Proteomic Profiling Identifies Specific Leukemic Stem Cell-Associated Protein Expression Patterns in Pediatric AML Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Fluorescence-Activated Cell Sorting
2.3. Proteomics
2.4. Statistical Analysis
3. Results
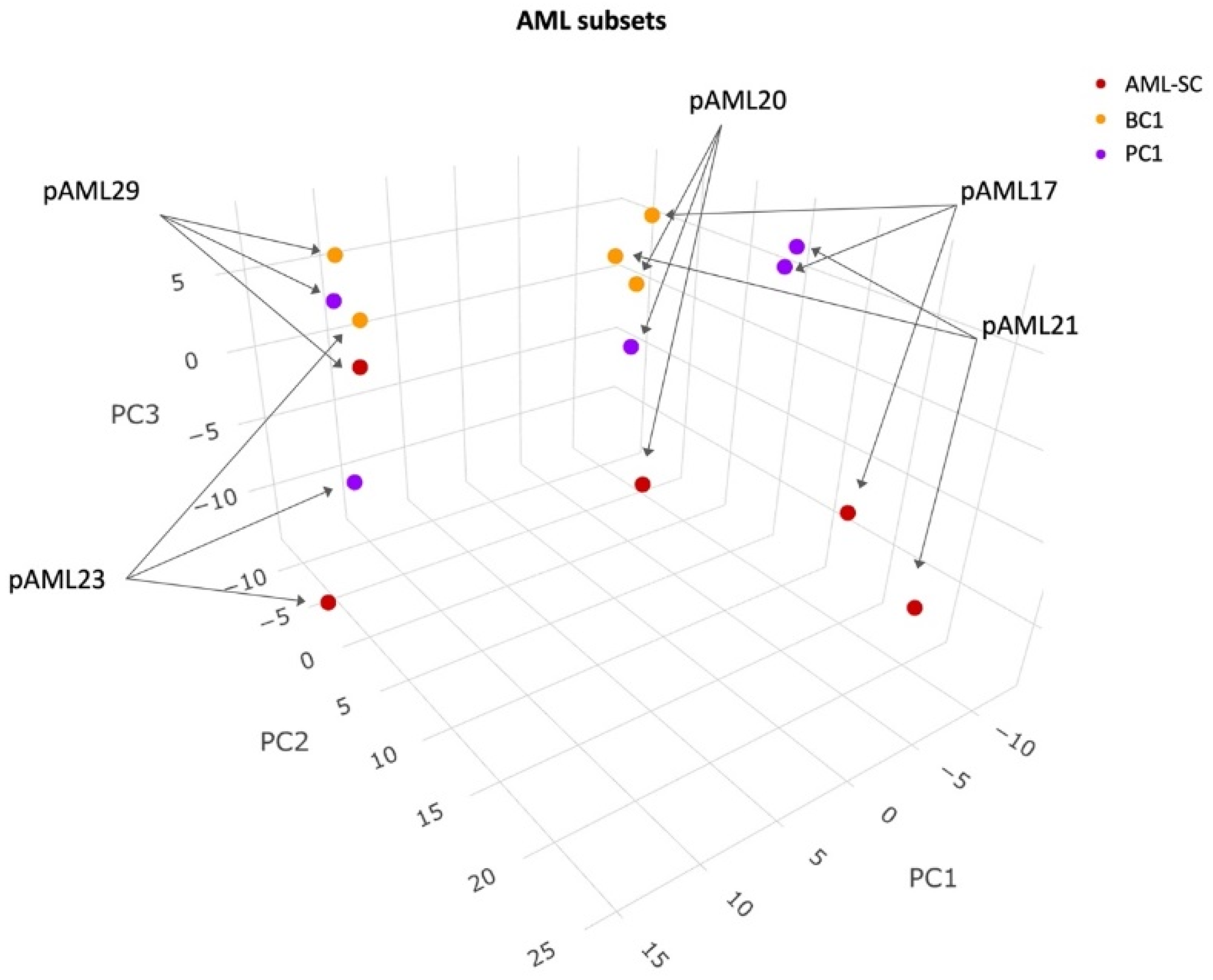
3.1. Protein Profiles of Leukemic Cell Subsets from Pediatric AML Are Partly Patient Specific and Partly Cell Subset Specific
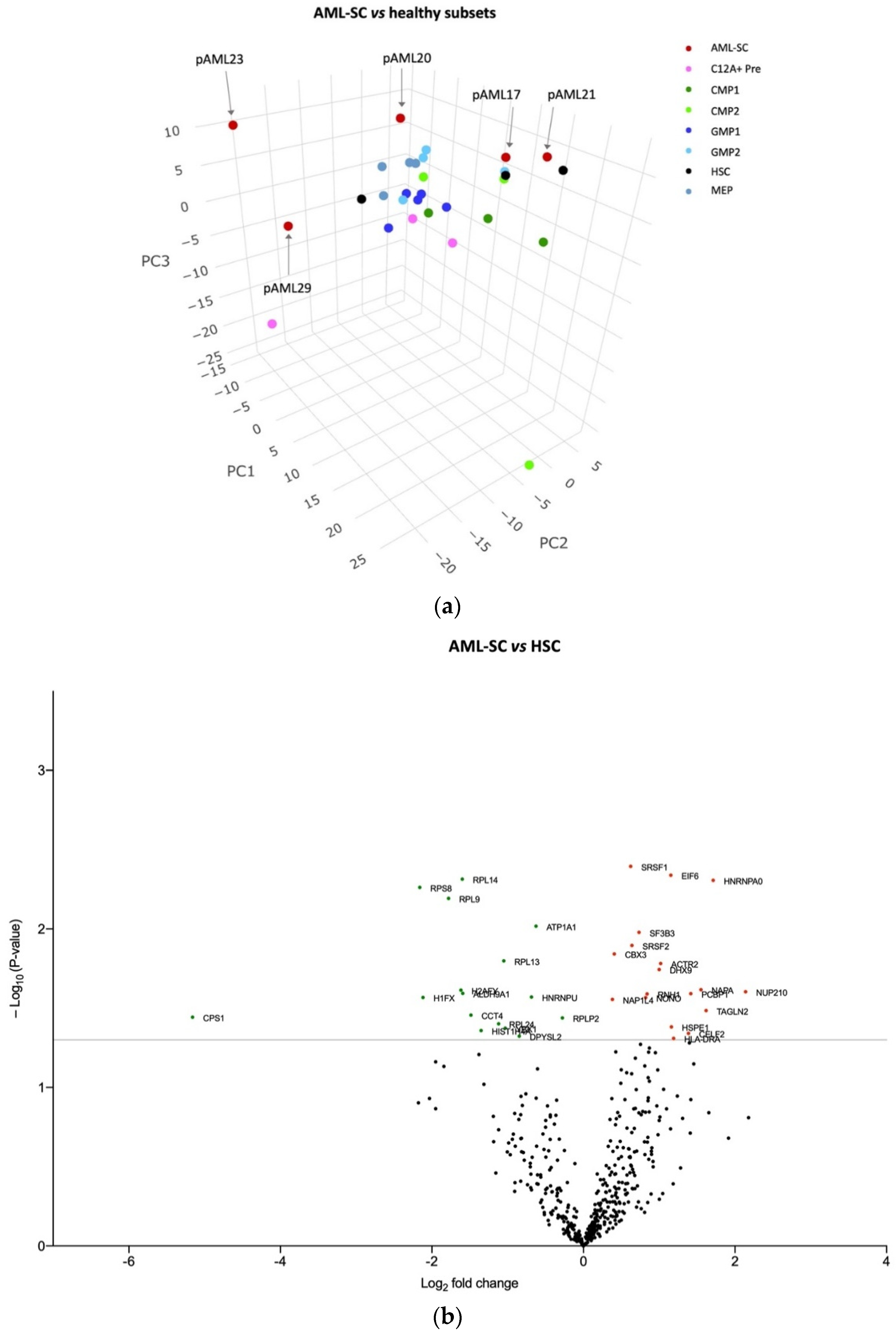
3.2. The AML-SC Protein Profile Differs in CD34 Negative and CD34 Positive AML compared with HSCs and HPCs
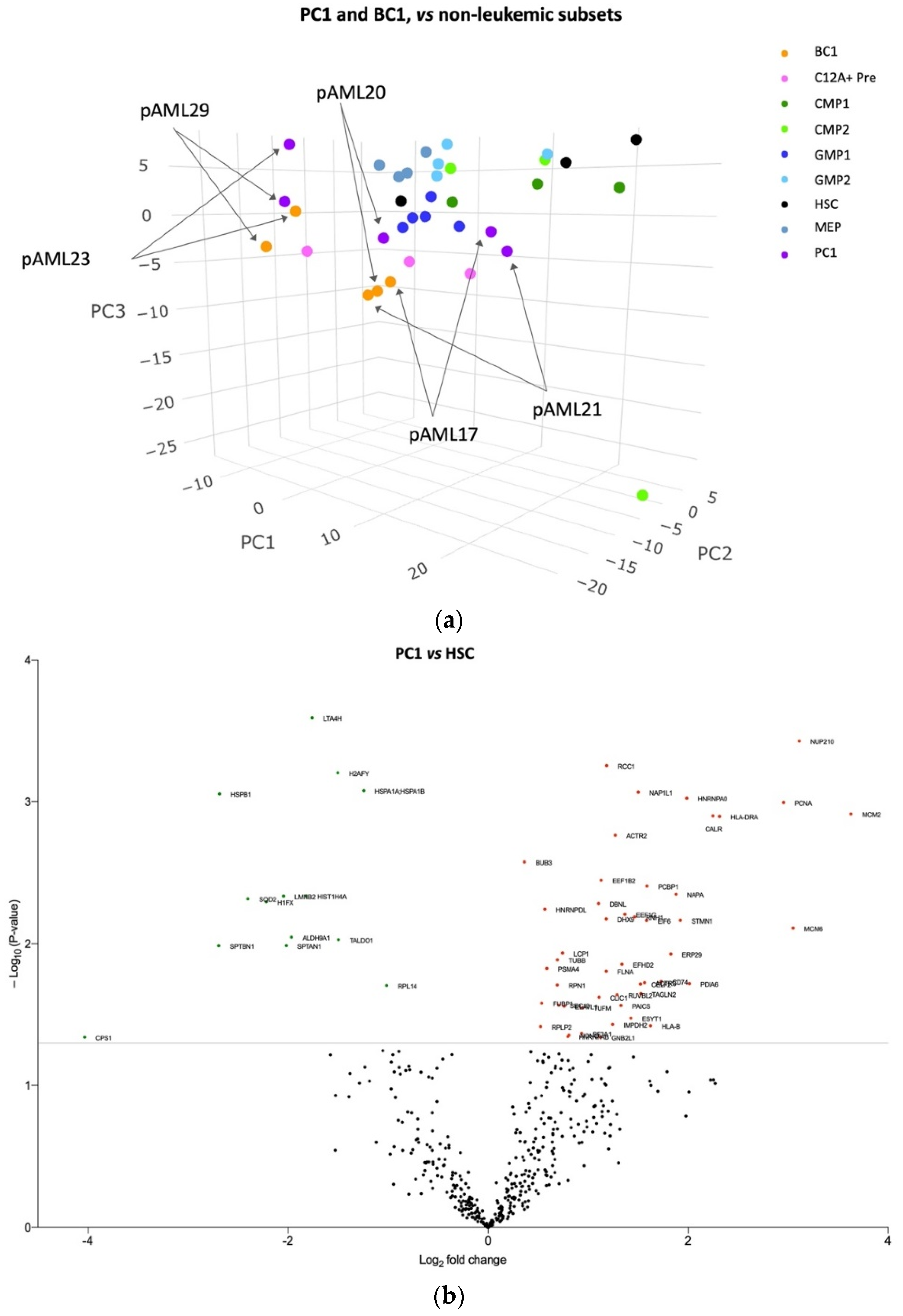
3.3. The Blast Proteome Resembles Healthy Progenitors in CD34 Negative AML but Is Unique in CD34 Positive AML
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zwaan, C.M.; Kolb, E.A.; Reinhardt, D.; Abrahamsson, J.; Adachi, S.; Aplenc, R.; De Bont, E.S.J.M.; De Moerloose, B.; Dworzak, M.; Gibson, B.E.S.; et al. Collaborative Efforts Driving Progress in Pediatric Acute Myeloid Leukemia. J. Clin. Oncol. 2015, 33, 2949–2962. [Google Scholar] [CrossRef] [PubMed]
- Hasle, H.; Kaspers, G.J.L. Strategies for Reducing the Treatment-Related Physical Burden of Childhood Acute Myeloid Leukaemia—A Review. Br. J. Haematol. 2017, 176, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Klein, K.; de Haas, V.; Kaspers, G.J.L. Clinical Challenges in de Novo Pediatric Acute Myeloid Leukemia. Expert Rev. Anticancer Ther. 2018, 18, 277–293. [Google Scholar] [CrossRef]
- Hoffman, A.E.; Schoonmade, L.J.; Kaspers, G.J.L. Pediatric Relapsed Acute Myeloid Leukemia: A Systematic Review. Expert Rev. Anticancer Ther. 2021, 21, 45–52. [Google Scholar] [CrossRef]
- Sørensen, G.V.; Winther, J.F.; de Fine Licht, S.; Andersen, K.K.; Holmqvist, A.S.; Madanat-Harjuoja, L.; Tryggvadottir, L.; Bautz, A.; Lash, T.L.; Hasle, H. Long-Term Risk of Hospitalization Among Five-Year Survivors of Childhood Leukemia in the Nordic Countries. JNCI J. Natl. Cancer Inst. 2019, 111, 943–951. [Google Scholar] [CrossRef]
- Perna, F.; Berman, S.H.; Soni, R.K.; Mansilla-Soto, J.; Eyquem, J.; Hamieh, M.; Hendrickson, R.C.; Brennan, C.W.; Sadelain, M. Integrating Proteomics and Transcriptomics for Systematic Combinatorial Chimeric Antigen Receptor Therapy of AML. Cancer Cell 2017, 32, 506–519.e5. [Google Scholar] [CrossRef] [PubMed]
- Haubner, S.; Perna, F.; Köhnke, T.; Schmidt, C.; Berman, S.; Augsberger, C.; Schnorfeil, F.M.; Krupka, C.; Lichtenegger, F.S.; Liu, X.; et al. Coexpression Profile of Leukemic Stem Cell Markers for Combinatorial Targeted Therapy in AML. Leukemia 2019, 33, 64–74. [Google Scholar] [CrossRef]
- Willier, S.; Rothämel, P.; Hastreiter, M.; Wilhelm, J.; Stenger, D.; Blaeschke, F.; Rohlfs, M.; Kaeuferle, T.; Schmid, I.; Albert, M.H.; et al. CLEC12A and CD33 Coexpression as a Preferential Target for Pediatric AML Combinatorial Immunotherapy. Blood 2021, 137, 1037–1049. [Google Scholar] [CrossRef]
- Herrmann, H.; Sadovnik, I.; Eisenwort, G.; Rülicke, T.; Blatt, K.; Herndlhofer, S.; Willmann, M.; Stefanzl, G.; Baumgartner, S.; Greiner, G.; et al. Delineation of Target Expression Profiles in CD34+/CD38− and CD34+/CD38+ Stem and Progenitor Cells in AML and CML. Blood Adv. 2020, 4, 5118–5132. [Google Scholar] [CrossRef]
- Ehninger, A.; Kramer, M.; Röllig, C.; Thiede, C.; Bornhäuser, M.; von Bonin, M.; Wermke, M.; Feldmann, A.; Bachmann, M.; Ehninger, G.; et al. Distribution and Levels of Cell Surface Expression of CD33 and CD123 in Acute Myeloid Leukemia. Blood Cancer J. 2014, 4, e218. [Google Scholar] [CrossRef]
- Tasian, S.K. Acute Myeloid Leukemia Chimeric Antigen Receptor T-Cell Immunotherapy: How Far up the Road Have We Traveled? Ther. Adv. Hematol. 2018, 9, 135–148. [Google Scholar] [CrossRef]
- Tasian, S.K.; Pollard, J.A.; Aplenc, R. Molecular Therapeutic Approaches for Pediatric Acute Myeloid Leukemia. Front. Oncol. 2014, 4, 55. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Stein, E.M.; de Botton, S.; Roboz, G.J.; Altman, J.K.; Mims, A.S.; Swords, R.; Collins, R.H.; Mannis, G.N.; Pollyea, D.A.; et al. Durable Remissions with Ivosidenib in IDH1 -Mutated Relapsed or Refractory AML. N. Engl. J. Med. 2018, 378, 2386–2398. [Google Scholar] [CrossRef] [PubMed]
- Testa, U.; Pelosi, E.; Castelli, G. CD123 as a Therapeutic Target in the Treatment of Hematological Malignancies. Cancers 2019, 11, 1358. [Google Scholar] [CrossRef]
- Bras, A.E.; Haas, V.; Stigt, A.; Jongen-Lavrencic, M.; Beverloo, H.B.; Marvelde, J.G.; Zwaan, C.M.; Dongen, J.J.M.; Leusen, J.H.W.; Velden, V.H.J. CD123 Expression Levels in 846 Acute Leukemia Patients Based on Standardized Immunophenotyping. Cytom. Part B Clin. Cytom. 2019, 96, 134–142. [Google Scholar] [CrossRef]
- Taussig, D.C. Hematopoietic Stem Cells Express Multiple Myeloid Markers: Implications for the Origin and Targeted Therapy of Acute Myeloid Leukemia. Blood 2005, 106, 4086–4092. [Google Scholar] [CrossRef]
- Van Rhenen, A.; van Dongen, G.A.M.S.; Kelder, A.; Rombouts, E.J.; Feller, N.; Moshaver, B.; Walsum, M.S.; Zweegman, S.; Ossenkoppele, G.J.; Jan Schuurhuis, G. The Novel AML Stem Cell–Associated Antigen CLL-1 Aids in Discrimination between Normal and Leukemic Stem Cells. Blood 2007, 110, 2659–2666. [Google Scholar] [CrossRef] [PubMed]
- Bill, M.; Aggerholm, A.; Kjeldsen, E.; Roug, A.S.; Hokland, P.; Nederby, L. Revisiting CLEC12A as Leukaemic Stem Cell Marker in AML: Highlighting the Necessity of Precision Diagnostics in Patients Eligible for Targeted Therapy. Br. J. Haematol. 2019, 184, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Roug, A.S.; Larsen, H.Ø.; Nederby, L.; Just, T.; Brown, G.; Nyvold, C.G.; Ommen, H.B.; Hokland, P. HMICL and CD123 in Combination with a CD45/CD34/CD117 Backbone—A Universal Marker Combination for the Detection of Minimal Residual Disease in Acute Myeloid Leukaemia. Br. J. Haematol. 2014, 164, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Bill, M.; Niekerk, P.B.V.K.; Woll, P.S.; Herborg, L.L.; Roug, A.S.; Hokland, P.; Nederby, L. Mapping the CLEC12A Expression on Myeloid Progenitors in Normal Bone Marrow; Implications for Understanding CLEC12A-Related Cancer Stem Cell Biology. J. Cell. Mol. Med. 2018, 22, 2311–2318. [Google Scholar] [CrossRef]
- Arvindam, U.S.; van Hauten, P.M.M.; Schirm, D.; Schaap, N.; Hobo, W.; Blazar, B.R.; Vallera, D.A.; Dolstra, H.; Felices, M.; Miller, J.S. A Trispecific Killer Engager Molecule against CLEC12A Effectively Induces NK-Cell Mediated Killing of AML Cells. Leukemia 2020, 35, 1586–1596. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute Myeloid Leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef] [PubMed]
- Creutzig, U.; Zimmermann, M.; Reinhardt, D.; Rasche, M.; von Neuhoff, C.; Alpermann, T.; Dworzak, M.; Perglerová, K.; Zemanova, Z.; Tchinda, J.; et al. Changes in Cytogenetics and Molecular Genetics in Acute Myeloid Leukemia from Childhood to Adult Age Groups. Cancer 2016, 122, 3821–3830. [Google Scholar] [CrossRef] [PubMed]
- Bolouri, H.; Farrar, J.E.; Triche, T.; Ries, R.E.; Lim, E.L.; Alonzo, T.A.; Ma, Y.; Moore, R.; Mungall, A.J.; Marra, M.A.; et al. The Molecular Landscape of Pediatric Acute Myeloid Leukemia Reveals Recurrent Structural Alterations and Age-Specific Mutational Interactions. Nat. Med. 2018, 24, 103–112. [Google Scholar] [CrossRef]
- Dores, G.M.; Devesa, S.S.; Curtis, R.E.; Linet, M.S.; Morton, L.M. Acute Leukemia Incidence and Patient Survival among Children and Adults in the United States, 2001–2007. Blood 2012, 119, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Pang, W.W.; Price, E.A.; Sahoo, D.; Beerman, I.; Maloney, W.J.; Rossi, D.J.; Schrier, S.L.; Weissman, I.L. Human Bone Marrow Hematopoietic Stem Cells Are Increased in Frequency and Myeloid-Biased with Age. Proc. Natl. Acad. Sci. USA 2011, 108, 20012–20017. [Google Scholar] [CrossRef]
- Hennrich, M.L.; Romanov, N.; Horn, P.; Jaeger, S.; Eckstein, V.; Steeples, V.; Ye, F.; Ding, X.; Poisa-Beiro, L.; Lai, M.C.; et al. Cell-Specific Proteome Analyses of Human Bone Marrow Reveal Molecular Features of Age-Dependent Functional Decline. Nat. Commun. 2018, 9, 4004. [Google Scholar] [CrossRef]
- Schmidt, J.; Rücker-Braun, E.; Heidrich, K.; von Bonin, M.; Stölzel, F.; Thiede, C.; Middeke, J.; Ehninger, G.; Bornhäuser, M.; Schetelig, J.; et al. Pilot Study on Mass Spectrometry–Based Analysis of the Proteome of CD34+CD123+ Progenitor Cells for the Identification of Potential Targets for Immunotherapy in Acute Myeloid Leukemia. Proteomes 2018, 6, 11. [Google Scholar] [CrossRef]
- Raffel, S.; Klimmeck, D.; Falcone, M.; Demir, A.; Pouya, A.; Zeisberger, P.; Lutz, C.; Tinelli, M.; Bischel, O.; Bullinger, L.; et al. Quantitative Proteomics Reveals Specific Metabolic Features of Acute Myeloid Leukemia Stem Cells. Blood 2020, 136, 1507–1519. [Google Scholar] [CrossRef]
- Bonnet, D.; Dick, J.E. Human Acute Myeloid Leukemia Is Organized as a Hierarchy That Originates from a Primitive Hematopoietic Cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef]
- Hope, K.J.; Jin, L.; Dick, J.E. Acute Myeloid Leukemia Originates from a Hierarchy of Leukemic Stem Cell Classes That Differ in Self-Renewal Capacity. Nat. Immunol. 2004, 5, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Shlush, L.I.; Mitchell, A.; Heisler, L.; Abelson, S.; Ng, S.W.K.; Trotman-Grant, A.; Medeiros, J.J.F.; Rao-Bhatia, A.; Jaciw-Zurakowsky, I.; Marke, R.; et al. Tracing the Origins of Relapse in Acute Myeloid Leukaemia to Stem Cells. Nature 2017, 547, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Taussig, D.C.; Vargaftig, J.; Miraki-Moud, F.; Griessinger, E.; Sharrock, K.; Luke, T.; Lillington, D.; Oakervee, H.; Cavenagh, J.; Agrawal, S.G.; et al. Leukemia-Initiating Cells from Some Acute Myeloid Leukemia Patients with Mutated Nucleophosmin Reside in the CD34− Fraction. Blood 2010, 115, 1976–1984. [Google Scholar] [CrossRef] [PubMed]
- Zeijlemaker, W.; Kelder, A.; Wouters, R.; Valk, P.J.M.; Witte, B.I.; Cloos, J.; Ossenkoppele, G.J.; Schuurhuis, G.J. Absence of Leukaemic CD34 + Cells in Acute Myeloid Leukaemia Is of High Prognostic Value: A Longstanding Controversy Deciphered. Br. J. Haematol. 2015, 171, 227–238. [Google Scholar] [CrossRef]
- Quek, L.; Otto, G.W.; Garnett, C.; Lhermitte, L.; Karamitros, D.; Stoilova, B.; Lau, I.-J.; Doondeea, J.; Usukhbayar, B.; Kennedy, A.; et al. Genetically Distinct Leukemic Stem Cells in Human CD34− Acute Myeloid Leukemia Are Arrested at a Hemopoietic Precursor-like Stage. J. Exp. Med. 2016, 213, 1513–1535. [Google Scholar] [CrossRef]
- Terwijn, M.; Zeijlemaker, W.; Kelder, A.; Rutten, A.P.; Snel, A.N.; Scholten, W.J.; Pabst, T.; Verhoef, G.; Löwenberg, B.; Zweegman, S.; et al. Leukemic Stem Cell Frequency: A Strong Biomarker for Clinical Outcome in Acute Myeloid Leukemia. PLoS ONE 2014, 9, e107587. [Google Scholar] [CrossRef]
- Sarry, J.-E.; Murphy, K.; Perry, R.; Sanchez, P.V.; Secreto, A.; Keefer, C.; Swider, C.R.; Strzelecki, A.-C.; Cavelier, C.; Récher, C.; et al. Human Acute Myelogenous Leukemia Stem Cells Are Rare and Heterogeneous When Assayed in NOD/SCID/IL2Rγc-Deficient Mice. J. Clin. Investig. 2011, 121, 384–395. [Google Scholar] [CrossRef]
- Doulatov, S.; Notta, F.; Eppert, K.; Nguyen, L.T.; Ohashi, P.S.; Dick, J.E. Revised Map of the Human Progenitor Hierarchy Shows the Origin of Macrophages and Dendritic Cells in Early Lymphoid Development. Nat. Immunol. 2010, 11, 585–593. [Google Scholar] [CrossRef]
- Petersen, M.A.; Rosenberg, C.A.; Brøndum, R.F.; Aggerholm, A.; Kjeldsen, E.; Rahbek, O.; Ludvigsen, M.; Hasle, H.; Roug, A.S.; Bill, M. Immunophenotypically Defined Stem Cell Subsets in Paediatric AML Are Highly Heterogeneous and Demonstrate Differences in BCL-2 Expression by Cytogenetic Subgroups. Br. J. Haematol. 2022, 197, 452–466. [Google Scholar] [CrossRef]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, Encapsulated Proteomic-Sample Processing Applied to Copy-Number Estimation in Eukaryotic Cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef]
- Ludvigsen, M.; Thorlacius-Ussing, L.; Vorum, H.; Moyer, M.P.; Stender, M.T.; Thorlacius-Ussing, O.; Honoré, B. Proteomic Characterization of Colorectal Cancer Cells versus Normal-Derived Colon Mucosa Cells: Approaching Identification of Novel Diagnostic Protein Biomarkers in Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 3466. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J.; Tugendreich, S. Causal Analysis Approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Kimball, S.R. Eukaryotic Initiation Factor EIF2. Int. J. Biochem. Cell Biol. 1999, 31, 25–29. [Google Scholar] [CrossRef]
- Van Galen, P.; Mbong, N.; Kreso, A.; Schoof, E.M.; Wagenblast, E.; Ng, S.W.K.; Krivdova, G.; Jin, L.; Nakauchi, H.; Dick, J.E. Integrated Stress Response Activity Marks Stem Cells in Normal Hematopoiesis and Leukemia. Cell Rep. 2018, 25, 1109–1117.e5. [Google Scholar] [CrossRef] [PubMed]
- Conneely, S.E.; Rau, R.E. The Genomics of Acute Myeloid Leukemia in Children. Cancer Metastasis Rev. 2020, 39, 189–209. [Google Scholar] [CrossRef]
- Chatterjee, S.; Burns, T.F. Targeting Heat Shock Proteins in Cancer: A Promising Therapeutic Approach. Int. J. Mol. Sci. 2017, 18, 1978. [Google Scholar] [CrossRef]
- De Necochea-Campion, R.; Shouse, G.P.; Zhou, Q.; Mirshahidi, S.; Chen, C.-S. Aberrant Splicing and Drug Resistance in AML. J. Hematol. Oncol. 2016, 9, 85. [Google Scholar] [CrossRef][Green Version]
- Thomas, X.; Campos, L.; Mounier, C.; Cornillon, J.; Flandrin, P.; Le, Q.-H.; Piselli, S.; Guyotat, D. Expression of Heat-Shock Proteins Is Associated with Major Adverse Prognostic Factors in Acute Myeloid Leukemia. Leuk. Res. 2005, 29, 1049–1058. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Zhang, H.; Du, C.; Liu, X.; Zhu, S.; Zhang, W.; Li, Z.; Gao, C.; Zhao, X.; Mei, M.; et al. Correlation of SRSF1 and PRMT1 Expression with Clinical Status of Pediatric Acute Lymphoblastic Leukemia. J. Hematol. Oncol. 2012, 5, 42. [Google Scholar] [CrossRef]
- Klampfl, T.; Gisslinger, H.; Harutyunyan, A.S.; Nivarthi, H.; Rumi, E.; Milosevic, J.D.; Them, N.C.C.; Berg, T.; Gisslinger, B.; Pietra, D.; et al. Somatic Mutations of Calreticulin in Myeloproliferative Neoplasms. N. Engl. J. Med. 2013, 369, 2379–2390. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Buqué, A.; Kepp, O.; Zitvogel, L.; Kroemer, G. Immunogenic Cell Death in Cancer and Infectious Disease. Nat. Rev. Immunol. 2017, 17, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Chao, M.P.; Jaiswal, S.; Weissman-Tsukamoto, R.; Alizadeh, A.A.; Gentles, A.J.; Volkmer, J.; Weiskopf, K.; Willingham, S.B.; Raveh, T.; Park, C.Y.; et al. Calreticulin Is the Dominant Pro-Phagocytic Signal on Multiple Human Cancers and Is Counterbalanced by CD47. Sci. Transl. Med. 2010, 2, 63ra94. [Google Scholar] [CrossRef] [PubMed]
- Fucikova, J.; Truxova, I.; Hensler, M.; Becht, E.; Kasikova, L.; Moserova, I.; Vosahlikova, S.; Klouckova, J.; Church, S.E.; Cremer, I.; et al. Calreticulin Exposure by Malignant Blasts Correlates with Robust Anticancer Immunity and Improved Clinical Outcome in AML Patients. Blood 2016, 128, 3113–3124. [Google Scholar] [CrossRef] [PubMed]
- Wemeau, M.; Kepp, O.; Tesnière, A.; Panaretakis, T.; Flament, C.; De Botton, S.; Zitvogel, L.; Kroemer, G.; Chaput, N. Calreticulin Exposure on Malignant Blasts Predicts a Cellular Anticancer Immune Response in Patients with Acute Myeloid Leukemia. Cell Death Dis. 2010, 1, e104. [Google Scholar] [CrossRef] [PubMed]
- Kornblau, S.M.; Qutub, A.; Yao, H.; York, H.; Qiu, Y.H.; Graber, D.; Ravandi, F.; Cortes, J.; Andreeff, M.; Zhang, N.; et al. Proteomic Profiling Identifies Distinct Protein Patterns in Acute Myelogenous Leukemia CD34+CD38- Stem-Like Cells. PLoS ONE 2013, 8, e78453. [Google Scholar] [CrossRef]
- Raffel, S.; Falcone, M.; Kneisel, N.; Hansson, J.; Wang, W.; Lutz, C.; Bullinger, L.; Poschet, G.; Nonnenmacher, Y.; Barnert, A.; et al. BCAT1 Restricts AKG Levels in AML Stem Cells Leading to IDHmut-like DNA Hypermethylation. Nature 2017, 551, 384–388. [Google Scholar] [CrossRef]
- Hoff, F.W.; Hu, C.W.; Qiu, Y.; Ligeralde, A.; Yoo, S.-Y.; Mahmud, H.; de Bont, E.S.J.M.; Qutub, A.A.; Horton, T.M.; Kornblau, S.M. Recognition of Recurrent Protein Expression Patterns in Pediatric Acute Myeloid Leukemia Identified New Therapeutic Targets. Mol. Cancer Res. 2018, 16, 1275–1286. [Google Scholar] [CrossRef]
- Braoudaki, M.; Tzortzatou-Stathopoulou, F.; Anagnostopoulos, A.K.; Papathanassiou, C.; Vougas, K.; Karamolegou, K.; Tsangaris, G.T. Proteomic Analysis of Childhood de Novo Acute Myeloid Leukemia and Myelodysplastic Syndrome/AML: Correlation to Molecular and Cytogenetic Analyses. Amino Acids 2011, 40, 943–951. [Google Scholar] [CrossRef]
- Hoff, F.W.; Van Dijk, A.D.; Qiu, Y.; Hu, C.W.; Ries, R.E.; Ligeralde, A.; Jenkins, G.N.; Gerbing, R.B.; Gamis, A.S.; Aplenc, R.; et al. Clinical Relevance of Proteomic Profiling in de Novo Pediatric Acute Myeloid Leukemia: A Children’s Oncology Group Study. Haematologica 2022. [Google Scholar] [CrossRef]
- Nguyen, N.H.K.; Wu, H.; Tan, H.; Peng, J.; Rubnitz, J.E.; Cao, X.; Pounds, S.; Lamba, J.K. Global Proteomic Profiling of Pediatric Aml: A Pilot Study. Cancers 2021, 13, 3161. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]


| Sample ID | Age | Sex | FAB Type | CD34 Category (%) # | Karyotype | Known Mutations | CD34+CD38– Mutational Status | Events |
|---|---|---|---|---|---|---|---|---|
| pAML17 | 2 | female | M5 | CD34 negative (0.07%) | 46, XX, inv(6)(p12q16), t(9;11)(p22;q23) [25] | none | t(9;11) not detected | Induction death |
| pAML20 | 12 | male | M4 | CD34 positive (8.6%) | 46, XY | none | unknown | Resistant disease to death |
| pAML21 | 4 | female | M5 | CD34 negative (0.12%) | 46, XX, t(9;11)(p22;q23) [16]/47, idem, +9 [9] | none | t(9;11) not detected | Death after relapse |
| pAML23 | 11 | female | M4 | CD34 positive (71.5%) | 46, XX | none | unknown | Induction death |
| pAML29 | 7 | female | M5 | CD34 positive (37.4%) | 46, XX, inv(16)(p13q22) [25] | FLT3-TKD | Inv(16) FLT3-TKD | Induction death |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petersen, M.A.; Rosenberg, C.A.; Bill, M.; Enemark, M.B.; Rahbek, O.; Roug, A.S.; Hasle, H.; Honoré, B.; Ludvigsen, M. Proteomic Profiling Identifies Specific Leukemic Stem Cell-Associated Protein Expression Patterns in Pediatric AML Patients. Cancers 2022, 14, 3567. https://doi.org/10.3390/cancers14153567
Petersen MA, Rosenberg CA, Bill M, Enemark MB, Rahbek O, Roug AS, Hasle H, Honoré B, Ludvigsen M. Proteomic Profiling Identifies Specific Leukemic Stem Cell-Associated Protein Expression Patterns in Pediatric AML Patients. Cancers. 2022; 14(15):3567. https://doi.org/10.3390/cancers14153567
Chicago/Turabian StylePetersen, Marianne Agerlund, Carina Agerbo Rosenberg, Marie Bill, Marie Beck Enemark, Ole Rahbek, Anne Stidsholt Roug, Henrik Hasle, Bent Honoré, and Maja Ludvigsen. 2022. "Proteomic Profiling Identifies Specific Leukemic Stem Cell-Associated Protein Expression Patterns in Pediatric AML Patients" Cancers 14, no. 15: 3567. https://doi.org/10.3390/cancers14153567
APA StylePetersen, M. A., Rosenberg, C. A., Bill, M., Enemark, M. B., Rahbek, O., Roug, A. S., Hasle, H., Honoré, B., & Ludvigsen, M. (2022). Proteomic Profiling Identifies Specific Leukemic Stem Cell-Associated Protein Expression Patterns in Pediatric AML Patients. Cancers, 14(15), 3567. https://doi.org/10.3390/cancers14153567






