Presence of Concurrent TP53 Mutations Is Necessary to Predict Poor Outcomes within the SMAD4 Mutated Subgroup of Metastatic Colorectal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Population
2.2. Genomic Analysis
2.3. Statistical Analysis
3. Results
3.1. Study Population
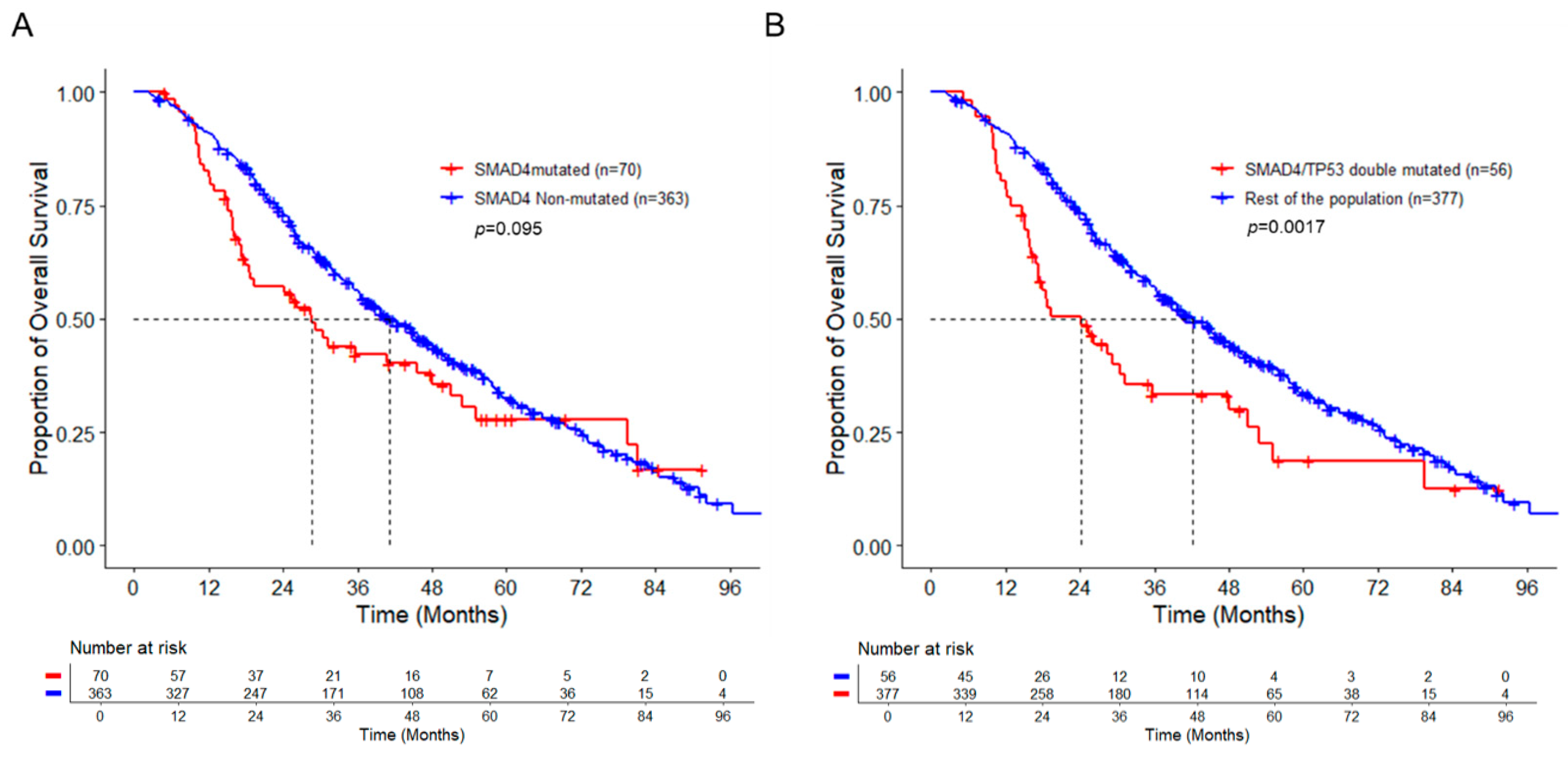
3.2. SAMD4 Mutation Alone Does Not Correlate with Poor Prognosis
3.3. Co-Mutation of SMAD4 with TP53 Is Associated with Worse Overall Survival
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA A Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Markowitz, S.D.; Bertagnolli, M.M. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 2009, 361, 2449–2460. [Google Scholar] [CrossRef] [Green Version]
- Linnekamp, J.F.; Wang, X.; Medema, J.P.; Vermeulen, L. Colorectal Cancer Heterogeneity and Targeted Therapy: A Case for Molecular Disease Subtypes. Cancer Res. 2015, 75, 245. [Google Scholar] [CrossRef] [Green Version]
- Fakih, M.G. Metastatic colorectal cancer: Current state and future directions. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 1809–1824. [Google Scholar] [CrossRef]
- Sandhu, J.; Lavingia, V.; Fakih, M. Systemic treatment for metastatic colorectal cancer in the era of precision medicine. J. Surg. Oncol. 2019, 119, 564–582. [Google Scholar] [CrossRef]
- Derynck, R.; Zhang, Y.; Feng, X.H. Smads: Transcriptional activators of TGF-beta responses. Cell 1998, 95, 737–740. [Google Scholar] [CrossRef] [Green Version]
- Fleming, N.I.; Jorissen, R.N.; Mouradov, D.; Christie, M.; Sakthianandeswaren, A.; Palmieri, M.; Day, F.; Li, S.; Tsui, C.; Lipton, L.; et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013, 73, 725–735. [Google Scholar] [CrossRef] [Green Version]
- Mehrvarz Sarshekeh, A.; Advani, S.; Overman, M.J.; Manyam, G.; Kee, B.K.; Fogelman, D.R.; Dasari, A.; Raghav, K.; Vilar, E.; Manuel, S.; et al. Association of SMAD4 mutation with patient demographics, tumor characteristics, and clinical outcomes in colorectal cancer. PLoS ONE 2017, 12, e0173345. [Google Scholar] [CrossRef] [Green Version]
- Yan, P.; Klingbiel, D.; Saridaki, Z.; Ceppa, P.; Curto, M.; McKee, T.A.; Roth, A.; Tejpar, S.; Delorenzi, M.; Bosman, F.T.; et al. Reduced Expression of SMAD4 Is Associated with Poor Survival in Colon Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 3037–3047. [Google Scholar] [CrossRef] [Green Version]
- Wasserman, I.; Lee, L.H.; Ogino, S.; Marco, M.R.; Wu, C.; Chen, X.; Datta, J.; Sadot, E.; Szeglin, B.; Guillem, J.G.; et al. SMAD4 Loss in Colorectal Cancer Patients Correlates with Recurrence, Loss of Immune Infiltrate, and Chemoresistance. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 1948–1956. [Google Scholar] [CrossRef]
- Kozak, M.M.; von Eyben, R.; Pai, J.; Vossler, S.R.; Limaye, M.; Jayachandran, P.; Anderson, E.M.; Shaffer, J.L.; Longacre, T.; Pai, R.K.; et al. Smad4 inactivation predicts for worse prognosis and response to fluorouracil-based treatment in colorectal cancer. J. Clin. Pathol. 2015, 68, 341–345. [Google Scholar] [CrossRef]
- Yaeger, R.; Chatila, W.K.; Lipsyc, M.D.; Hechtman, J.F.; Cercek, A.; Sanchez-Vega, F.; Jayakumaran, G.; Middha, S.; Zehir, A.; Donoghue, M.T.A.; et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 2018, 33, 125–136.e123. [Google Scholar]
- Chun, Y.S.; Passot, G.; Yamashita, S.; Nusrat, M.; Katsonis, P.; Loree, J.M.; Conrad, C.; Tzeng, C.-W.D.; Xiao, L.; Aloia, T.A.; et al. Deleterious Effect of RAS and Evolutionary High-risk TP53 Double Mutation in Colorectal Liver Metastases. Ann. Surg. 2019, 269, 917–923. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Kopetz, S.; Newhook, T.E.; De Bellis, M.; Chun, Y.S.; Tzeng, C.-W.D.; Aloia, T.A.; Vauthey, J.-N. Mutation Status of RAS, TP53, and SMAD4 is Superior to Mutation Status of RAS Alone for Predicting Prognosis after Resection of Colorectal Liver Metastases. Clin. Cancer Res. 2019, 25, 5843. [Google Scholar] [CrossRef]
- Datta, J.; Smith, J.J.; Chatila, W.K.; McAuliffe, J.C.; Kandoth, C.; Vakiani, E.; Frankel, T.L.; Ganesh, K.; Wasserman, I.; Lipsyc-Sharf, M.; et al. Coaltered Ras/B-raf and TP53 Is Associated with Extremes of Survivorship and Distinct Patterns of Metastasis in Patients with Metastatic Colorectal Cancer. Clin. Cancer Res. 2020, 26, 1077. [Google Scholar] [CrossRef]
- Shi, Y.; Massagué, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef] [Green Version]
- ten Dijke, P.; Hill, C.S. New insights into TGF-beta-Smad signalling. Trends Biochem. Sci. 2004, 29, 265–273. [Google Scholar] [CrossRef]
- Levy, L.; Hill, C.S. Smad4 dependency defines two classes of transforming growth factor {beta} (TGF-{beta}) target genes and distinguishes TGF-{beta}-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol. Cell. Biol. 2005, 25, 8108–8125. [Google Scholar] [CrossRef] [Green Version]
- Alazzouzi, H.; Alhopuro, P.; Salovaara, R.; Sammalkorpi, H.; Järvinen, H.; Mecklin, J.P.; Hemminki, A.; Schwartz, S., Jr.; Aaltonen, L.A.; Arango, D. SMAD4 as a prognostic marker in colorectal cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005, 11, 2606–2611. [Google Scholar] [CrossRef] [Green Version]
- Roth, A.D.; Delorenzi, M.; Tejpar, S.; Yan, P.; Klingbiel, D.; Fiocca, R.; d’Ario, G.; Cisar, L.; Labianca, R.; Cunningham, D.; et al. Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. J. Natl. Cancer Inst. 2012, 104, 1635–1646. [Google Scholar] [CrossRef] [Green Version]
- Ahn, B.K.; Jang, S.H.; Paik, S.S.; Lee, K.H. Smad4 may help to identify a subset of colorectal cancer patients with early recurrence after curative therapy. Hepato-Gastroenterology 2011, 58, 1933–1936. [Google Scholar] [CrossRef]
- Toledo, F.; Wahl, G.M. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 2006, 6, 909–923. [Google Scholar] [CrossRef]
- Munro, A.J.; Lain, S.; Lane, D.P. P53 abnormalities and outcomes in colorectal cancer: A systematic review. Br. J. Cancer 2005, 92, 434–444. [Google Scholar] [CrossRef] [Green Version]
- Pilat, N.; Grünberger, T.; Längle, F.; Mittlböck, M.; Perisanidis, B.; Kappel, S.; Wolf, B.; Starlinger, P.; Kührer, I.; Mühlbacher, F.; et al. Assessing the TP53 marker type in patients treated with or without neoadjuvant chemotherapy for resectable colorectal liver metastases: A p53 Research Group study. Eur. J. Surg. Oncol. J. Eur. Soc. Surg. Oncol. Br. Assoc. Surg. Oncol. 2015, 41, 683–689. [Google Scholar] [CrossRef]
- Elsaleh, H.; Powell, B.; McCaul, K.; Grieu, F.; Grant, R.; Joseph, D.; Iacopetta, B. P53 alteration and microsatellite instability have predictive value for survival benefit from chemotherapy in stage III colorectal carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2001, 7, 1343–1349. [Google Scholar]
- Kandioler, D.; Mittlböck, M.; Kappel, S.; Puhalla, H.; Herbst, F.; Langner, C.; Wolf, B.; Tschmelitsch, J.; Schippinger, W.; Steger, G.; et al. TP53 Mutational Status and Prediction of Benefit from Adjuvant 5-Fluorouracil in Stage III Colon Cancer Patients. EBioMedicine 2015, 2, 825–830. [Google Scholar] [CrossRef] [Green Version]
- Seligmann, J.F.; Fisher, D.J.; Brown, L.C.; Adams, R.A.; Graham, J.; Quirke, P.; Richman, S.D.; Butler, R.; Domingo, E.; Blake, A.; et al. Inhibition of WEE1 Is Effective in TP53- and RAS-Mutant Metastatic Colorectal Cancer: A Randomized Trial (FOCUS4-C) Comparing Adavosertib (AZD1775) With Active Monitoring. J. Clin. Oncol. 2021, 39, 3705–3715. [Google Scholar] [CrossRef]
- Pan, M.; Jiang, C.; Tse, P.; Achacoso, N.; Alexeeff, S.; Solorzano, A.V.; Chung, E.; Hu, W.; Truong, T.-G.; Arora, A.; et al. TP53 Gain-of-Function and Non–Gain-of-Function Mutations Are Differentially Associated With Sidedness-Dependent Prognosis in Metastatic Colorectal Cancer. J. Clin. Oncol. 2021, 40, 171–179. [Google Scholar] [CrossRef]
- Cordenonsi, M.; Dupont, S.; Maretto, S.; Insinga, A.; Imbriano, C.; Piccolo, S. Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell 2003, 113, 301–314. [Google Scholar] [CrossRef]
- Elston, R.; Inman, G.J. Crosstalk between p53 and TGF-β Signalling. J. Signal Transduct. 2012, 2012, 294097. [Google Scholar] [CrossRef] [Green Version]
- Adorno, M.; Cordenonsi, M.; Montagner, M.; Dupont, S.; Wong, C.; Hann, B.; Solari, A.; Bobisse, S.; Rondina, M.B.; Guzzardo, V.; et al. A Mutant-p53/Smad Complex Opposes p63 to Empower TGFβ-Induced Metastasis. Cell 2009, 137, 87–98. [Google Scholar] [CrossRef]
- Wang, C.; Ouyang, C.; Cho, M.; Ji, J.; Sandhu, J.; Goel, A.; Kahn, M.; Fakih, M. Wild-type APC Is Associated with Poor Survival in Metastatic Microsatellite Stable Colorectal Cancer. Oncologist 2021, 26, 208–214. [Google Scholar] [CrossRef]
- Kang, J.-K.; Heo, S.; Kim, H.-P.; Song, S.-H.; Yun, H.; Han, S.-W.; Kang, G.H.; Bang, D.; Kim, T.-Y. Liquid biopsy-based tumor profiling for metastatic colorectal cancer patients with ultra-deep targeted sequencing. PLoS ONE 2020, 15, e0232754. [Google Scholar] [CrossRef]
- Gupta, R.; Othman, T.; Chen, C.; Sandhu, J.; Ouyang, C.; Fakih, M. Guardant360 Circulating Tumor DNA Assay Is Concordant with FoundationOne Next-Generation Sequencing in Detecting Actionable Driver Mutations in Anti-EGFR Naive Metastatic Colorectal Cancer. Oncologist 2020, 25, 235–243. [Google Scholar] [CrossRef] [Green Version]
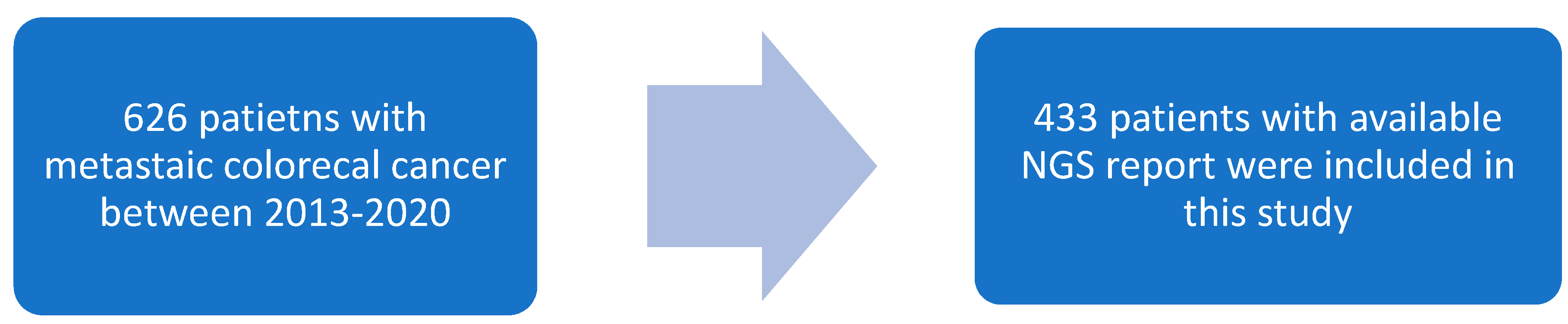
| Characteristics | Total (%) | SMAD4-WT | SMAD4-MT | p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| (n = 433) | (n = 363) | (n = 70) | ||||||
| Age at Diagnosis (Median, Range) | 55 (16–90) | 55 (20–90) | 56 (16–77) | 0.78 | ||||
| Gender | ||||||||
| Male | 251 | 58% | 212 | 58% | 39 | 56% | 0.69 | |
| Female | 182 | 42% | 151 | 42% | 31 | 44% | ||
| Sidedness * | ||||||||
| Left | 309 | 72% | 264 | 73% | 45 | 64% | 0.19 | |
| Right | 121 | 28% | 97 | 27% | 24 | 34% | ||
| RAS | ||||||||
| Mutated | 207 | 48% | 169 | 47% | 38 | 54% | 0.24 | |
| Non-mutated | 226 | 52% | 194 | 53% | 32 | 46% | ||
| BRAFV600E | ||||||||
| Mutated | 32 | 7% | 24 | 7% | 8 | 11% | 0.21 | |
| Non-mutated | 401 | 93% | 339 | 93% | 62 | 89% | ||
| APC | ||||||||
| Mutated | 311 | 72% | 270 | 74% | 41 | 59% | 0.01 | |
| Non-mutated | 122 | 28% | 93 | 26% | 29 | 41% | ||
| TP53 | ||||||||
| Mutated | 346 | 80% | 290 | 80% | 56 | 80% | 1.00 | |
| Non-mutated | 87 | 20% | 73 | 20% | 14 | 20% | ||
| Clinicopathogenic Variables | COH Cohort (n = 433) | |||
|---|---|---|---|---|
| 95% CI | ||||
| HR | Lower | Upper | p-Value | |
| Age at diagnosis (years) | ||||
| ≥65 vs. <65 | 0.98 | 0.74 | 1.30 | 0.88 |
| Gender | ||||
| Male vs. Female | 1.19 | 0.93 | 1.52 | 0.17 |
| Sidedness | ||||
| Right vs. Left | 1.33 | 0.99 | 1.79 | 0.06 |
| RAS | ||||
| Mutated vs. Non-mutated | 1.23 | 0.95 | 1.60 | 0.11 |
| BRAFV600E | ||||
| Mutated vs. Non-mutated | 1.43 | 0.86 | 2.39 | 0.17 |
| APC | ||||
| Mutated vs. Non-mutated | 0.76 | 0.58 | 1.01 | 0.06 |
| TP53 | ||||
| Mutated vs. Non-mutated | 1.36 | 0.99 | 1.85 | 0.06 |
| SMAD4 | ||||
| Mutated vs. Non-mutated | 1.25 | 0.90 | 1.73 | 0.18 |
| Clinicopathogenic Variables | SMAD4-MT Cohort (n = 70) | |||
|---|---|---|---|---|
| 95% CI | ||||
| HR | Lower | Upper | p-Value | |
| RAS | ||||
| Mutated vs. Non-mutated | 1.06 | 0.58 | 1.96 | 0.85 |
| BRAFV600E | ||||
| Mutated vs. Non-mutated | 1.54 | 0.61 | 3.87 | 0.36 |
| APC | ||||
| Mutated vs. Non-mutated | 0.61 | 0.33 | 1.13 | 0.12 |
| TP53 | ||||
| Mutated vs. Non-mutated | 3.20 | 1.31 | 7.80 | 0.01 |
| APC, BRAFV600E, RAS, SMAD4, and TP53 Mutation Status | Reference | Multivariable HR | 95% CI | p |
|---|---|---|---|---|
| SMAD4-MT + TP53-MT | vs. rest of the population | 2.5 | 1.44–4.36 | 0.001 |
| SMAD4-MT + RAS-MT | vs. rest of the population | 0.95 | 0.54–1.67 | 0.853 |
| SMAD4-MT + BRAFV600E-MT | vs. rest of the population | 1.5 | 0.64–3.47 | 0.353 |
| SMAD4-MT + APC-MT | vs. rest of the population | 0.51 | 0.29–0.91 | 0.022 |
| RAS-MT + TP53-MT | vs. rest of the population | 1.2 | 0.95–1.60 | 0.111 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Sandhu, J.; Tsao, A.; Fakih, M. Presence of Concurrent TP53 Mutations Is Necessary to Predict Poor Outcomes within the SMAD4 Mutated Subgroup of Metastatic Colorectal Cancer. Cancers 2022, 14, 3644. https://doi.org/10.3390/cancers14153644
Wang C, Sandhu J, Tsao A, Fakih M. Presence of Concurrent TP53 Mutations Is Necessary to Predict Poor Outcomes within the SMAD4 Mutated Subgroup of Metastatic Colorectal Cancer. Cancers. 2022; 14(15):3644. https://doi.org/10.3390/cancers14153644
Chicago/Turabian StyleWang, Chongkai, Jaideep Sandhu, Amber Tsao, and Marwan Fakih. 2022. "Presence of Concurrent TP53 Mutations Is Necessary to Predict Poor Outcomes within the SMAD4 Mutated Subgroup of Metastatic Colorectal Cancer" Cancers 14, no. 15: 3644. https://doi.org/10.3390/cancers14153644
APA StyleWang, C., Sandhu, J., Tsao, A., & Fakih, M. (2022). Presence of Concurrent TP53 Mutations Is Necessary to Predict Poor Outcomes within the SMAD4 Mutated Subgroup of Metastatic Colorectal Cancer. Cancers, 14(15), 3644. https://doi.org/10.3390/cancers14153644






