Apigenin Targets MicroRNA-155, Enhances SHIP-1 Expression, and Augments Anti-Tumor Responses in Pancreatic Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Pancreatic Ductal Adenocarcinoma Cancer Cell Lines
2.2. Bioflavonoid
2.3. Cell Viability and Apoptosis Assays of Pancreatic Cancer Cells
2.4. Transfection of miR-155 Inhibitor into Pancreatic Cancer Cells
2.5. Pancreatic Cancer Murine Models
2.6. Flow Cytometry
2.7. miRNA Extraction and TaqMan miRNA Assay
2.8. RT-Quantitative PCR (RT-qPCR)
2.9. Western Blot (WB)
2.10. Akoya Codex
2.11. The Cancer Genome Atlas (TGCA) Database
2.12. Statistical Analysis
3. Results
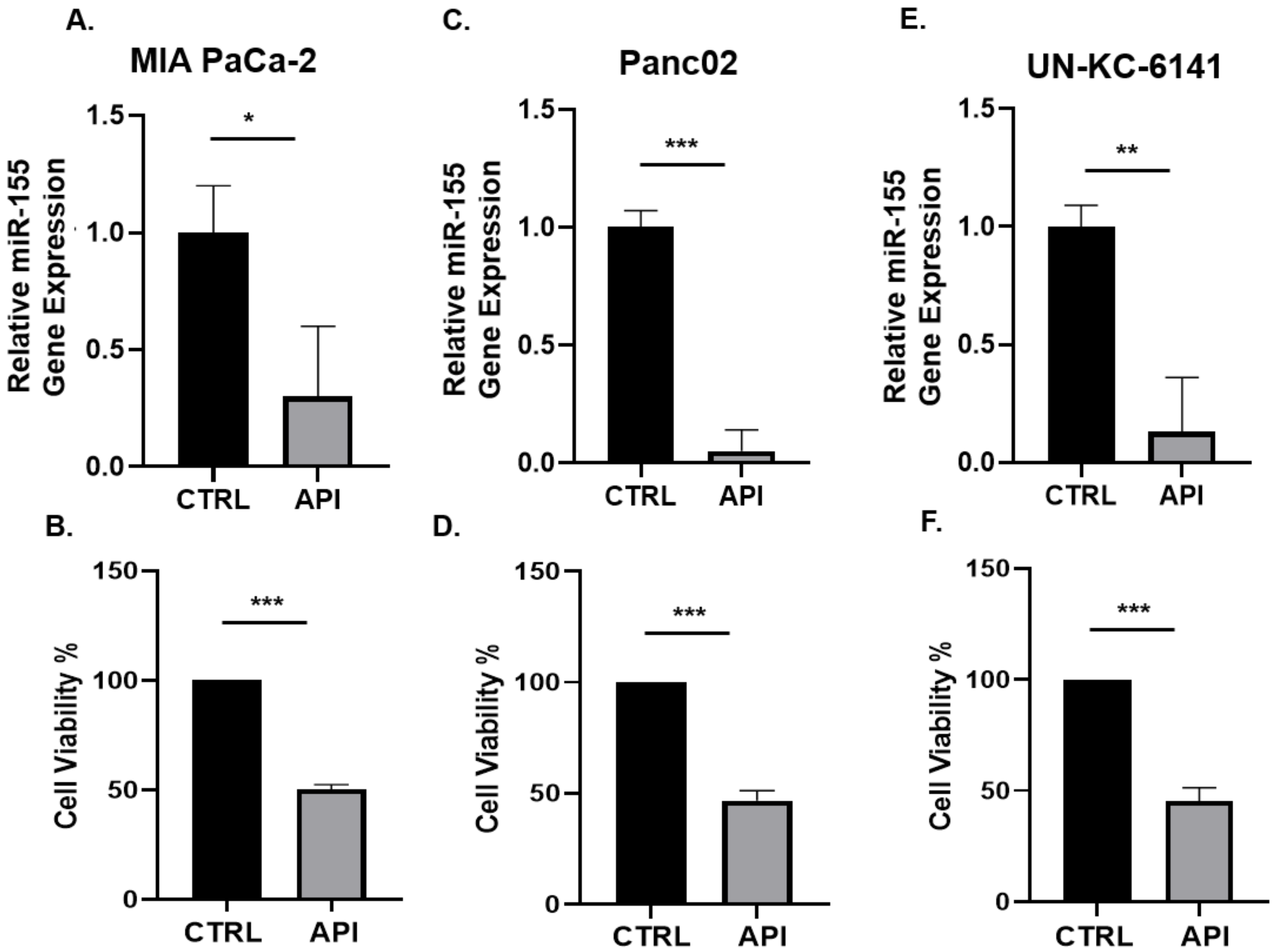
3.1. API Suppresses miR-155 Gene Expression and Inhibits the Viability of Pancreatic Cancer Cells In Vitro
3.2. Inhibition of miR-155 Decreases the Viability of PC Cells In Vitro
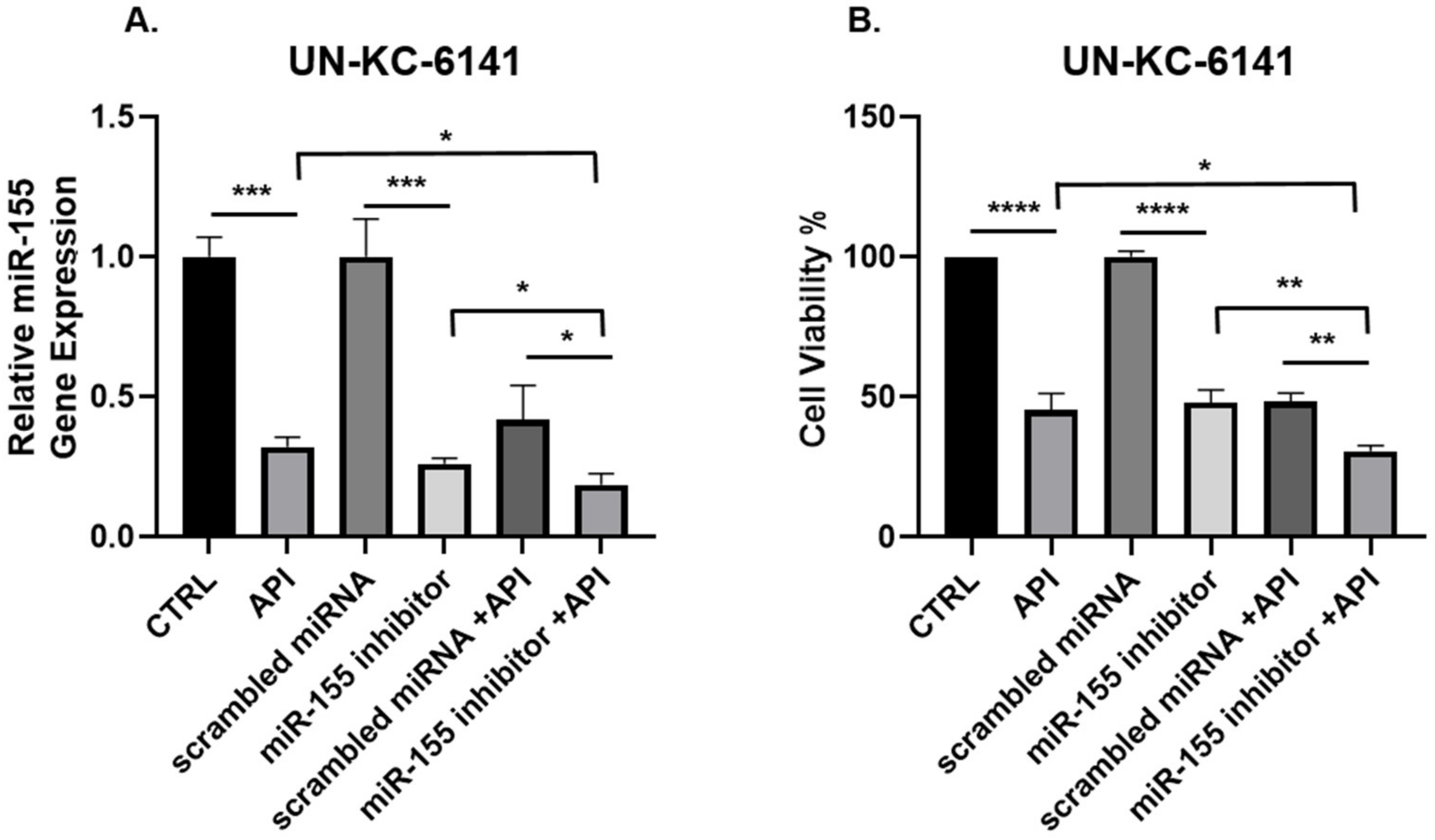
3.3. Concomitant API and miR-155 Inhibitor Treatment Synergistically Suppresses miR-155 and the Viability of PC Cells In Vitro
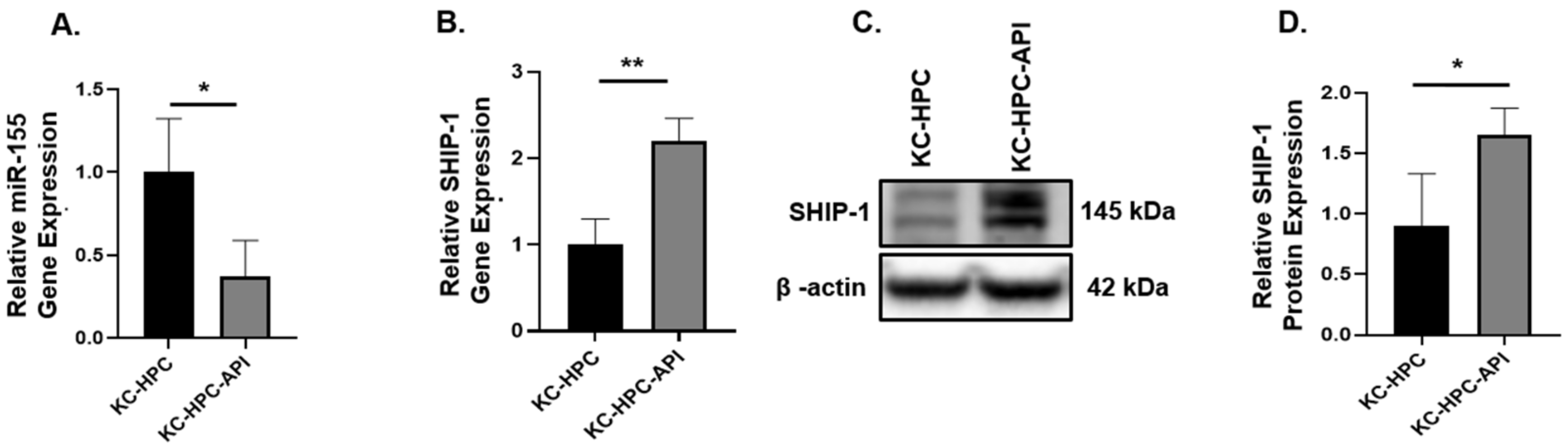
3.4. API Decreases the Production of miR-155 and This Correlates with an Increase in SHIP-1 Expression in HPC, OPC and KC-HPC Mouse Models
3.5. miR-155 Expression in SHIPKO-HPC Is Increased Compared to SHIPWT-HPC Mice
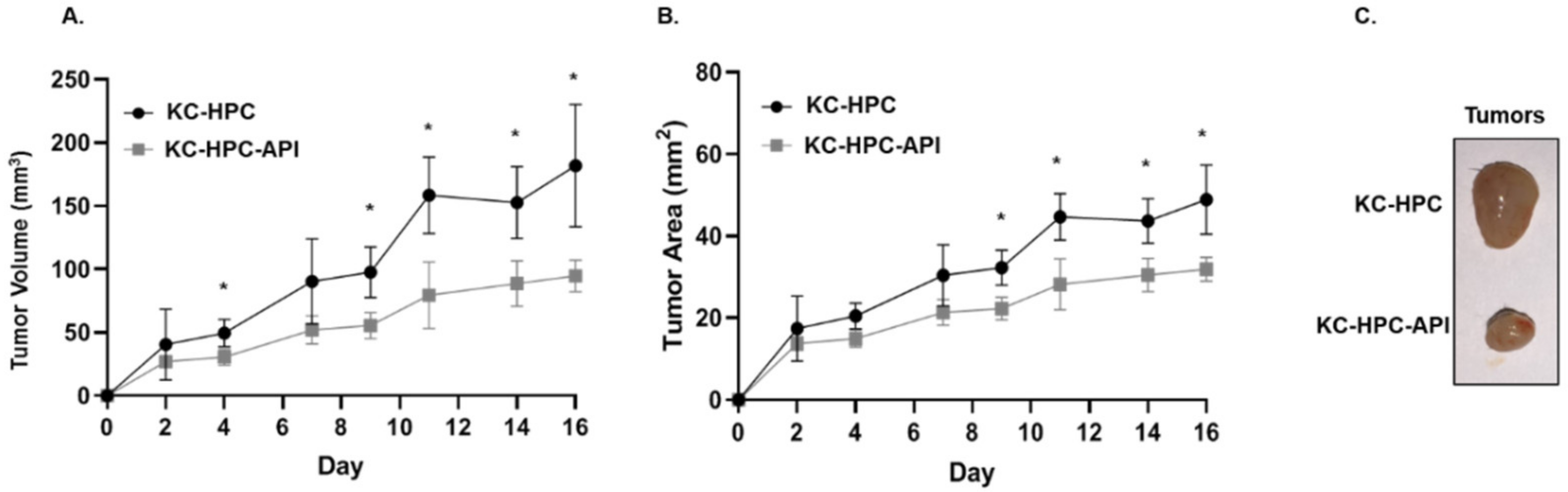
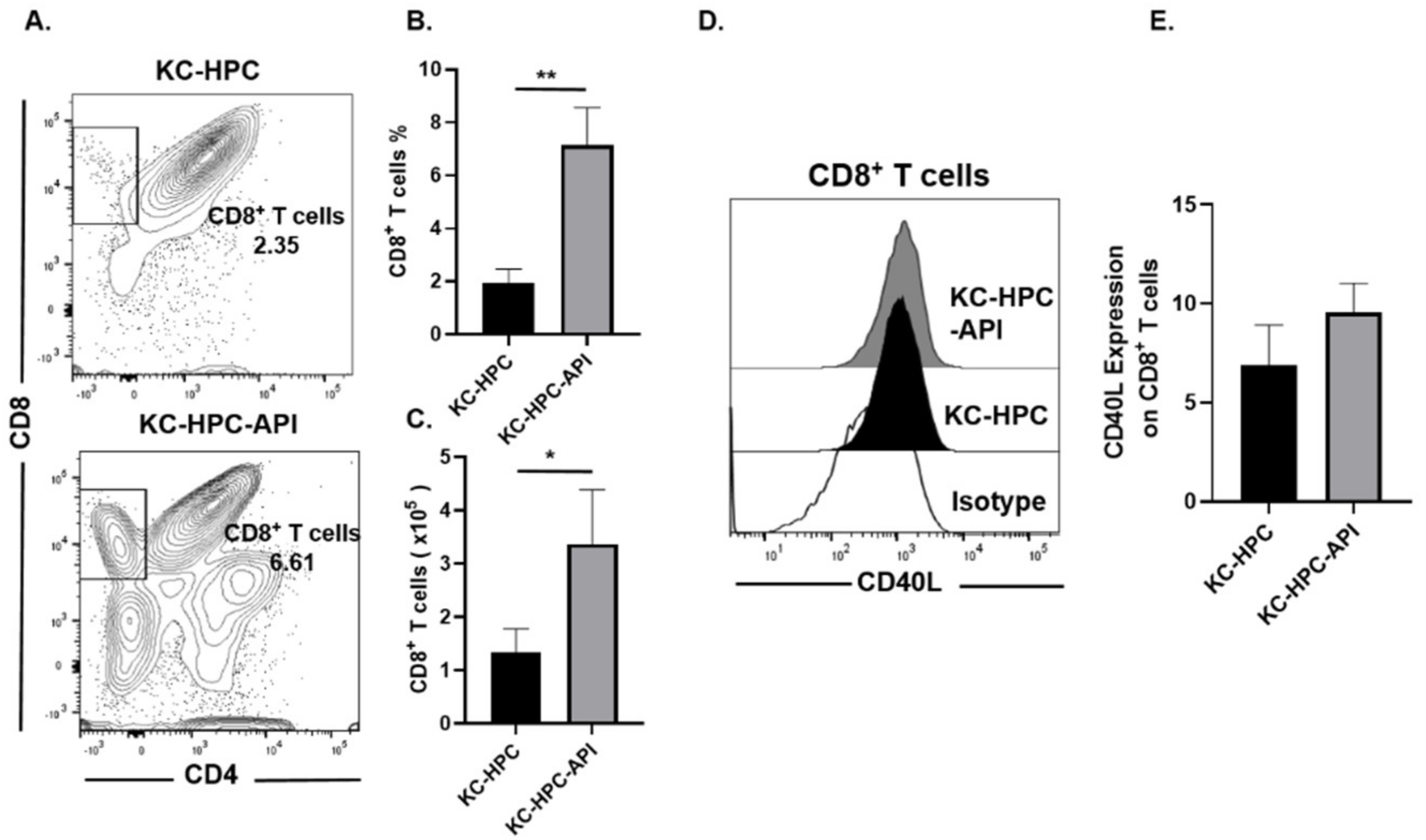
3.6. API Improves Myelopoiesis and Anti-Tumor Responses in KC-HPC Mice
3.7. Lower Survival Is Associated with High miR-155 and Low SHIP-1 Gene Expressions in PC Patients
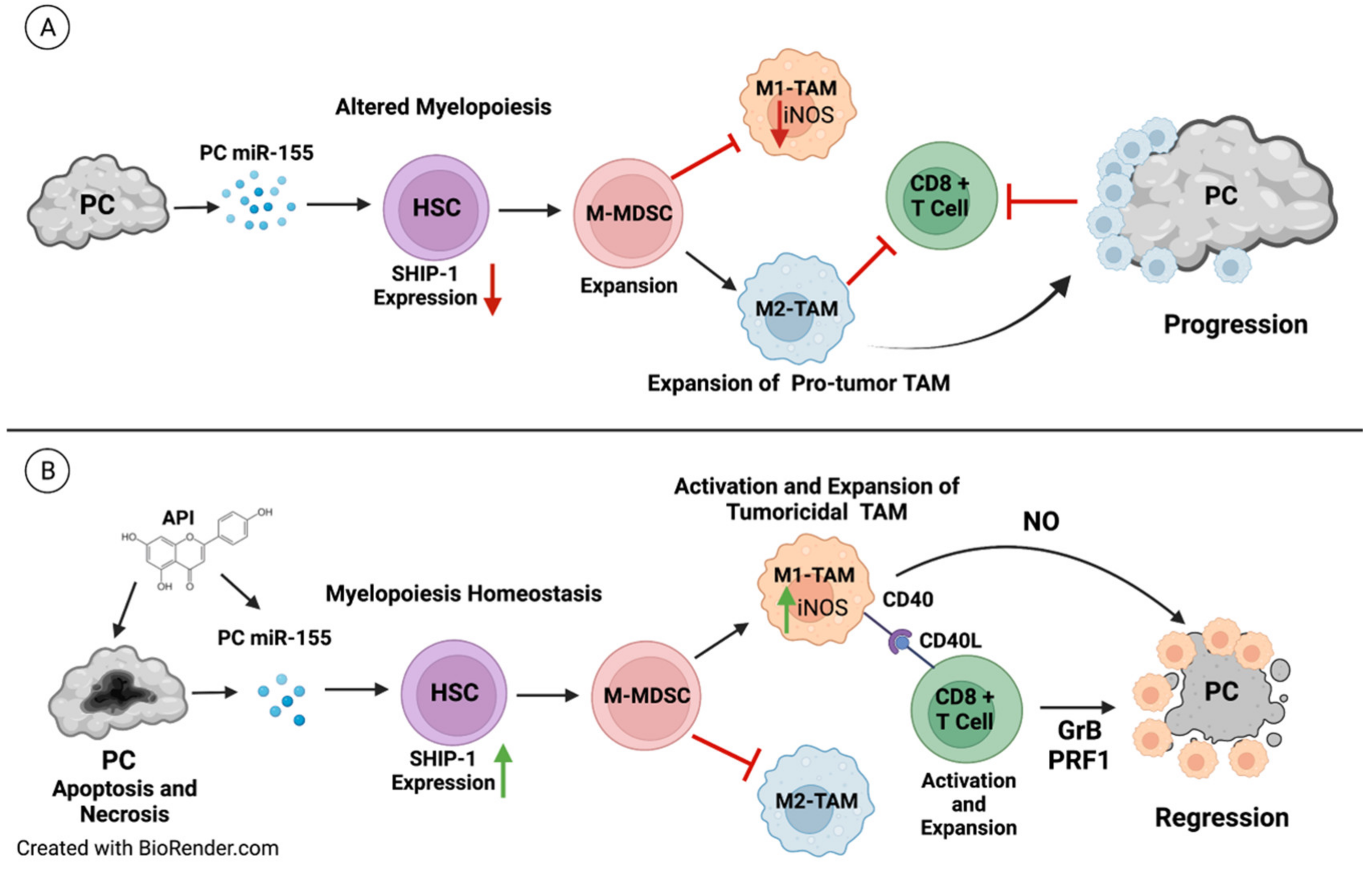
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Tavakkoli, A.; Singal, A.G.; Waljee, A.K.; Elmunzer, B.J.; Pruitt, S.L.; McKey, T.; Rubenstein, J.H.; Scheiman, J.M.; Murphy, C.C. Racial Disparities and Trends in Pancreatic Cancer Incidence and Mortality in the United States. Clin. Gastroenterol. Hepatol. 2020, 18, 171–178.e110. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic cancer. Lancet 2020, 395, 2008–2020. [Google Scholar] [CrossRef]
- Zeng, S.; Pottler, M.; Lan, B.; Grutzmann, R.; Pilarsky, C.; Yang, H. Chemoresistance in Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 4504. [Google Scholar] [CrossRef] [PubMed]
- Looi, C.K.; Chung, F.F.; Leong, C.O.; Wong, S.F.; Rosli, R.; Mai, C.W. Therapeutic challenges and current immunomodulatory strategies in targeting the immunosuppressive pancreatic tumor microenvironment. J. Exp. Clin. Cancer Res. 2019, 38, 162. [Google Scholar] [CrossRef] [PubMed]
- Schmiechen, Z.C.; Stromnes, I.M. Mechanisms Governing Immunotherapy Resistance in Pancreatic Ductal Adenocarcinoma. Front. Immunol. 2020, 11, 613815. [Google Scholar] [CrossRef]
- Padoan, A.; Plebani, M.; Basso, D. Inflammation and pancreatic cancer: Focus on metabolism, cytokines, and immunity. Int. J. Mol. Sci. 2019, 20, 676. [Google Scholar] [CrossRef]
- Draghiciu, O.; Lubbers, J.; Nijman, H.W.; Daemen, T. Myeloid derived suppressor cells—An overview of combat strategies to increase immunotherapy efficacy. OncoImmunology 2015, 4, e954829. [Google Scholar] [CrossRef]
- Porembka, M.R.; Mitchem, J.B.; Belt, B.A.; Hsieh, C.-S.; Lee, H.-M.; Herndon, J.; Gillanders, W.E.; Linehan, D.C.; Goedegebuure, P.J.C.I. Immunotherapy. Pancreatic adenocarcinoma induces bone marrow mobilization of myeloid-derived suppressor cells which promote primary tumor growth. Cancer Immunol. Immunother. 2012, 61, 1373–1385. [Google Scholar] [CrossRef]
- Pilon-Thomas, S.; Nelson, N.; Vohra, N.; Jerald, M.; Pendleton, L.; Szekeres, K.; Ghansah, T. Murine pancreatic adenocarcinoma dampens SHIP-1 expression and alters MDSC homeostasis and function. PLoS ONE 2011, 6, e27729. [Google Scholar] [CrossRef] [PubMed]
- Villalobos-Ayala, K.; Ortiz Rivera, I.; Alvarez, C.; Husain, K.; DeLoach, D.; Krystal, G.; Hibbs, M.L.; Jiang, K.; Ghansah, T. Apigenin Increases SHIP-1 Expression, Promotes Tumoricidal Macrophages and Anti-Tumor Immune Responses in Murine Pancreatic Cancer. Cancers 2020, 12, 3631. [Google Scholar] [CrossRef] [PubMed]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Mielgo, A.; Schmid, M.C. Impact of tumour associated macrophages in pancreatic cancer. BMB Rep. 2013, 46, 131–138. [Google Scholar] [CrossRef]
- Farajzadeh Valilou, S.; Keshavarz-Fathi, M.; Silvestris, N.; Argentiero, A.; Rezaei, N. The role of inflammatory cytokines and tumor associated macrophages (TAMs) in microenvironment of pancreatic cancer. Cytokine Growth Factor Rev. 2018, 39, 46–61. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Garzon, R.; Fabbri, M.; Cimmino, A.; Calin, G.A.; Croce, C.M. MicroRNA expression and function in cancer. Trends Mol. Med. 2006, 12, 580–587. [Google Scholar] [CrossRef]
- Gilles, M.E.; Slack, F.J. Let-7 microRNA as a potential therapeutic target with implications for immunotherapy. Expert Opin. Ther. Targets 2018, 22, 929–939. [Google Scholar] [CrossRef]
- Yu, Z.; Li, Y.; Fan, H.; Liu, Z.; Pestell, R.G. miRNAs regulate stem cell self-renewal and differentiation. Front. Genet. 2012, 3, 191. [Google Scholar] [CrossRef]
- Galasso, M.; Sandhu, S.K.; Volinia, S. MicroRNA expression signatures in solid malignancies. Cancer J. 2012, 18, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Eis, P.S.; Tam, W.; Sun, L.; Chadburn, A.; Li, Z.; Gomez, M.F.; Lund, E.; Dahlberg, J.E. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 2005, 102, 3627–3632. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, Y.G.; Lucas, A.L. MicroRNA in pancreatic ductal adenocarcinoma and its precursor lesions. World J. Gastrointest. Oncol. 2016, 8, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Wei, C.; Cui, M.Y.; Xia, Q.Q.; Wang, S.B.; Zhang, Y. Prognostic value of microRNAs in pancreatic cancer: A meta-analysis. Aging (Albany N. Y.) 2020, 12, 9380–9404. [Google Scholar] [CrossRef] [PubMed]
- Rawat, M.; Kadian, K.; Gupta, Y.; Kumar, A.; Chain, P.S.; Kovbasnjuk, O.; Kumar, S.; Parasher, G. MicroRNA in pancreatic cancer: From biology to therapeutic potential. Genes 2019, 10, 752. [Google Scholar] [CrossRef]
- Lee, D.W.; Futami, M.; Carroll, M.; Feng, Y.; Wang, Z.; Fernandez, M.; Whichard, Z.; Chen, Y.; Kornblau, S.; Shpall, E.J.; et al. Loss of SHIP-1 protein expression in high-risk myelodysplastic syndromes is associated with miR-210 and miR-155. Oncogene 2012, 31, 4085–4094. [Google Scholar] [CrossRef]
- Baradaran, B.; Shahbazi, R.; Khordadmehr, M. Dysregulation of key microRNAs in pancreatic cancer development. Biomed. Pharmacother. 2019, 109, 1008–1015. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Chaudhuri, A.A.; Rao, D.S.; Baltimore, D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. USA 2009, 106, 7113–7118. [Google Scholar] [CrossRef]
- Ghansah, T.; Paraiso, K.H.; Highfill, S.; Desponts, C.; May, S.; McIntosh, J.K.; Wang, J.W.; Ninos, J.; Brayer, J.; Cheng, F.; et al. Expansion of myeloid suppressor cells in SHIP-deficient mice represses allogeneic T cell responses. J. Immunol. 2004, 173, 7324–7330. [Google Scholar] [CrossRef]
- Hamilton, M.J.; Ho, V.W.; Kuroda, E.; Ruschmann, J.; Antignano, F.; Lam, V.; Krystal, G. Role of SHIP in cancer. Exp. Hematol. 2011, 39, 2–13. [Google Scholar] [CrossRef]
- Hibbs, M.L.; Raftery, A.L.; Tsantikos, E. Regulation of hematopoietic cell signaling by SHIP-1 inositol phosphatase: Growth factors and beyond. Growth Factors 2018, 36, 213–231. [Google Scholar] [CrossRef] [PubMed]
- Sly, L.M.; Ho, V.; Antignano, F.; Ruschmann, J.; Hamilton, M.; Lam, V.; Rauh, M.J.; Krystal, G. The role of SHIP in macrophages. Front. Biosci. 2007, 12, 2836–2848. [Google Scholar] [CrossRef] [PubMed]
- Kalesnikoff, J.; Sly, L.M.; Hughes, M.R.; Buchse, T.; Rauh, M.J.; Cao, L.P.; Lam, V.; Mui, A.; Huber, M.; Krystal, G. The role of SHIP in cytokine-induced signaling. Rev. Physiol. Biochem. Pharmacol. 2003, 149, 87–103. [Google Scholar] [CrossRef]
- Rauh, M.J.; Ho, V.; Pereira, C.; Sham, A.; Sly, L.M.; Lam, V.; Huxham, L.; Minchinton, A.I.; Mui, A.; Krystal, G. SHIP represses the generation of alternatively activated macrophages. Immunity 2005, 23, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Ghansah, T. A novel strategy for modulation of MDSC to enhance cancer immunotherapy. Oncoimmunology 2012, 1, 984–985. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Silverman, D.T.; Swanson, C.A.; Gridley, G.; Wacholder, S.; Greenberg, R.S.; Brown, L.M.; Hayes, R.B.; Swanson, G.M.; Schoenberg, J.B.; Pottern, L.M.; et al. Dietary and nutritional factors and pancreatic cancer: A case-control study based on direct interviews. J. Natl. Cancer Inst. 1998, 90, 1710–1719. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.R.; Greenwald, P. Nutritional interventions in cancer prevention. J. Clin. Oncol. 2005, 23, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Van den Brandt, P.A.; Goldbohm, R.A. Nutrition in the prevention of gastrointestinal cancer. Best Pract. Res. Clin. Gastroenterol. 2006, 20, 589–603. [Google Scholar] [CrossRef]
- Perrott, K.M.; Wiley, C.D.; Desprez, P.Y.; Campisi, J. Apigenin suppresses the senescence-associated secretory phenotype and paracrine effects on breast cancer cells. Geroscience 2017, 39, 161–173. [Google Scholar] [CrossRef]
- Qin, Y.; Zhao, D.; Zhou, H.G.; Wang, X.H.; Zhong, W.L.; Chen, S.; Gu, W.G.; Wang, W.; Zhang, C.H.; Liu, Y.R.; et al. Apigenin inhibits NF-kappaB and snail signaling, EMT and metastasis in human hepatocellular carcinoma. Oncotarget 2016, 7, 41421–41431. [Google Scholar] [CrossRef]
- Shukla, S.; Bhaskaran, N.; Babcook, M.A.; Fu, P.; Maclennan, G.T.; Gupta, S. Apigenin inhibits prostate cancer progression in TRAMP mice via targeting PI3K/Akt/FoxO pathway. Carcinogenesis 2014, 35, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Yang, Z.; Yang, Z.; Zhou, S.; Zhang, H.; Zang, L. Effect of apigenin on proliferation and apoptosis of human lung cancer NCI-H460 cells. Nan Fang Yi Ke Da Xue Xue Bao 2013, 33, 1137–1140. [Google Scholar]
- Lee, Y.; Sung, B.; Kang, Y.J.; Kim, D.H.; Jang, J.Y.; Hwang, S.Y.; Kim, M.; Lim, H.S.; Yoon, J.H.; Chung, H.Y.; et al. Apigenin-induced apoptosis is enhanced by inhibition of autophagy formation in HCT116 human colon cancer cells. Int. J. Oncol. 2014, 44, 1599–1606. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Han, X.; Cheng, W.; Ni, J.; Zhang, Y.; Lin, J.; Song, Z.J.O.R. Apigenin inhibits proliferation and invasion, and induces apoptosis and cell cycle arrest in human melanoma cells. Oncol. Rep. 2017, 37, 2277–2285. [Google Scholar] [CrossRef]
- Angulo, P.; Kaushik, G.; Subramaniam, D.; Dandawate, P.; Neville, K.; Chastain, K.; Anant, S. Natural compounds targeting major cell signaling pathways: A novel paradigm for osteosarcoma therapy. J. Hematol. Oncol. 2017, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Ashrafizadeh, M.; Bakhoda, M.R.; Bahmanpour, Z.; Ilkhani, K.; Zarrabi, A.; Makvandi, P.; Khan, H.; Mazaheri, S.; Darvish, M.; Mirzaei, H.J.F.i.C. Apigenin as tumor suppressor in cancers: Biotherapeutic activity, nanodelivery, and mechanisms with emphasis on pancreatic cancer. Front. Chem. 2020, 8, 829. [Google Scholar] [CrossRef] [PubMed]
- Ujiki, M.B.; Ding, X.-Z.; Salabat, M.R.; Bentrem, D.J.; Golkar, L.; Milam, B.; Talamonti, M.S.; Bell, R.H.; Iwamura, T.; Adrian, T.E. Apigenin inhibits pancreatic cancer cell proliferation through G2/M cell cycle arrest. Mol. Cancer 2006, 5, 76. [Google Scholar] [CrossRef]
- Yan, X.; Qi, M.; Li, P.; Zhan, Y.; Shao, H. Apigenin in cancer therapy: Anti-cancer effects and mechanisms of action. Cell Biosci. 2017, 7, 50. [Google Scholar] [CrossRef]
- Ahmed, S.A.; Parama, D.; Daimari, E.; Girisa, S.; Banik, K.; Harsha, C.; Dutta, U.; Kunnumakkara, A.B. Rationalizing the therapeutic potential of apigenin against cancer. Life Sci. 2021, 267, 118814. [Google Scholar] [CrossRef]
- Johnson, J.L.; de Mejia, E.G. Interactions between dietary flavonoids apigenin or luteolin and chemotherapeutic drugs to potentiate anti-proliferative effect on human pancreatic cancer cells, in vitro. Food Chem. Toxicol. 2013, 60, 83–91. [Google Scholar] [CrossRef]
- Jang, J.Y.; Sung, B.; Kim, N.D. Role of Induced Programmed Cell Death in the Chemopreventive Potential of Apigenin. Int. J. Mol. Sci. 2022, 23, 3757. [Google Scholar] [CrossRef] [PubMed]
- Oishi, M.; Iizumi, Y.; Taniguchi, T.; Goi, W.; Miki, T.; Sakai, T. Apigenin sensitizes prostate cancer cells to Apo2L/TRAIL by targeting adenine nucleotide translocase-2. PLoS ONE 2013, 8, e55922. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, X.; Zha, D.; Cai, F.; Zhang, W.; He, Y.; Huang, Q.; Zhuang, H.; Hua, Z.-C. Apigenin potentiates TRAIL therapy of non-small cell lung cancer via upregulating DR4/DR5 expression in a p53-dependent manner. Sci. Rep. 2016, 6, 35468. [Google Scholar] [CrossRef]
- Shukla, S.; Gupta, S. Apigenin: A promising molecule for cancer prevention. Pharm. Res. 2010, 27, 962–978. [Google Scholar] [CrossRef]
- Nelson, N.; Szekeres, K.; Iclozan, C.; Rivera, I.O.; McGill, A.; Johnson, G.; Nwogu, O.; Ghansah, T. Apigenin: Selective CK2 inhibitor increases Ikaros expression and improves T cell homeostasis and function in murine pancreatic cancer. PLoS ONE 2017, 12, e0170197. [Google Scholar] [CrossRef] [PubMed]
- Corbett, T.; Roberts, B.; Leopold, W.; Peckham, J.; Wilkoff, L.; Griswold, D.; Schabel, F. Induction and chemotherapeutic response of two transplantable ductal adenocarcinomas of the pancreas in C57BL/6 mice. Cancer Res. 1984, 44, 717–726. [Google Scholar]
- Torres, M.P.; Rachagani, S.; Souchek, J.J.; Mallya, K.; Johansson, S.L.; Batra, S.K. Novel pancreatic cancer cell lines derived from genetically engineered mouse models of spontaneous pancreatic adenocarcinoma: Applications in diagnosis and therapy. PLoS ONE 2013, 8, e80580. [Google Scholar] [CrossRef]
- Husain, K.; Francois, R.A.; Yamauchi, T.; Perez, M.; Sebti, S.M.; Malafa, M.P. Vitamin E delta-tocotrienol augments the antitumor activity of gemcitabine and suppresses constitutive NF-kappaB activation in pancreatic cancer. Mol. Cancer Ther. 2011, 10, 2363–2372. [Google Scholar] [CrossRef]
- Chai, M.G.; Kim-Fuchs, C.; Angst, E.; Sloan, E.K. Bioluminescent orthotopic model of pancreatic cancer progression. J. Vis. Exp. 2013, 76, e50395. [Google Scholar] [CrossRef] [PubMed]
- Tsantikos, E.; Lau, M.; Castelino, C.M.; Maxwell, M.J.; Passey, S.L.; Hansen, M.J.; McGregor, N.E.; Sims, N.A.; Steinfort, D.P.; Irving, L.B.; et al. Granulocyte-CSF links destructive inflammation and comorbidities in obstructive lung disease. J. Clin. Investig. 2018, 128, 2406–2418. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, K.; Wang, C.; Gong, W.; Yoshimura, T.; Wang, J.M.; Liu, M. Isolation of mouse tumor-infiltrating leukocytes by percoll gradient centrifugation. Bio-Protocol 2013, 3, e892. [Google Scholar] [CrossRef]
- Jiang, B.-H.; Qiu, J.-G.; Wang, L.; Liu, W.-J.; Wang, J.-F.; Zhao, E.-J.; Zhou, F.-M.; Ji, X.-B.; Wang, L.-H.; Xia, Z.-K. Apigenin inhibits IL-6 transcription and suppresses esophageal carcinogenesis. Front. Pharmacol. 2019, 10, 1002. [Google Scholar]
- Shukla, S.; Gupta, S. Apigenin-induced prostate cancer cell death is initiated by reactive oxygen species and p53 activation. Free Radic. Biol. Med. 2008, 44, 1833–1845. [Google Scholar] [CrossRef]
- Cao, H.H.; Chu, J.H.; Kwan, H.Y.; Su, T.; Yu, H.; Cheng, C.Y.; Fu, X.Q.; Guo, H.; Li, T.; Tse, A.K.; et al. Inhibition of the STAT3 signaling pathway contributes to apigenin-mediated anti-metastatic effect in melanoma. Sci. Rep. 2016, 6, 21731. [Google Scholar] [CrossRef]
- Dai, J.; Van Wie, P.G.; Fai, L.Y.; Kim, D.; Wang, L.; Poyil, P.; Luo, J.; Zhang, Z. Downregulation of NEDD9 by apigenin suppresses migration, invasion, and metastasis of colorectal cancer cells. Toxicol. Appl. Pharmacol. 2016, 311, 106–112. [Google Scholar] [CrossRef]
- Masuelli, L.; Benvenuto, M.; Mattera, R.; Di Stefano, E.; Zago, E.; Taffera, G.; Tresoldi, I.; Giganti, M.G.; Frajese, G.V.; Berardi, G.; et al. In Vitro and In Vivo Anti-tumoral Effects of the Flavonoid Apigenin in Malignant Mesothelioma. Front. Pharmacol. 2017, 8, 373. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Zhu, Y.; Li, J.F.; Wang, X.; Liang, Z.; Li, S.Q.; Xu, X.; Chen, H.; Liu, B.; Zheng, X.Y.; et al. Apigenin inhibits renal cell carcinoma cell proliferation. Oncotarget 2017, 8, 19834–19842. [Google Scholar] [CrossRef]
- Tseng, T.H.; Chien, M.H.; Lin, W.L.; Wen, Y.C.; Chow, J.M.; Chen, C.K.; Kuo, T.C.; Lee, W.J. Inhibition of MDA-MB-231 breast cancer cell proliferation and tumor growth by apigenin through induction of G2/M arrest and histone H3 acetylation-mediated p21(WAF1/CIP1) expression. Environ. Toxicol. 2017, 32, 434–444. [Google Scholar] [CrossRef]
- Qiu, W.; Su, G.H. Development of orthotopic pancreatic tumor mouse models. Methods Mol. Biol. 2013, 980, 215–223. [Google Scholar] [CrossRef]
- Arango, D.; Diosa-Toro, M.; Rojas-Hernandez, L.S.; Cooperstone, J.L.; Schwartz, S.J.; Mo, X.; Jiang, J.; Schmittgen, T.D.; Doseff, A.I. Dietary apigenin reduces LPS-induced expression of miR-155 restoring immune balance during inflammation. Mol. Nutr. Food Res. 2015, 59, 763–772. [Google Scholar] [CrossRef]
- Chumsakul, O.; Wakayama, K.; Tsuhako, A.; Baba, Y.; Takai, Y.; Kurose, T.; Honma, Y.; Watanabe, S. Apigenin Regulates Activation of Microglia and Counteracts Retinal Degeneration. J. Ocul. Pharmacol. Ther. 2020, 36, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Thyagarajan, A.; Alshehri, M.S.A.; Miller, K.L.; Sherwin, C.M.; Travers, J.B.; Sahu, R.P. Myeloid-derived suppressor cells and pancreatic cancer: Implications in novel therapeutic approaches. Cancers 2019, 11, 1627. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yu, X.; Cao, Q.; Wang, Y.; Zheng, G.; Tan, T.K.; Zhao, H.; Zhao, Y.; Wang, Y.; Harris, D.C. Characterization of murine macrophages from bone marrow, spleen and peritoneum. BMC Immunol. 2013, 14, 6. [Google Scholar] [CrossRef]
- Balli, D.; Rech, A.J.; Stanger, B.Z.; Vonderheide, R.H. Immune Cytolytic Activity Stratifies Molecular Subsets of Human Pancreatic Cancer. Clin. Cancer Res. 2017, 23, 3129–3138. [Google Scholar] [CrossRef]
- Stromnes, I.M.; Brockenbrough, J.S.; Izeradjene, K.; Carlson, M.A.; Cuevas, C.; Simmons, R.M.; Greenberg, P.D.; Hingorani, S.R. Targeted depletion of an MDSC subset unmasks pancreatic ductal adenocarcinoma to adaptive immunity. Gut 2014, 63, 1769–1781. [Google Scholar] [CrossRef] [PubMed]
- Torphy, R.J.; Zhu, Y.; Schulick, R.D. Immunotherapy for pancreatic cancer: Barriers and breakthroughs. Ann. Gastroenterol. Surg. 2018, 2, 274–281. [Google Scholar] [CrossRef]
- Zhu, Y.; Knolhoff, B.L.; Meyer, M.A.; Nywening, T.M.; West, B.L.; Luo, J.; Wang-Gillam, A.; Goedegebuure, S.P.; Linehan, D.C.; DeNardo, D.G. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 2014, 74, 5057–5069. [Google Scholar] [CrossRef]
- Balachandran, V.P.; Beatty, G.L.; Dougan, S.K. Broadening the Impact of Immunotherapy to Pancreatic Cancer: Challenges and Opportunities. Gastroenterology 2019, 156, 2056–2072. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.A.; Bednar, F.; Zhang, Y.; Brisset, J.-C.; Galbán, S.; Galbán, C.J.; Rakshit, S.; Flannagan, K.S.; Adsay, N.V.; di Magliano, M.P. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J. Clin. Investig. 2012, 122, 639–653. [Google Scholar] [CrossRef]
- Totiger, T.M.; Srinivasan, S.; Jala, V.R.; Lamichhane, P.; Dosch, A.R.; Gaidarski, A.A.; Joshi, C.; Rangappa, S.; Castellanos, J.; Vemula, P.K. Urolithin A, a novel natural compound to target PI3K/AKT/mTOR pathway in pancreatic cancer. Mol. Cancer Ther. 2019, 18, 301–311. [Google Scholar] [CrossRef]
- Mattiske, S.; Suetani, R.J.; Neilsen, P.M.; Callen, D.F. The Oncogenic Role of miR-155 in Breast Cancer. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1236–1243. [Google Scholar] [CrossRef] [PubMed]
- Zadeh, M.M.; Ranji, N.; Motamed, N. Deregulation of miR-21 and miR-155 and their putative targets after silibinin treatment in T47D breast cancer cells. Iran. J. Basic Med. Sci. 2015, 18, 1209. [Google Scholar] [PubMed]
- Ravishankar, D.; Rajora, A.K.; Greco, F.; Osborn, H.M. Flavonoids as prospective compounds for anti-cancer therapy. Int. J. Biochem. Cell Biol. 2013, 45, 2821–2831. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- Cote, G.A.; Gore, A.J.; McElyea, S.D.; Heathers, L.E.; Xu, H.; Sherman, S.; Korc, M. A pilot study to develop a diagnostic test for pancreatic ductal adenocarcinoma based on differential expression of select miRNA in plasma and bile. Am. J. Gastroenterol. 2014, 109, 1942–1952. [Google Scholar] [CrossRef]
- Mikamori, M.; Yamada, D.; Eguchi, H.; Hasegawa, S.; Kishimoto, T.; Tomimaru, Y.; Asaoka, T.; Noda, T.; Wada, H.; Kawamoto, K.J. MicroRNA-155 controls exosome synthesis and promotes gemcitabine resistance in pancreatic ductal adenocarcinoma. Sci. Rep. 2017, 7, 42339. [Google Scholar] [CrossRef]
- Rakhmilevich, A.L.; Alderson, K.L.; Sondel, P.M. T-cell-independent antitumor effects of CD40 ligation. Int. Rev. Immunol. 2012, 31, 267–278. [Google Scholar] [CrossRef]
- Schoenberger, S.P.; Toes, R.E.; Van Der Voort, E.I.; Offringa, R.; Melief, C.J. T-cell help for cytotoxic T lymphocytes is mediated by CD40–CD40L interactions. Nature 1998, 393, 480–483. [Google Scholar] [CrossRef]
- Zhang, J.Q.; Zeng, S.; Vitiello, G.A.; Seifert, A.M.; Medina, B.D.; Beckman, M.J.; Loo, J.K.; Santamaria-Barria, J.; Maltbaek, J.H.; Param, N.J. Macrophages and CD8+ T cells mediate the antitumor efficacy of combined CD40 ligation and imatinib therapy in gastrointestinal stromal tumors. Cancer Immunol. Res. 2018, 6, 434–447. [Google Scholar] [CrossRef]
- Li, L.; Zhang, J.; Diao, W.; Wang, D.; Wei, Y.; Zhang, C.Y.; Zen, K. MicroRNA-155 and MicroRNA-21 promote the expansion of functional myeloid-derived suppressor cells. J. Immunol. 2014, 192, 1034–1043. [Google Scholar] [CrossRef]
- Faget, J.; Peters, S.; Quantin, X.; Meylan, E.; Bonnefoy, N. Neutrophils in the era of immune checkpoint blockade. J. Immunother. Cancer 2021, 9, e002242. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Kim, H.S.; Shi, J. Neutrophil in the Pancreatic Tumor Microenvironment. Biomolecules 2021, 11, 1170. [Google Scholar] [CrossRef] [PubMed]
- Rauh, M.J.; Sly, L.M.; Kalesnikoff, J.; Hughes, M.R.; Cao, L.P.; Lam, V.; Krystal, G. The role of SHIP1 in macrophage programming and activation. Biochem. Soc. Trans. 2004, 32, 785–788. [Google Scholar] [CrossRef] [PubMed]
- Krystal, G.; Hamilton, M.J.; Bennewith, K.L. SHIP prevents metastasis. Aging 2016, 8, 837–838. [Google Scholar] [CrossRef][Green Version]
- Lakhanpal, G.K.; Vecchiarelli-Federico, L.M.; Li, Y.-J.; Cui, J.-W.; Bailey, M.L.; Spaner, D.E.; Dumont, D.J.; Barber, D.L.; Ben-David, Y. The inositol phosphatase SHIP-1 is negatively regulated by Fli-1 and its loss accelerates leukemogenesis. Blood 2010, 116, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Alinikula, J.; Kohonen, P.; Nera, K.P.; Lassila, O.J. Concerted action of Helios and Ikaros controls the expression of the inositol 5-phosphatase SHIP. Eur. J. Immunol. 2010, 40, 2599–2607. [Google Scholar] [CrossRef]
- Pauls, S.D.; Marshall, A.J. Regulation of immune cell signaling by SHIP1: A phosphatase, scaffold protein, and potential therapeutic target. Eur. J. Immunol. 2017, 47, 932–945. [Google Scholar] [CrossRef]
- Singh, U.P.; Murphy, A.E.; Enos, R.T.; Shamran, H.A.; Singh, N.P.; Guan, H.; Hegde, V.L.; Fan, D.; Price, R.L.; Taub, D.D.; et al. MiR-155 deficiency protects mice from experimental colitis by reducing T helper type 1/type 17 responses. Immunology 2014, 143, 478–489. [Google Scholar] [CrossRef]
- Wang, D.; Tang, M.; Zong, P.; Liu, H.; Zhang, T.; Liu, Y.; Zhao, Y. MiRNA-155 regulates the Th17/Treg ratio by targeting SOCS1 in severe acute pancreatitis. Front. Physiol. 2018, 9, 686. [Google Scholar] [CrossRef]
- Huang, C.; Li, H.; Wu, W.; Jiang, T.; Qiu, Z. Regulation of miR-155 affects pancreatic cancer cell invasiveness and migration by modulating the STAT3 signaling pathway through SOCS1. Oncol. Rep. 2013, 30, 1223–1230. [Google Scholar] [CrossRef]
- Kurowska-Stolarska, M.; Alivernini, S.; Ballantine, L.E.; Asquith, D.L.; Millar, N.L.; Gilchrist, D.S.; Reilly, J.; Ierna, M.; Fraser, A.R.; Stolarski, B.; et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc. Natl. Acad. Sci. USA 2011, 108, 11193–11198. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Negri, E.; Lagiou, P.; Talamini, R.; Dal Maso, L.; Montella, M.; Franceschi, S.; La Vecchia, C. Flavonoids and ovarian cancer risk: A case-control study in Italy. Int. J. Cancer 2008, 123, 895–898. [Google Scholar] [CrossRef] [PubMed]
- Bosetti, C.; Spertini, L.; Parpinel, M.; Gnagnarella, P.; Lagiou, P.; Negri, E.; Franceschi, S.; Montella, M.; Peterson, J.; Dwyer, J.; et al. Flavonoids and breast cancer risk in Italy. Cancer Epidemiol. Biomark. Prev. 2005, 14, 805–808. [Google Scholar] [CrossRef]
- Knekt, P.; Jarvinen, R.; Seppanen, R.; Hellovaara, M.; Teppo, L.; Pukkala, E.; Aromaa, A. Dietary flavonoids and the risk of lung cancer and other malignant neoplasms. Am. J. Epidemiol. 1997, 146, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Hoensch, H.; Groh, B.; Edler, L.; Kirch, W. Prospective cohort comparison of flavonoid treatment in patients with resected colorectal cancer to prevent recurrence. World J. Gastroenterol. 2008, 14, 2187–2193. [Google Scholar] [CrossRef]
- De Font-Reaulx Rojas, E.; Dorazco-Barragan, G. Clinical stabilisation in neurodegenerative diseases: Clinical study in phase II. Rev. Neurol. 2010, 50, 520–528. [Google Scholar]





| Qiagen miRCURY LNA miRNA Inhibitors | Sequence 5′–3′ |
|---|---|
| MMU-MIR-155-5P (mouse) | CCCCTATCACAATTAGCATT |
| HSA-MIR-155-5P (human) | CCTATCACGATTAGCATT |
| Survival Analysis Type | Time Period | Two Proportion Test p-Value |
|---|---|---|
| Overall survival | 10 months | 0.006 |
| Disease specific | 10 months | 0.078 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Husain, K.; Villalobos-Ayala, K.; Laverde, V.; Vazquez, O.A.; Miller, B.; Kazim, S.; Blanck, G.; Hibbs, M.L.; Krystal, G.; Elhussin, I.; et al. Apigenin Targets MicroRNA-155, Enhances SHIP-1 Expression, and Augments Anti-Tumor Responses in Pancreatic Cancer. Cancers 2022, 14, 3613. https://doi.org/10.3390/cancers14153613
Husain K, Villalobos-Ayala K, Laverde V, Vazquez OA, Miller B, Kazim S, Blanck G, Hibbs ML, Krystal G, Elhussin I, et al. Apigenin Targets MicroRNA-155, Enhances SHIP-1 Expression, and Augments Anti-Tumor Responses in Pancreatic Cancer. Cancers. 2022; 14(15):3613. https://doi.org/10.3390/cancers14153613
Chicago/Turabian StyleHusain, Kazim, Krystal Villalobos-Ayala, Valentina Laverde, Oscar A. Vazquez, Bradley Miller, Samra Kazim, George Blanck, Margaret L. Hibbs, Gerald Krystal, Isra Elhussin, and et al. 2022. "Apigenin Targets MicroRNA-155, Enhances SHIP-1 Expression, and Augments Anti-Tumor Responses in Pancreatic Cancer" Cancers 14, no. 15: 3613. https://doi.org/10.3390/cancers14153613
APA StyleHusain, K., Villalobos-Ayala, K., Laverde, V., Vazquez, O. A., Miller, B., Kazim, S., Blanck, G., Hibbs, M. L., Krystal, G., Elhussin, I., Mori, J., Yates, C., & Ghansah, T. (2022). Apigenin Targets MicroRNA-155, Enhances SHIP-1 Expression, and Augments Anti-Tumor Responses in Pancreatic Cancer. Cancers, 14(15), 3613. https://doi.org/10.3390/cancers14153613







