioxide Inhibits Viability of Pancreatic Cancer Stem Cells in Culture and in a Xenograft Model via Binding to SHH-Gli. Onco Targets Ther. 2013, 6, 1129–1138. [Google Scholar] [CrossRef]
- Mazumdar, T.; Devecchio, J.; Agyeman, A.; Shi, T.; Houghton, J.A. The GLI Genes as the Molecular Switch in Disrupting Hedgehog Signaling in Colon Cancer. Oncotarget 2011, 2, 638–645. [Google Scholar] [CrossRef]
- Wickström, M.; Dyberg, C.; Shimokawa, T.; Milosevic, J.; Baryawno, N.; Fuskevåg, O.M.; Larsson, R.; Kogner, P.; Zaphiropoulos, P.G.; Johnsen, J.I. Targeting the Hedgehog Signal Transduction Pathway at the Level of GLI Inhibits Neuroblastoma Cell Growth in Vitro and in Vivo. Int. J. Cancer 2013, 132, 1516–1524. [Google Scholar] [CrossRef]
- Miyazaki, Y.; Matsubara, S.; Ding, Q.; Tsukasa, K.; Yoshimitsu, M.; Kosai, K.; Takao, S. Efficient Elimination of Pancreatic Cancer Stem Cells by Hedgehog/GLI Inhibitor GANT61 in Combination with MTOR Inhibition. Mol. Cancer 2016, 15, 49. [Google Scholar] [CrossRef]
- Santini, R.; Vinci, M.C.; Pandolfi, S.; Penachioni, J.Y.; Montagnani, V.; Olivito, B.; Gattai, R.; Pimpinelli, N.; Gerlini, G.; Borgognoni, L.; et al. HEDGEHOG-GLI Signaling Drives Self-Renewal and Tumorigenicity of Human Melanoma-Initiating Cells. Stem Cells 2012, 30, 1808–1818. [Google Scholar] [CrossRef]
- Peer, E.; Tesanovic, S.; Aberger, F. Next-Generation Hedgehog/GLI Pathway Inhibitors for Cancer Therapy. Cancers 2019, 11, 538. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, A.; Chaudhary, S.C.; Rana, M.; Elmets, C.A.; Athar, M. Basal Cell Carcinoma Pathogenesis and Therapy Involving Hedgehog Signaling and Beyond. Mol. Carcinog. 2017, 56, 2543–2557. [Google Scholar] [CrossRef]
- Wessler, S.; Krisch, L.M.; Elmer, D.P.; Aberger, F. From Inflammation to Gastric Cancer—The Importance of Hedgehog/GLI Signaling in Helicobacter Pylori-Induced Chronic Inflammatory and Neoplastic Diseases. Cell Commun. Signal. 2017, 15, 15. [Google Scholar] [CrossRef]
- Akyala, A.I.; Peppelenbosch, M.P. Gastric Cancer and Hedgehog Signaling Pathway: Emerging New Paradigms. Genes Cancer 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Tsavachidou, D.; Coleman, M.L.; Athanasiadis, G.; Li, S.; Licht, J.D.; Olson, M.F.; Weber, B.L. SPRY2 Is an Inhibitor of the Ras/Extracellular Signal-Regulated Kinase Pathway in Melanocytes and Melanoma Cells with Wild-Type BRAF but Not with the V599E Mutant. Cancer Res. 2004, 64, 5556–5559. [Google Scholar] [CrossRef]
- Chen, L.; Wang, G.; Luo, Y.; Wang, Y.; Xie, C.; Jiang, W.; Xiao, Y.; Qian, G.; Wang, X. Downregulation of LAPTM5 Suppresses Cell Proliferation and Viability Inducing Cell Cycle Arrest at G0/G1 Phase of Bladder Cancer Cells. Int. J. Oncol. 2017, 50, 263–271. [Google Scholar] [CrossRef]

| Gene | Function According to Gene Cards | Role in Cancer | Reference |
|---|---|---|---|
| KRT16 | type I keratin that regulates innate immunity in response to skin barrier breach | regulates immune response, metastasis, cancer stemness and drug resistance in melanoma, SCC, and breast cancer | [33,34,35,36] |
| KRT17 | type I keratin involved in regulation of protein synthesis and epithelial cell growth | regulates therapy resistance, proliferation, migration and invasion in CRC, pancreatic cancer, and NSCLC | [37,38,39,40] |
| S100A7 | member of the S100 family of proteins involved in the regulation of cell cycle and differentiation | regulates tumor invasion, angiogenesis, migration, EMT and chemoresistance in melanoma, cervical cancer, and ovarian cancer | [41,42,43,44,45] |
| S100A9 | calcium- and zinc-binding protein involved in immune response | regulates chemoresistance, cell invasion and metastasis in melanoma, cervical carcinoma, and prostate cancer | [46,47,48,49] |
| GH1 | member of the somatotropin/prolactin family of hormones important for growth control | dysregulates MAPK pathway and blocks cell motility in colon cancer | [50] |
| SOX9 | transcription factor important for differentiation and skeletal development | regulates metastasis, cell invasion, migration and stemness in melanoma, CRC, and esophageal cancer | [51,52,53,54] |
| MRAS | Ras GTPase that functions as signal transducer in cell growth and differentiation | regulates MAPK pathway and drives tumorigenesis in gastric cancer and prostate cancer | [55,56,57] |
| BIRC7 | member of the inhibitor of apoptosis protein family associated with cancer progression and chemotherapy resistance | regulates chemoresistance and can serve as a biomarker in prostate cancer, melanoma, and lung cancer | [58,59,60,61,62,63,64,65,66] |
| IL1R2 | cytokine receptor that belongs to the interleukin 1 receptor family | regulates proliferation, angiogenesis and tumorigenesis initiation in breast cancer, melanoma, gastric cancer, and CRC | [67,68,69,70,71] |
| RET | receptor tyrosine-protein kinase involved in cell proliferation, migration, and differentiation | RET fusion are associated with tumorigenesis of chronic myelomonocytic leukemia | [72] |
| HES1 | transcriptional repressor involved in cell differentiation, cell cycle, apoptosis, and self-renewal | regulates cell proliferation, invasion and self-renewal in CRC, breast cancer and glioblastoma | [73,74,75,76] |
| EBI3 | secretory glycoprotein belonging to the hematopoietin receptor family involved in IL-27 formation | EBI3 overexpression is associated with poor prognosis of breast and cervical cancer and impaired immune response in melanoma | [77,78,79,80] |
| FLG | intermediate filament-associated protein that aggregates keratin intermediate filaments in mammalian epidermis | regulates growth and angiogenesis and can be valuable in prognosis and treatment of melanoma | [81,82] |
| RAB34 | small GTPase involved in protein transport and ciliogenesis pathways | regulates cell adhesion, migration and invasion in breast cancer and correlates with tumor progression of HCC and glioma | [83,84,85] |
| SAMMSON | lncRNA with crucial role in cell survival and mitochondrial metabolism | regulates therapy response and mitochondrial function in melanoma | [86,87] |
| LAPTM5 | transmembrane receptor associated with lysosomes | potential biomarker for HCC, glioblastoma, and testicular cancer | [88,89,90] |
| LY6D | marker at earliest stage specification of lymphocytes between B-and T-cell development | therapy outcome and survival prediction in BCC, prostate cancer, NSCLC, laryngeal cancer, and breast cancer | [91,92,93,94,95] |
| CACNA2D2 | alpha-2/delta subunit of the voltage-dependent calcium channel complex | regulates cell proliferation and angiogenesis in prostate cancer, while CACAN2D2 inhibition induces NSCLC tumorigenesis | [96,97] |
| RDH10 | retinol dehydrogenase essential for organ development | RDH10 overexpression has an antiproliferative effect in hepatocellular carcinoma | [98] |
| STK32C | serine/threonine protein kinase | STK32C overexpression in bladder cancer contributes to tumor progression | [99] |
| SPRY2 | inhibitor of RTK signaling proteins activity | SPRY2 inhibits cell growth and therapy resistance occurrence via MAPK pathway in melanoma and hepatocellular carcinoma | [100,101,102] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurtović, M.; Piteša, N.; Bartoniček, N.; Ozretić, P.; Musani, V.; Čonkaš, J.; Petrić, T.; King, C.; Sabol, M. RNA-seq and ChIP-seq Identification of Unique and Overlapping Targets of GLI Transcription Factors in Melanoma Cell Lines. Cancers 2022, 14, 4540. https://doi.org/10.3390/cancers14184540
Kurtović M, Piteša N, Bartoniček N, Ozretić P, Musani V, Čonkaš J, Petrić T, King C, Sabol M. RNA-seq and ChIP-seq Identification of Unique and Overlapping Targets of GLI Transcription Factors in Melanoma Cell Lines. Cancers. 2022; 14(18):4540. https://doi.org/10.3390/cancers14184540
Chicago/Turabian StyleKurtović, Matea, Nikolina Piteša, Nenad Bartoniček, Petar Ozretić, Vesna Musani, Josipa Čonkaš, Tina Petrić, Cecile King, and Maja Sabol. 2022. "RNA-seq and ChIP-seq Identification of Unique and Overlapping Targets of GLI Transcription Factors in Melanoma Cell Lines" Cancers 14, no. 18: 4540. https://doi.org/10.3390/cancers14184540
APA StyleKurtović, M., Piteša, N., Bartoniček, N., Ozretić, P., Musani, V., Čonkaš, J., Petrić, T., King, C., & Sabol, M. (2022). RNA-seq and ChIP-seq Identification of Unique and Overlapping Targets of GLI Transcription Factors in Melanoma Cell Lines. Cancers, 14(18), 4540. https://doi.org/10.3390/cancers14184540










