A Novel Risk Score Model of Lactate Metabolism for Predicting over Survival and Immune Signature in Lung Adenocarcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Data and Method
2.1. Data Acquisition and Top Genes Identification
2.2. Method
2.2.1. The Basic Genetic Landscape of Lactate-Related Genes
2.2.2. Pairwise Correlation of Lactate Metabolism-Associated Gene and Its Relationship with OS
2.2.3. Immune Cells Infiltration
2.2.4. GSVA (Hallmark)
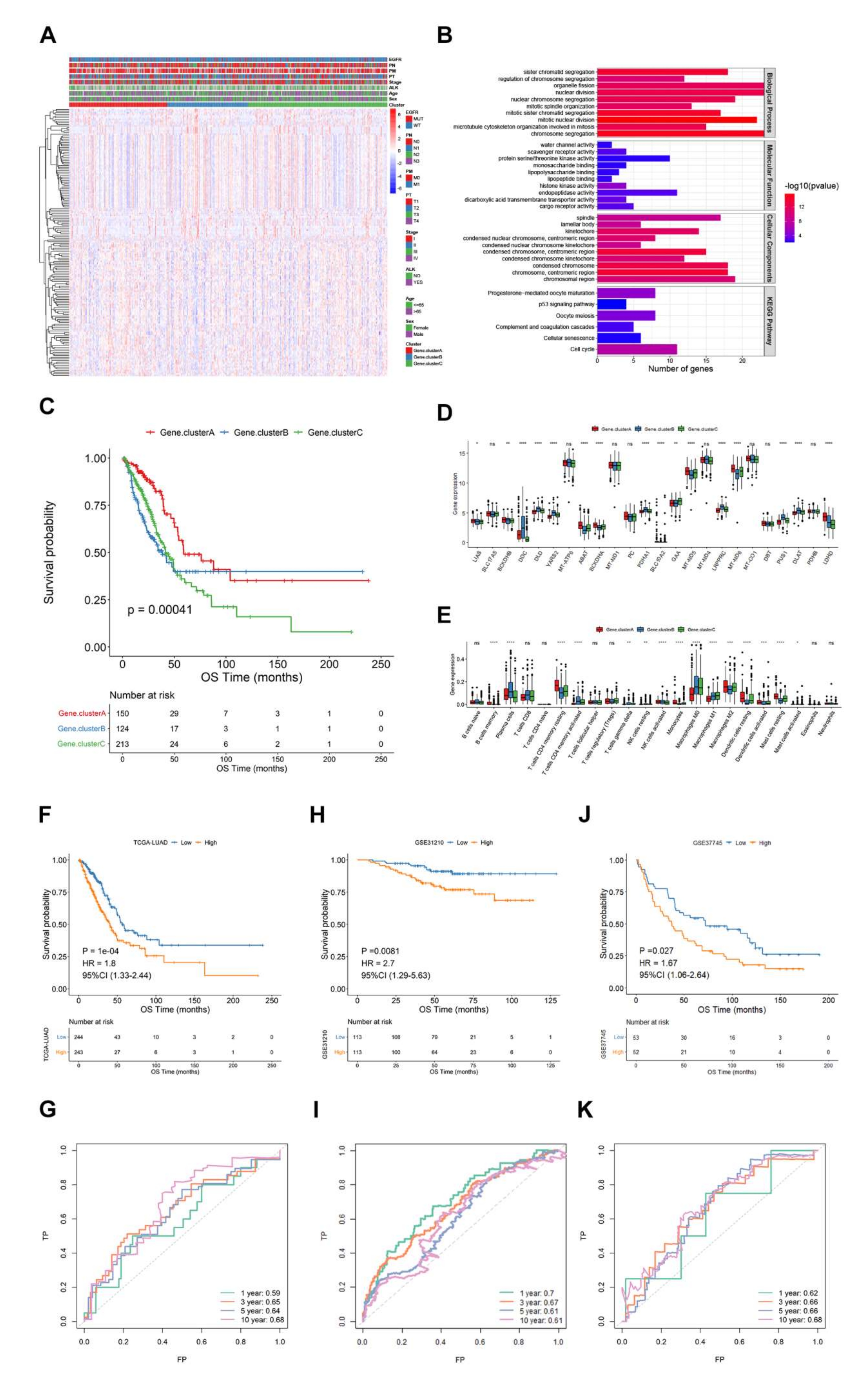
2.2.5. Tumor Samples Were Classified Based on Lactate Metabolism-Associated Gene
2.2.6. Genotyping of Tumor Samples Based on Differential Genes
2.2.7. Construction and Validation of Prognostic Models
2.2.8. Evaluation of Proportion of Immune Infiltrating Cells
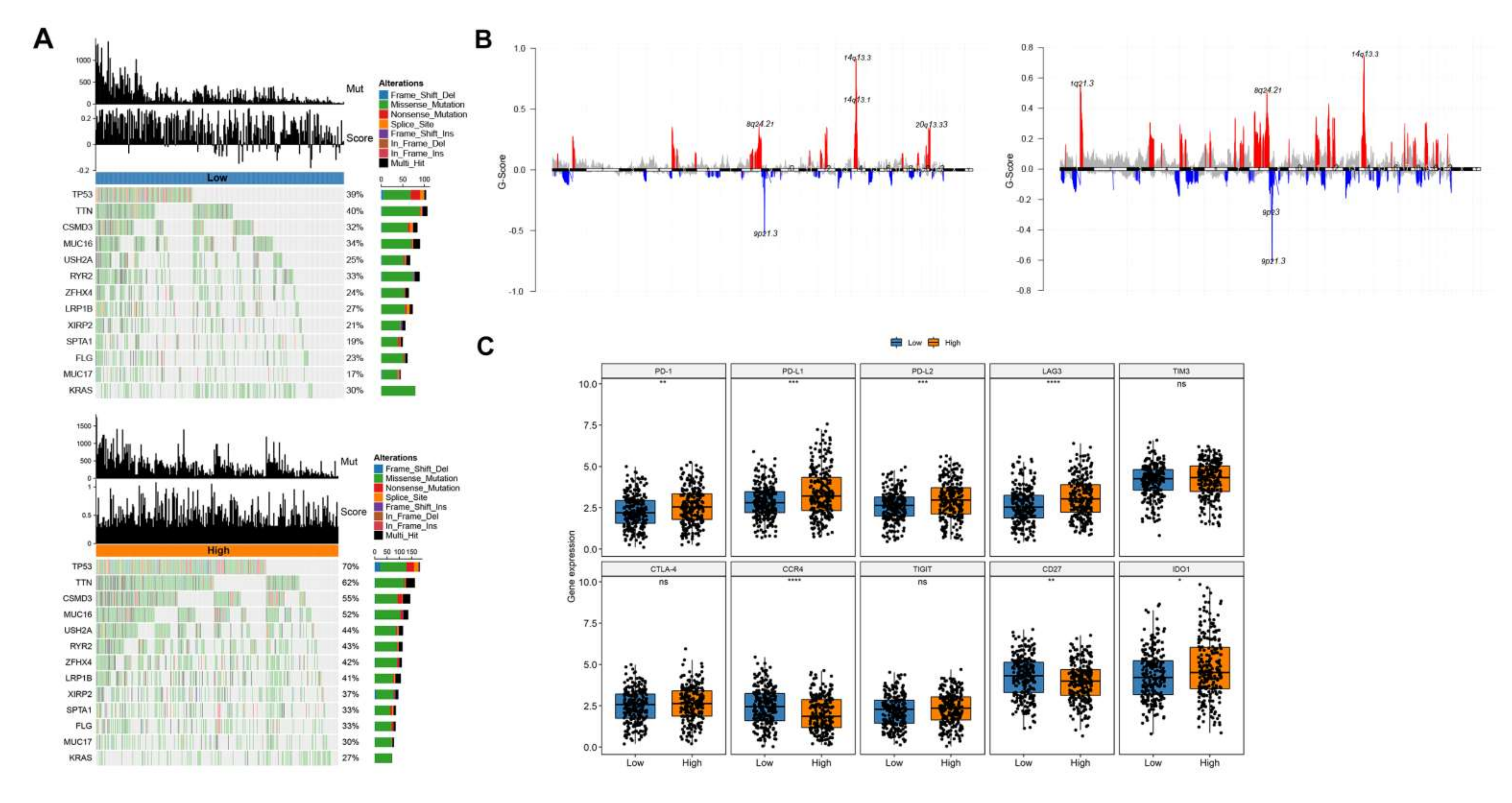
2.2.9. Mutations and CNV Differences in High and Low Risk Group
2.2.10. Validation by Clinical Immunotherapy Data
2.2.11. Statistical Analysis
3. Results
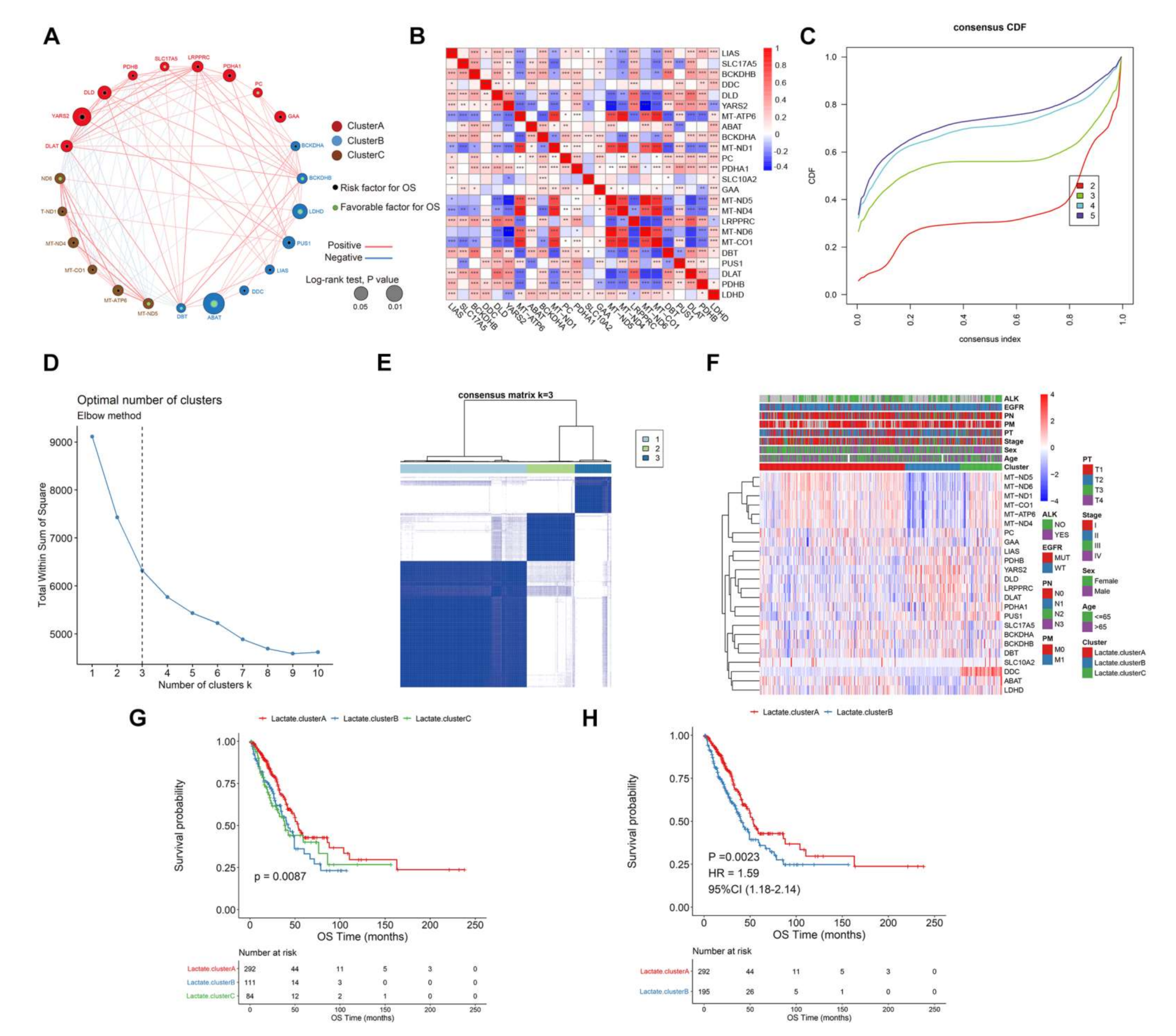
3.1. Landscape of Genetic Variation and Immune Infiltration of Lactic Genes in LUAD
3.2. Survival Patterns Classified by 24 Lactate Metabolism-Associated Gene
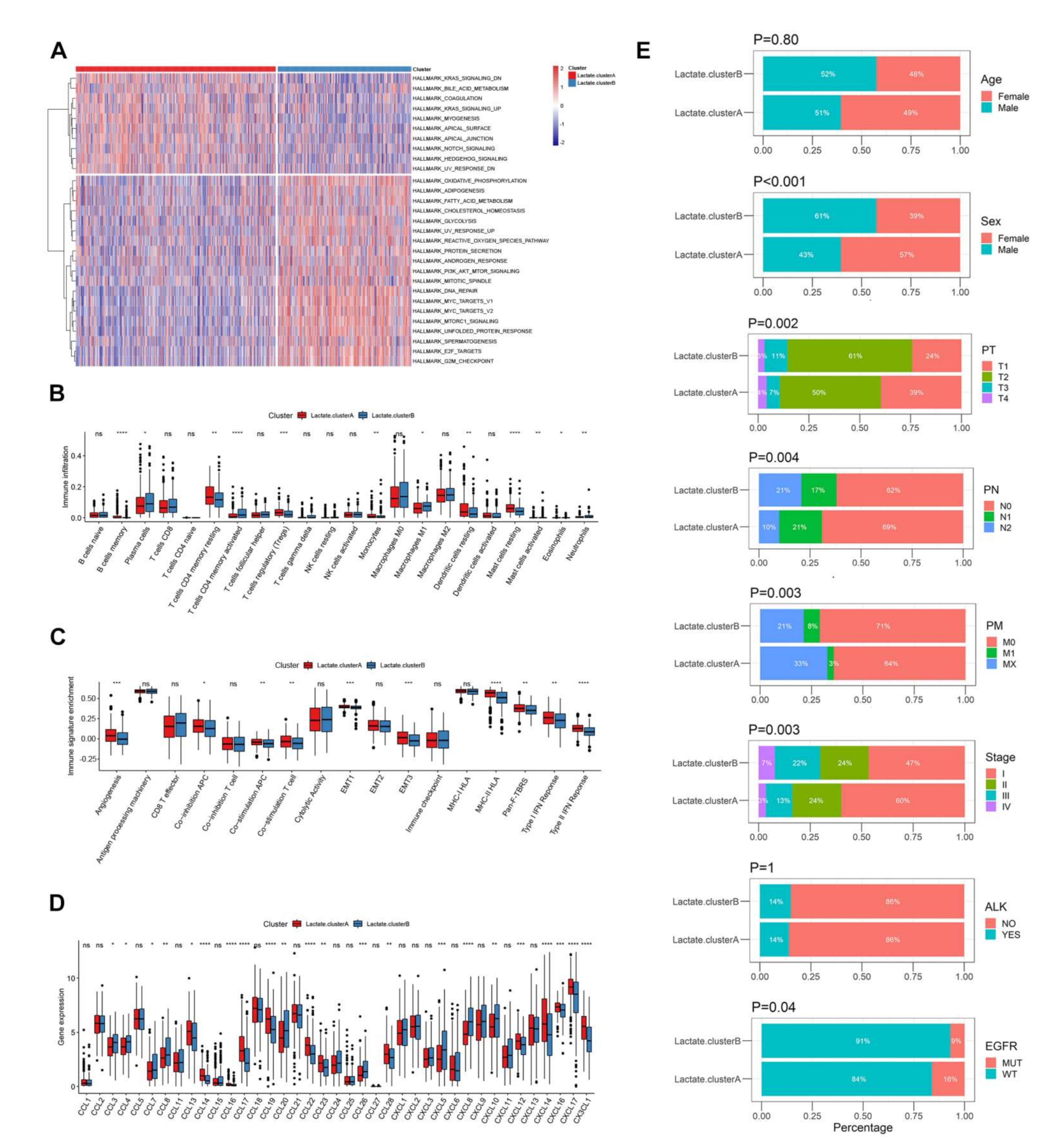
3.3. Biological Function and TME Cell Infiltration Characteristics in Two Lactate Metabolism-Associated Gene Patterns
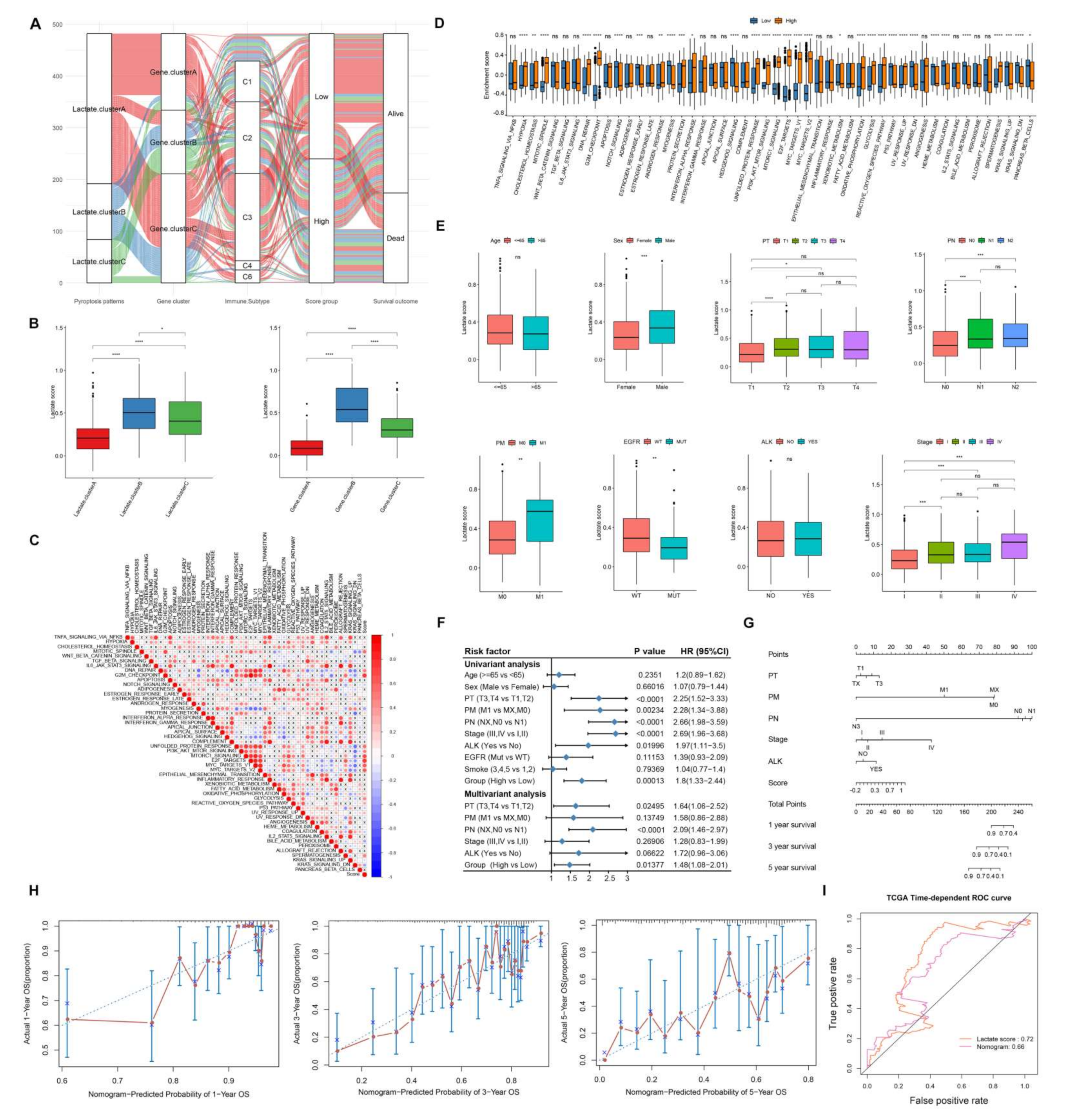
3.4. Lactate Related Score Model Construction and Verification
3.5. Characteristics of Lactate Related Score Risk Model
3.6. Genes of Model in the Role of Anti-PD-1/L1 Immunotherapy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siska, P.J.; Singer, K.; Evert, K.; Renner, K.; Kreutz, M. The immunological Warburg effect: Can a metabolic-tumor-stroma score (MeTS) guide cancer immunotherapy? Immunol. Rev. 2020, 295, 187–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbet, C.; Feron, O. Tumour acidosis: From the passenger to the driver’s seat. Nat. Rev. Cancer 2017, 17, 577–593. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Lin, C.; He, Y.; Lu, L.; Xu, K.; Tao, B.; Xia, Z.; Zeng, R.; Mao, Y.; Luo, Z.; et al. Engineering of Cascade-Responsive Nanoplatform to Inhibit Lactate Efflux for Enhanced Tumor Chemo-Immunotherapy. ACS Nano 2020, 14, 14164–14180. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Franco, F.; Tsui, Y.C.; Xie, X.; Trefny, M.P.; Zappasodi, R.; Mohmood, S.R.; Fernandez-Garcia, J.; Tsai, C.H.; Schulze, I.; et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat. Immunol. 2020, 21, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.G.; Stromnes, I.M.; Greenberg, P.D. Obstacles Posed by the Tumor Microenvironment to T cell Activity: A Case for Synergistic Therapies. Cancer Cell 2017, 31, 311–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, P.J. On macrophage diversity and inflammatory metabolic timers. Nat. Rev. Immunol. 2020, 20, 89–90. [Google Scholar] [CrossRef]
- Marin, E.; Bouchet-Delbos, L.; Renoult, O.; Louvet, C.; Nerriere-Daguin, V.; Managh, A.J.; Even, A.; Giraud, M.; Vu Manh, T.P.; Aguesse, A.; et al. Human Tolerogenic Dendritic Cells Regulate Immune Responses through Lactate Synthesis. Cell Metab. 2019, 30, 1075–1090.e8. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Zhao, Y.Y.; Shen, J.; Sun, X.; Liu, Y.; Liu, H.; Wang, Y.; Wang, J. Nanoenabled Modulation of Acidic Tumor Microenvironment Reverses Anergy of Infiltrating T Cells and Potentiates Anti-PD-1 Therapy. Nano Lett. 2019, 19, 2774–2783. [Google Scholar] [CrossRef]
- Watson, M.J.; Vignali, P.D.A.; Mullett, S.J.; Overacre-Delgoffe, A.E.; Peralta, R.M.; Grebinoski, S.; Menk, A.V.; Rittenhouse, N.L.; DePeaux, K.; Whetstone, R.D.; et al. Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature 2021, 591, 645–651. [Google Scholar] [CrossRef]
- Wu, Q.; Liang, X.; Wang, K.; Lin, J.; Wang, X.; Wang, P.; Zhang, Y.; Nie, Q.; Liu, H.; Zhang, Z.; et al. Intestinal hypoxia-inducible factor 2alpha regulates lactate levels to shape the gut microbiome and alter thermogenesis. Cell Metab. 2021, 33, 1988–2003.e7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, G.; Xu, Z.G.; Tu, H.; Hu, F.; Dai, J.; Chang, Y.; Chen, Y.; Lu, Y.; Zeng, H.; et al. Lactate Is a Natural Suppressor of RLR Signaling by Targeting MAVS. Cell 2019, 178, 176–189.e115. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.; Zhang, P.; Wang, B.; Yang, D.; Duan, X.; Jiang, Y.; Xu, T.; Jiang, Y.; Shi, J.; Ding, C.; et al. Aurora-A mediated phosphorylation of LDHB promotes glycolysis and tumor progression by relieving the substrate-inhibition effect. Nat. Commun. 2019, 10, 5566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, S.J.; Tyler, B.M.; Gould, T.; Veal, G.J.; Gorelick, N.; Rowlinson, J.; Serra, R.; Ritchie, A.; Berry, P.; Otto, A.; et al. Overall Survival in Malignant Glioma Is Significantly Prolonged by Neurosurgical Delivery of Etoposide and Temozolomide from a Thermo-Responsive Biodegradable Paste. Clin. Cancer Res. 2019, 25, 5094–5106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundo, K.; Trauelsen, M.; Pedersen, S.F.; Schwartz, T.W. Why Warburg Works: Lactate Controls Immune Evasion through GPR81. Cell Metab. 2020, 31, 666–668. [Google Scholar] [CrossRef]
- Li, X.; Jiang, Y.; Meisenhelder, J.; Yang, W.; Hawke, D.H.; Zheng, Y.; Xia, Y.; Aldape, K.; He, J.; Hunter, T.; et al. Mitochondria-Translocated PGK1 Functions as a Protein Kinase to Coordinate Glycolysis and the TCA Cycle in Tumorigenesis. Mol. Cell 2016, 61, 705–719. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.T.; Li, H.; Borch Jensen, M.; Maksoud, E.; Borneo, J.; Liang, Y.; Quake, S.R.; Luo, L.; Haghighi, P.; Jasper, H. Gut cytokines modulate olfaction through metabolic reprogramming of glia. Nature 2021, 596, 97–102. [Google Scholar] [CrossRef]
- Walton, Z.E.; Patel, C.H.; Brooks, R.C.; Yu, Y.; Ibrahim-Hashim, A.; Riddle, M.; Porcu, A.; Jiang, T.; Ecker, B.L.; Tameire, F.; et al. Acid Suspends the Circadian Clock in Hypoxia through Inhibition of mTOR. Cell 2018, 174, 72–87.e32. [Google Scholar] [CrossRef] [Green Version]
- Certo, M.; Tsai, C.H.; Pucino, V.; Ho, P.C.; Mauro, C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat. Rev. Immunol. 2021, 21, 151–161. [Google Scholar] [CrossRef]
- Tekpli, X.; Lien, T.; Rossevold, A.H.; Nebdal, D.; Borgen, E.; Ohnstad, H.O.; Kyte, J.A.; Vallon-Christersson, J.; Fongaard, M.; Due, E.U.; et al. An independent poor-prognosis subtype of breast cancer defined by a distinct tumor immune microenvironment. Nat. Commun. 2019, 10, 5499. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Tang, Y.; Liu, W.L.; Zou, M.Z.; Huang, C.; Liu, C.J.; Zhang, X.Z. Intra/Extracellular Lactic Acid Exhaustion for Synergistic Metabolic Therapy and Immunotherapy of Tumors. Adv. Mater. 2019, 31, e1904639. [Google Scholar] [CrossRef]
- Locatelli, S.L.; Careddu, G.; Serio, S.; Consonni, F.M.; Maeda, A.; Viswanadha, S.; Vakkalanka, S.; Castagna, L.; Santoro, A.; Allavena, P.; et al. Targeting Cancer Cells and Tumor Microenvironment in Preclinical and Clinical Models of Hodgkin Lymphoma Using the Dual PI3Kdelta/gamma Inhibitor RP6530. Clin. Cancer Res. 2019, 25, 1098–1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Huo, M.; Xu, Z.; Wang, Y.; Huang, L. Nanoparticle delivery of CDDO-Me remodels the tumor microenvironment and enhances vaccine therapy for melanoma. Biomaterials 2015, 68, 54–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, G.A. The Science and Translation of Lactate Shuttle Theory. Cell Metab. 2018, 27, 757–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burr, S.P.; Costa, A.S.; Grice, G.L.; Timms, R.T.; Lobb, I.T.; Freisinger, P.; Dodd, R.B.; Dougan, G.; Lehner, P.J.; Frezza, C.; et al. Mitochondrial Protein Lipoylation and the 2-Oxoglutarate Dehydrogenase Complex Controls HIF1alpha Stability in Aerobic Conditions. Cell Metab. 2016, 24, 740–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Batran, R.; Gopal, K.; Capozzi, M.E.; Chahade, J.J.; Saleme, B.; Tabatabaei-Dakhili, S.A.; Greenwell, A.A.; Niu, J.; Almutairi, M.; Byrne, N.J.; et al. Pimozide Alleviates Hyperglycemia in Diet-Induced Obesity by Inhibiting Skeletal Muscle Ketone Oxidation. Cell Metab. 2020, 31, 909–919.e8. [Google Scholar] [CrossRef]
- Li, Y.; Mo, H.; Wu, S.; Liu, X.; Tu, K. A Novel Lactate Metabolism-Related Gene Signature for Predicting Clinical Outcome and Tumor Microenvironment in Hepatocellular Carcinoma. Front. Cell Dev. Biol. 2021, 9, 801959. [Google Scholar] [CrossRef]
- Xia, L.; Oyang, L.; Lin, J.; Tan, S.; Han, Y.; Wu, N.; Yi, P.; Tang, L.; Pan, Q.; Rao, S.; et al. The cancer metabolic reprogramming and immune response. Mol. Cancer 2021, 20, 28. [Google Scholar] [CrossRef]
- Kumagai, S.; Koyama, S.; Itahashi, K.; Tanegashima, T.; Lin, Y.T.; Togashi, Y.; Kamada, T.; Irie, T.; Okumura, G.; Kono, H.; et al. Lactic acid promotes PD-1 expression in regulatory T cells in highly glycolytic tumor microenvironments. Cancer Cell 2022, 40, 201–218.e9. [Google Scholar] [CrossRef]
- Ippolito, L.; Comito, G.; Parri, M.; Iozzo, M.; Duatti, A.; Virgilio, F.; Lorito, N.; Bacci, M.; Pardella, E.; Sandrini, G.; et al. Lactate rewires lipid metabolism and sustains a metabolic-epigenetic axis in prostate cancer. Cancer Res. 2022, 82, 1267–1282. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e4. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Shang, X.; Li, J.; Zhang, S. The Prognosis and Immune Checkpoint Blockade Efficacy Prediction of Tumor-Infiltrating Immune Cells in Lung Cancer. Front. Cell Dev. Biol. 2021, 9, 707143. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Jeremias, I.; Wilde, S.; Leisegang, M.; Starck, L.; Mosetter, B.; Uckert, W.; Heemskerk, M.H.; Schendel, D.J.; Frankenberger, B. TCR-transgenic lymphocytes specific for HMMR/Rhamm limit tumor outgrowth in vivo. Blood 2012, 119, 3440–3449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevens, L.E.; Cheung, W.K.C.; Adua, S.J.; Arnal-Estape, A.; Zhao, M.; Liu, Z.; Brewer, K.; Herbst, R.S.; Nguyen, D.X. Extracellular Matrix Receptor Expression in Subtypes of Lung Adenocarcinoma Potentiates Outgrowth of Micrometastases. Cancer Res. 2017, 77, 1905–1917. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Mei, L.; Connell, M.; Maxwell, C.A. Hyaluronan Mediated Motility Receptor (HMMR) Encodes an Evolutionarily Conserved Homeostasis, Mitosis, and Meiosis Regulator Rather than a Hyaluronan Receptor. Cells 2020, 9, 819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, S.; Fu, L.; Zhang, J.; Xu, J.; Yuan, L.; Qin, J.; Zhang, W. Identification of a DNA Methylation-Driven Genes-Based Prognostic Model and Drug Targets in Breast Cancer: In silico Screening of Therapeutic Compounds and in vitro Characterization. Front. Immunol. 2021, 12, 761326. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Feng, J.; Chen, X.; Wang, D.; Wong, M.; Zhang, G.; Na, J.; Zhang, T.; Chen, Z.; Chen, Y.T.; et al. High levels of truncated RHAMM cooperate with dysfunctional p53 to accelerate the progression of pancreatic cancer. Cancer Lett. 2021, 514, 79–89. [Google Scholar] [CrossRef]
- Lee, K.S.; Burke, T.R., Jr.; Park, J.E.; Bang, J.K.; Lee, E. Recent Advances and New Strategies in Targeting Plk1 for Anticancer Therapy. Trends Pharmacol. Sci. 2015, 36, 858–877. [Google Scholar] [CrossRef] [Green Version]
- Schicht, M.; Rausch, F.; Finotto, S.; Mathews, M.; Mattil, A.; Schubert, M.; Koch, B.; Traxdorf, M.; Bohr, C.; Worlitzsch, D.; et al. SFTA3, a novel protein of the lung: Three-dimensional structure, characterisation and immune activation. Eur. Respir. J. 2014, 44, 447–456. [Google Scholar] [CrossRef] [Green Version]
- Weinberger, P.; Ponny, S.R.; Xu, H.; Bai, S.; Smallridge, R.; Copland, J.; Sharma, A. Cell Cycle M-Phase Genes Are Highly Upregulated in Anaplastic Thyroid Carcinoma. Thyroid 2017, 27, 236–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Han, L.; Zhao, J.; Wang, K.; Xu, M.; Shi, J.; Dong, Z. Tumor stemness and immune infiltration synergistically predict response of radiotherapy or immunotherapy and relapse in lung adenocarcinoma. Cancer Med. 2021, 10, 8944–8960. [Google Scholar] [CrossRef]
- Choreno-Parra, J.A.; Thirunavukkarasu, S.; Zuniga, J.; Khader, S.A. The protective and pathogenic roles of CXCL17 in human health and disease: Potential in respiratory medicine. Cytokine Growth Factor Rev. 2020, 53, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xie, X.; Wu, J. Mechanism of lung adenocarcinoma spine metastasis induced by CXCL17. Cell Oncol. 2020, 43, 311–320. [Google Scholar] [CrossRef] [PubMed]


| Database | Sample Size | Application |
|---|---|---|
| TCGA-LUAD | 487 T vs. 56 N | Model construction |
| GSE31210 | 226 | Model validation |
| GSE37745 | 105 | Model validation |
| Characteristics | TCGA-LUAD | GSE31210 | GSE37745 |
|---|---|---|---|
| Age | |||
| ≤65 | 230 | 176 | 230 |
| >65 | 247 | 50 | 209 |
| Sex | |||
| Female | 261 | 121 | 218 |
| Male | 226 | 105 | 221 |
| NA | 1 | ||
| PT | |||
| Tx | 3 | ||
| T1 | 162 | ||
| T2 | 263 | ||
| T3 | 41 | ||
| T4 | 18 | ||
| PM | |||
| M0 | 323 | ||
| M1 | 24 | ||
| MX | 136 | ||
| MA | 4 | ||
| PN | |||
| N0 | 314 | ||
| N1 | 92 | ||
| N2 | 78 | ||
| N3 | 2 | ||
| NX | 10 | ||
| NA | 1 | ||
| Stage | |||
| Stage I | 262 | 168 | |
| Stage II | 114 | 28 | |
| Stage III | 79 | ||
| Stage IV | 25 | ||
| NA | 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Z.; Luo, Y.; Zhang, L.; Li, H.; Pan, C.; Yang, H.; Cheng, T.; Chen, J. A Novel Risk Score Model of Lactate Metabolism for Predicting over Survival and Immune Signature in Lung Adenocarcinoma. Cancers 2022, 14, 3727. https://doi.org/10.3390/cancers14153727
Jiang Z, Luo Y, Zhang L, Li H, Pan C, Yang H, Cheng T, Chen J. A Novel Risk Score Model of Lactate Metabolism for Predicting over Survival and Immune Signature in Lung Adenocarcinoma. Cancers. 2022; 14(15):3727. https://doi.org/10.3390/cancers14153727
Chicago/Turabian StyleJiang, Zhou, Yongzhong Luo, Lemeng Zhang, Haitao Li, Changqie Pan, Hua Yang, Tianli Cheng, and Jianhua Chen. 2022. "A Novel Risk Score Model of Lactate Metabolism for Predicting over Survival and Immune Signature in Lung Adenocarcinoma" Cancers 14, no. 15: 3727. https://doi.org/10.3390/cancers14153727
APA StyleJiang, Z., Luo, Y., Zhang, L., Li, H., Pan, C., Yang, H., Cheng, T., & Chen, J. (2022). A Novel Risk Score Model of Lactate Metabolism for Predicting over Survival and Immune Signature in Lung Adenocarcinoma. Cancers, 14(15), 3727. https://doi.org/10.3390/cancers14153727





