High PANX1 Expression Leads to Neutrophil Recruitment and the Formation of a High Adenosine Immunosuppressive Tumor Microenvironment in Basal-like Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Clinical Specimen Collection
2.3. Cell Lines and Culture Conditions
2.4. RNA Sequencing
2.5. Bulk Transcription Data Analysis
2.6. Single Cell Transcription Data Analysis
2.7. Immunofluorescence Staining
2.8. Immunohistochemical Staining
2.9. Neutrophil Isolation
2.10. ShRNA Knockdown of PANX1
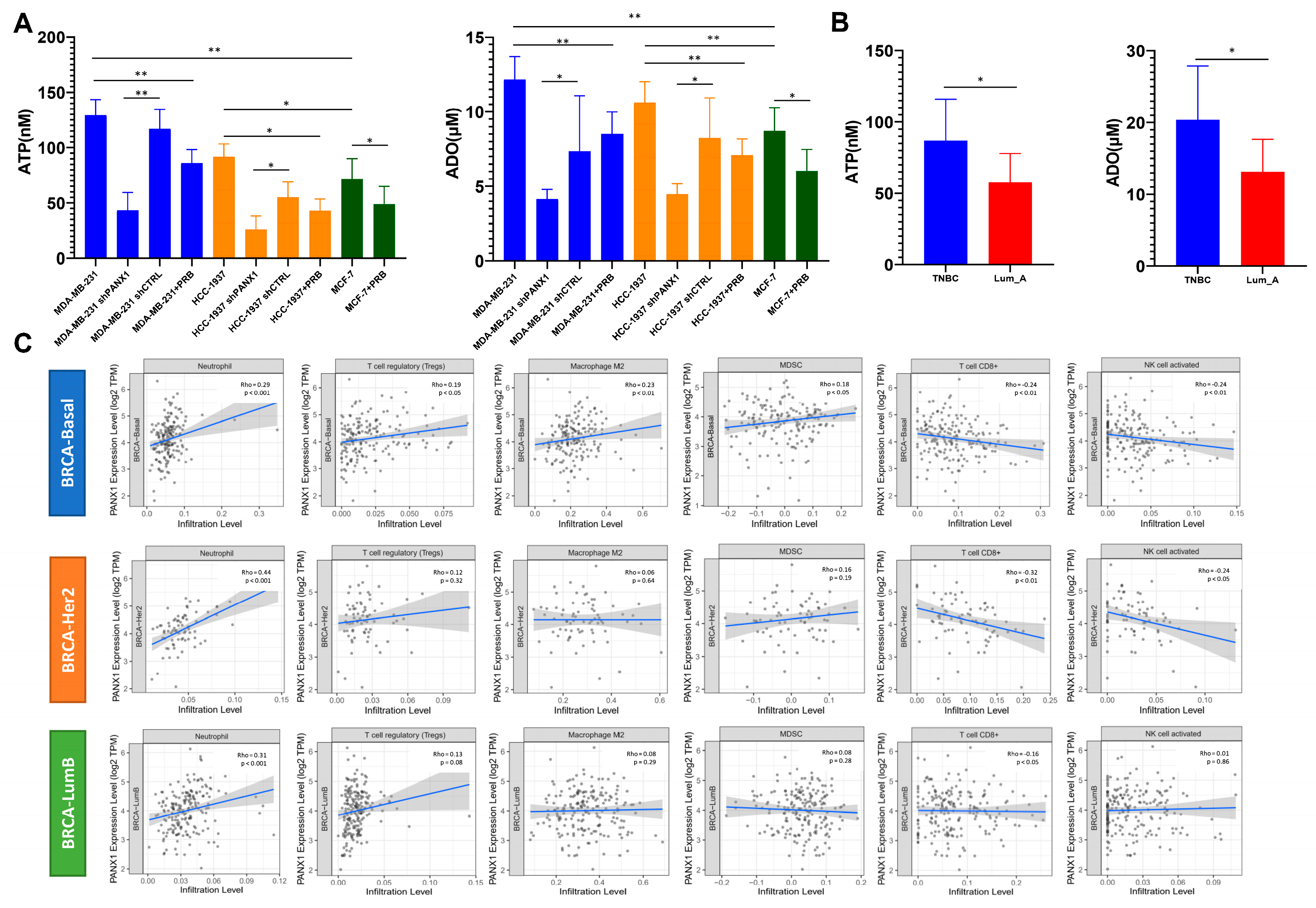
2.11. Extracellular ATP/ADO Assay
2.12. Statistical Analyses
3. Results
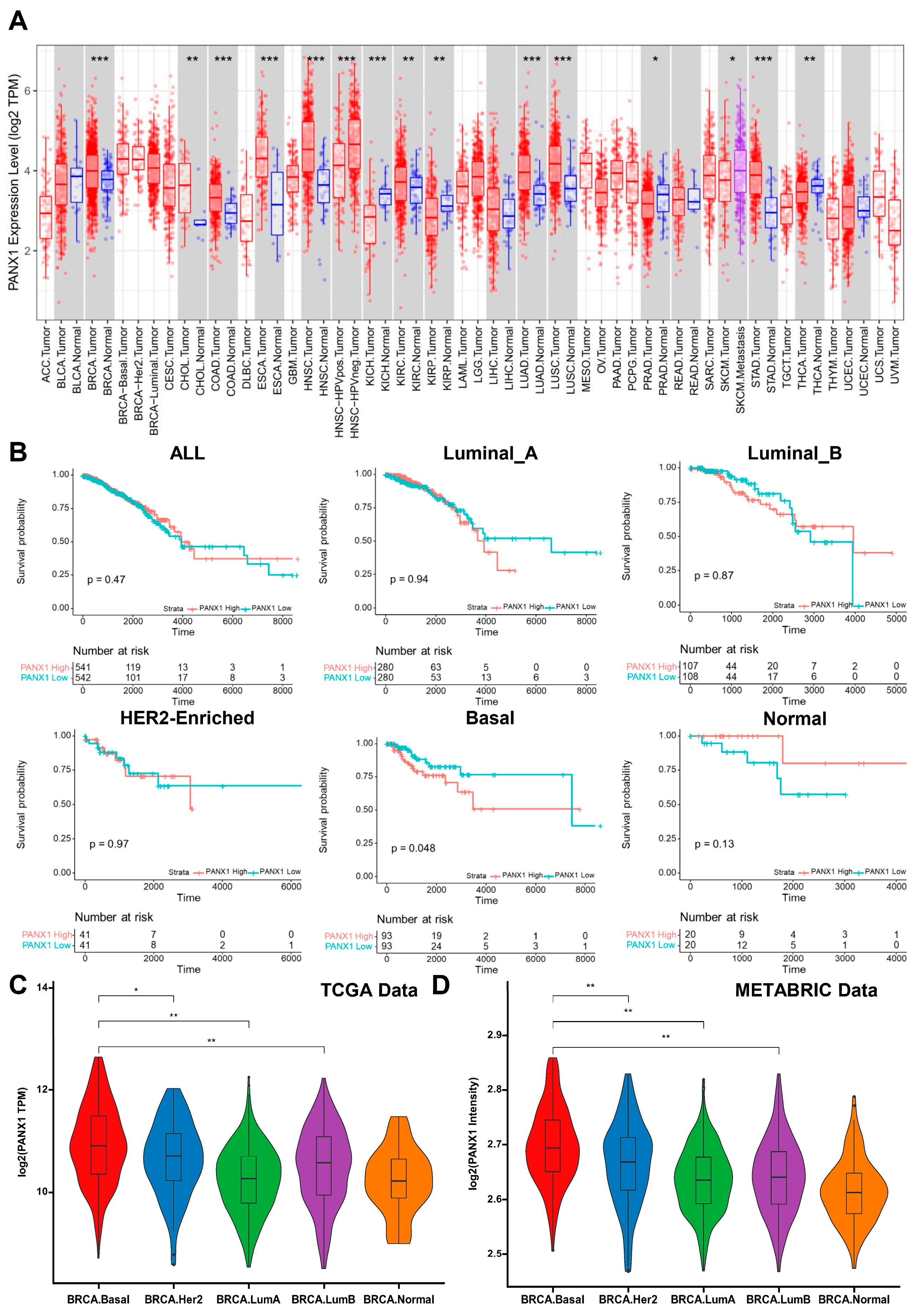
3.1. PANX1 Was Highly Expressed in Basal-like Breast Cancer
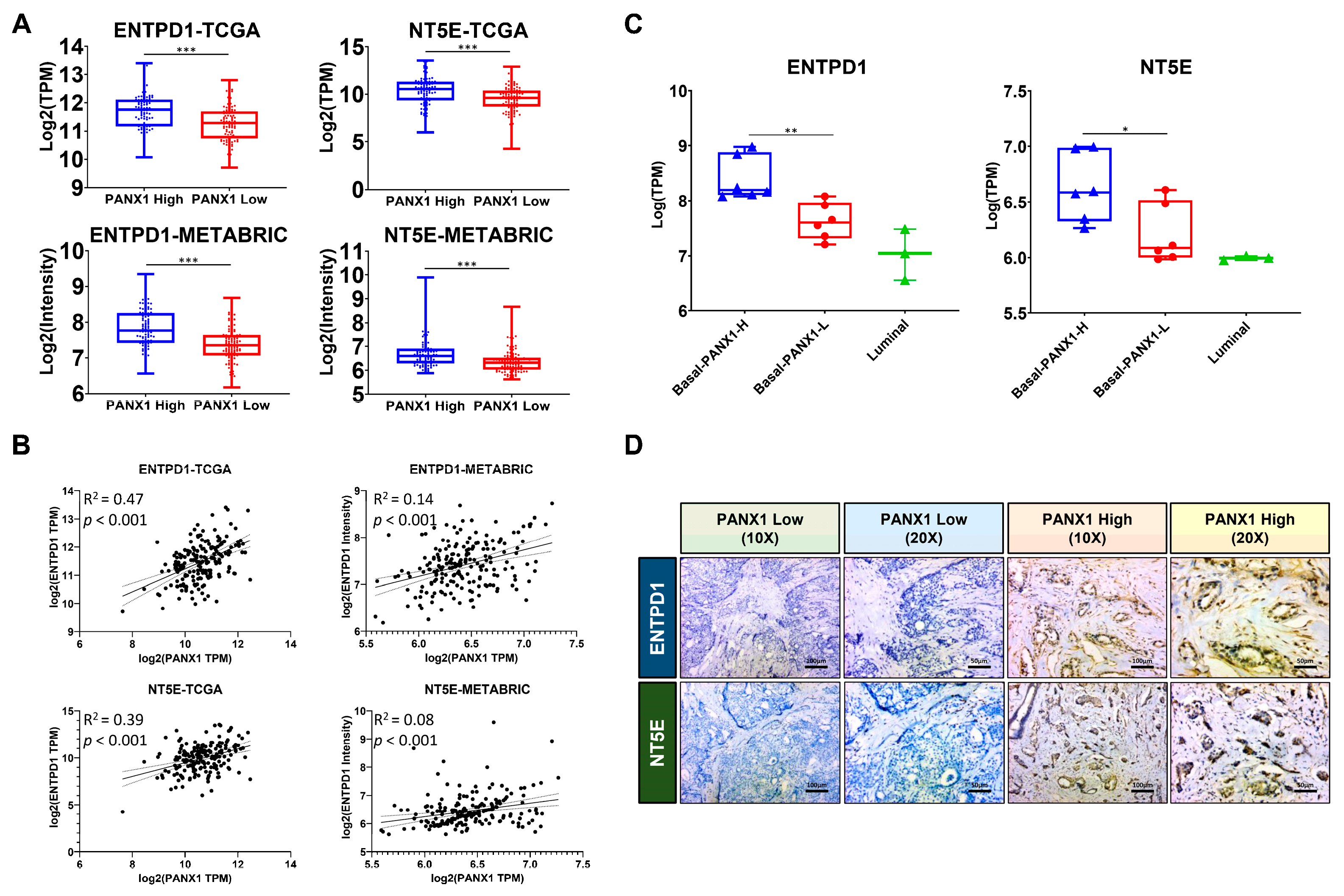
3.2. PANX1 Expression Positive Correlated with ENTPD1/NT5E Expression in the TME
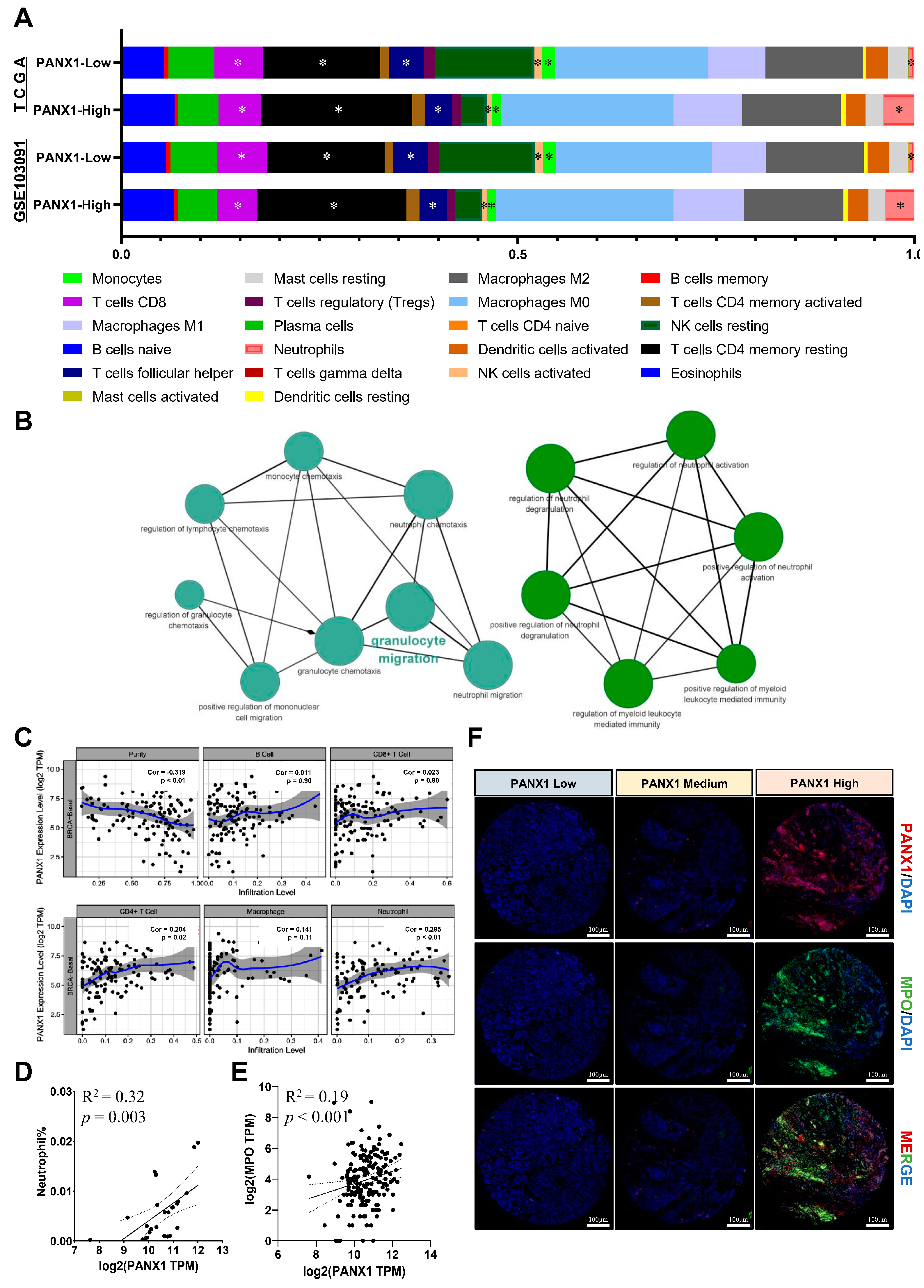
3.3. PANX1 Expression Was Positively Correlated with TAN Infiltration in Basal-like Breast Cancer
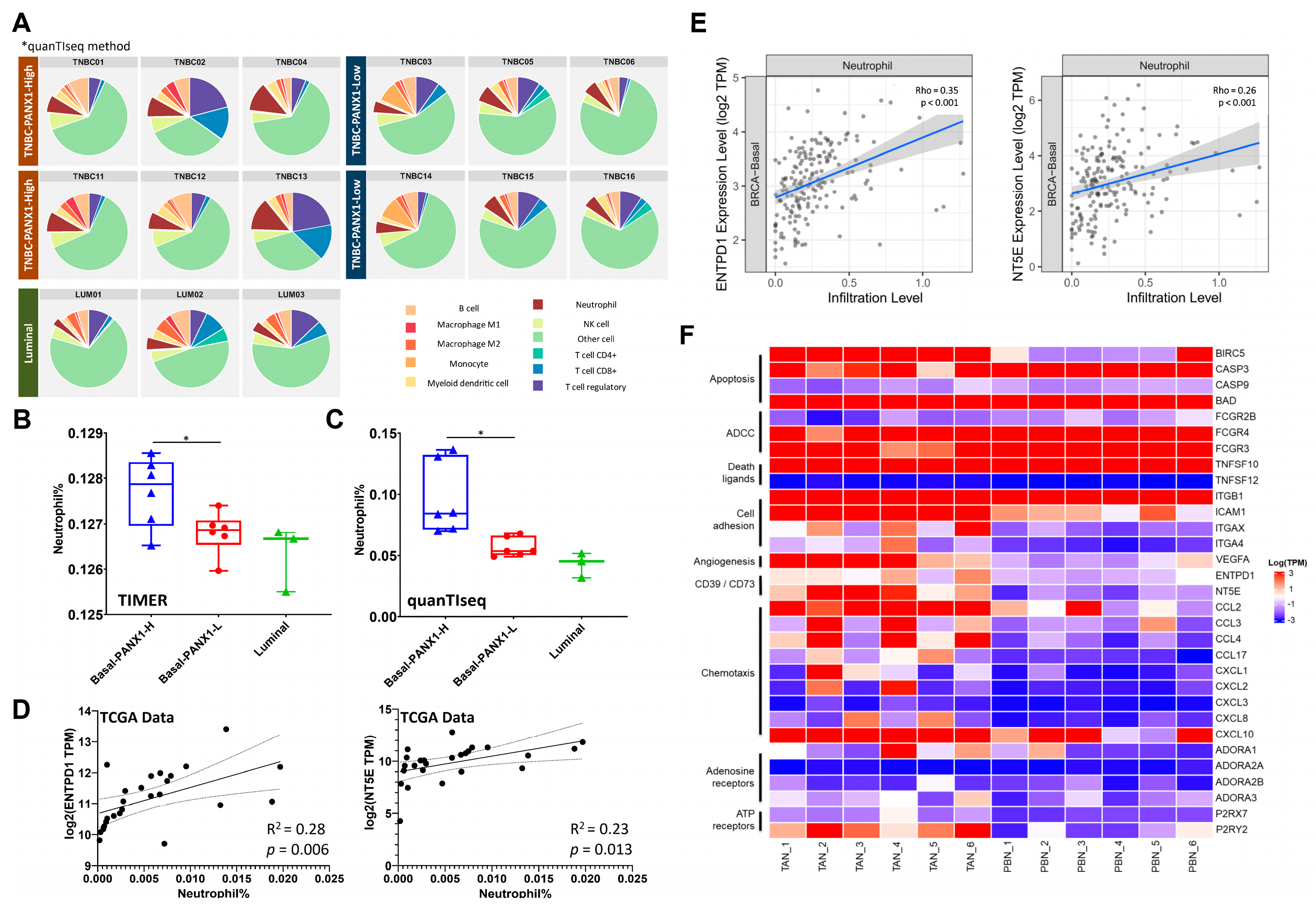
3.4. Immunosuppressive TANs Demonstrated More Infiltration in Basal-like Breast Cancer with High PANX1 Expression
3.5. High PANX1 Expression Induced a High exADO Immunosuppressive TME in Basal-like Breast Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alluri, P.; Newman, L.A. Basal-like and triple-negative breast cancers: Searching for positives among many negatives. Surg. Oncol. Clin. N. Am. 2014, 23, 567–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertucci, F.; Finetti, P.; Cervera, N.; Esterni, B.; Hermitte, F.; Viens, P.; Birnbaum, D. How basal are triple-negative breast cancers? Int. J. Cancer 2008, 123, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Abramson, V.G.; Lehmann, B.D.; Ballinger, T.J.; Pietenpol, J.A. Subtyping of triple-negative breast cancer: Implications for therapy. Cancer 2015, 121, 8–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badve, S.; Dabbs, D.J.; Schnitt, S.J.; Baehner, F.L.; Decker, T.; Eusebi, V.; Fox, S.B.; Ichihara, S.; Jacquemier, J.; Lakhani, S.R.; et al. Basal-like and triple-negative breast cancers: A critical review with an emphasis on the implications for pathologists and oncologists. Mod. Pathol. 2011, 24, 157–167. [Google Scholar] [CrossRef] [Green Version]
- Gao, G.; Wang, Z.; Qu, X.; Zhang, Z. Prognostic value of tumor-infiltrating lymphocytes in patients with triple-negative breast cancer: A systematic review and meta-analysis. BMC Cancer 2020, 20, 179. [Google Scholar] [CrossRef] [Green Version]
- Bayraktar, S.; Batoo, S.; Okuno, S.; Glück, S. Immunotherapy in breast cancer. J. Carcinog. 2019, 18, 2. [Google Scholar] [CrossRef]
- Lohman, A.W.; Leskov, I.L.; Butcher, J.T.; Johnstone, S.R.; Stokes, T.A.; Begandt, D.; DeLalio, L.J.; Best, A.K.; Penuela, S.; Leitinger, N.; et al. Pannexin 1 channels regulate leukocyte emigration through the venous endothelium during acute inflammation. Nat. Commun. 2015, 6, 7965. [Google Scholar] [CrossRef]
- Jacob, F.; Perez Novo, C.; Bachert, C.; Van Crombruggen, K. Purinergic signaling in inflammatory cells: P2 receptor expression, functional effects, and modulation of inflammatory responses. Purinergic Signal 2013, 9, 285–306. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; He, Y.; Muñoz-Planillo, R.; Liu, Q.; Núñez, G. Caspase-11 Requires the Pannexin-1 Channel and the Purinergic P2X7 Pore to Mediate Pyroptosis and Endotoxic Shock. Immunity 2015, 43, 923–932. [Google Scholar] [CrossRef] [Green Version]
- Vijayan, D.; Young, A.; Teng, M.W.L.; Smyth, M.J. Targeting immunosuppressive adenosine in cancer. Nat. Rev. Cancer 2017, 17, 709–724. [Google Scholar] [CrossRef]
- Feng, L.L.; Cai, Y.Q.; Zhu, M.C.; Xing, L.J.; Wang, X. The yin and yang functions of extracellular ATP and adenosine in tumor immunity. Cancer Cell Int. 2020, 20, 110. [Google Scholar] [CrossRef] [PubMed]
- Medina, C.B.; Chiu, Y.H.; Stremska, M.E.; Lucas, C.D.; Poon, I.; Tung, K.S.; Elliott, M.R.; Desai, B.; Lorenz, U.M.; Bayliss, D.A.; et al. Pannexin 1 channels facilitate communication between T cells to restrict the severity of airway inflammation. Immunity 2021, 54, 1715–1727.e7. [Google Scholar] [CrossRef]
- Jalaleddine, N.; El-Hajjar, L.; Dakik, H.; Shaito, A.; Saliba, J.; Safi, R.; Zibara, K.; El-Sabban, M. Pannexin1 Is Associated with Enhanced Epithelial-To-Mesenchymal Transition in Human Patient Breast Cancer Tissues and in Breast Cancer Cell Lines. Cancers 2019, 11, 1967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic. Acids. Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Mounir, M.; Lucchetta, M.; Silva, T.C.; Olsen, C.; Bontempi, G.; Chen, X.; Noushmehr, H.; Colaprico, A.; Papaleo, E. New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput. Biol. 2019, 15, e1006701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, T.C.; Colaprico, A.; Olsen, C.; D’Angelo, F.; Bontempi, G.; Ceccarelli, M.; Noushmehr, H. TCGA Workflow: Analyze cancer genomics and epigenomics data using Bioconductor packages. F1000Research 2016, 5, 1542. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, W.; Jin, X.; Ji, H.; Wang, H.; Glusman, G.; Robinson, M.; Liu, L.; Ruan, J.; Gao, S. NormExpression: An R Package to Normalize Gene Expression Data Using Evaluated Methods. Front. Genet. 2019, 10, 400. [Google Scholar] [CrossRef] [Green Version]
- Jézéquel, P.; Loussouarn, D.; Guérin-Charbonnel, C.; Campion, L.; Vanier, A.; Gouraud, W.; Lasla, H.; Guette, C.; Valo, I.; Verrièle, V.; et al. Gene-expression molecular subtyping of triple-negative breast cancer tumours: Importance of immune response. Breast Cancer Res. 2015, 17, 43. [Google Scholar] [CrossRef] [Green Version]
- Jézéquel, P.; Kerdraon, O.; Hondermarck, H.; Guérin-Charbonnel, C.; Lasla, H.; Gouraud, W.; Canon, J.L.; Gombos, A.; Dalenc, F.; Delaloge, S.; et al. Identification of three subtypes of triple-negative breast cancer with potential therapeutic implications. Breast Cancer Res. 2019, 21, 65. [Google Scholar] [CrossRef]
- Jørgensen, C.L.T.; Larsson, A.M.; Forsare, C.; Aaltonen, K.; Jansson, S.; Bradshaw, R.; Bendahl, P.O.; Rydén, L. PAM50 Intrinsic Subtype Profiles in Primary and Metastatic Breast Cancer Show a Significant Shift toward More Aggressive Subtypes with Prognostic Implications. Cancers 2021, 13, 1592. [Google Scholar] [CrossRef] [PubMed]
- Gendoo, D.M.; Ratanasirigulchai, N.; Schröder, M.S.; Paré, L.; Parker, J.S.; Prat, A.; Haibe-Kains, B. Genefu: An R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics 2016, 32, 1097–1099. [Google Scholar] [CrossRef] [Green Version]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; Van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Finotello, F.; Mayer, C.; Plattner, C.; Laschober, G.; Rieder, D.; Hackl, H.; Krogsdam, A.; Loncova, Z.; Posch, W.; Wilflingseder, D.; et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019, 11, 34. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, S.; Dunn, P.; Thomas, C.G.; Smith, B.; Schaefer, H.; Chen, J.; Hu, Z.; Zalocusky, K.A.; Shankar, R.D.; Shen-Orr, S.S.; et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data 2018, 5, 180015. [Google Scholar] [CrossRef]
- Chen, X.; Li, J.; Gray, W.H.; Lehmann, B.D.; Bauer, J.A.; Shyr, Y.; Pietenpol, J.A. TNBCtype: A Subtyping Tool for Triple-Negative Breast Cancer. Cancer Inform. 2012, 11, 147–156. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Z.; Roden, D.L.; Wang, C.; Holliday, H.; Harvey, K.; Cazet, A.S.; Murphy, K.J.; Pereira, B.; Al-Eryani, G.; Bartonicek, N.; et al. Stromal cell diversity associated with immune evasion in human triple-negative breast cancer. EMBO J. 2020, 39, e104063. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Crowe, A.R.; Yue, W. Semi-quantitative Determination of Protein Expression using Immunohistochemistry Staining and Analysis: An Integrated Protocol. Bio-Protocol 2019, 9, e3465. [Google Scholar] [CrossRef]
- Finotello, F.; Trajanoski, Z. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol. Immunother. 2018, 67, 1031–1040. [Google Scholar] [CrossRef]
- Velasquez, S.; Eugenin, E.A. Role of Pannexin-1 hemichannels and purinergic receptors in the pathogenesis of human diseases. Front. Physiol. 2014, 5, 96. [Google Scholar] [CrossRef] [Green Version]
- Dahl, G. ATP release through pannexon channels. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140191. [Google Scholar] [CrossRef]
- Bao, L.; Sun, K.; Zhang, X. PANX1 is a potential prognostic biomarker associated with immune infiltration in pancreatic adenocarcinoma: A pan-cancer analysis. Channels 2021, 15, 680–696. [Google Scholar] [CrossRef]
- Furlow, P.W.; Zhang, S.; Soong, T.D.; Halberg, N.; Goodarzi, H.; Mangrum, C.; Wu, Y.G.; Elemento, O.; Tavazoie, S.F. Mechanosensitive pannexin-1 channels mediate microvascular metastatic cell survival. Nat. Cell Biol. 2015, 17, 943–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, G.; Liu, C.; Yang, Y.; Song, L.; Liu, X.; Wang, C.; Peng, Z.; Li, H.; Zhong, L. Panx1 promotes invasion-metastasis cascade in hepatocellular carcinoma. J. Cancer 2019, 10, 5681–5688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Yuan, M.; Yao, Y.; Wu, D.; Dong, S.; Tong, X. In vitro effect of Pannexin 1 channel on the invasion and migration of I-10 testicular cancer cells via ERK1/2 signaling pathway. Biomed. Pharmacother 2019, 117, 109090. [Google Scholar] [CrossRef]
- Penuela, S.; Gyenis, L.; Ablack, A.; Churko, J.M.; Berger, A.C.; Litchfield, D.W.; Lewis, J.D.; Laird, D.W. Loss of pannexin 1 attenuates melanoma progression by reversion to a melanocytic phenotype. J. Biol. Chem. 2012, 287, 29184–29193. [Google Scholar] [CrossRef] [Green Version]
- Xiang, X.; Langlois, S.; St-Pierre, M.E.; Blinder, A.; Charron, P.; Graber, T.E.; Fowler, S.L.; Baird, S.D.; Bennett, S.A.L.; Alain, T.; et al. Identification of pannexin 1-regulated genes, interactome, and pathways in rhabdomyosarcoma and its tumor inhibitory interaction with AHNAK. Oncogene 2021, 40, 1868–1883. [Google Scholar] [CrossRef] [PubMed]
- Eruslanov, E.B. Phenotype and function of tumor-associated neutrophils and their subsets in early-stage human lung cancer. Cancer Immunol. Immunother. 2017, 66, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Shaul, M.E.; Fridlender, Z.G. Cancer-related circulating and tumor-associated neutrophils—subtypes, sources and function. FEBS J. 2018, 285, 4316–4342. [Google Scholar] [CrossRef] [PubMed]
- Di Virgilio, F.; Sarti, A.C.; Coutinho-Silva, R. Purinergic signaling, DAMPs, and inflammation. Am. J. Physiol. Cell Physiol. 2020, 318, C832–C835. [Google Scholar] [CrossRef] [Green Version]
- Karmakar, M.; Katsnelson, M.A.; Dubyak, G.R.; Pearlman, E. Neutrophil P2X7 receptors mediate NLRP3 inflammasome-dependent IL-1β secretion in response to ATP. Nat. Commun. 2016, 7, 10555. [Google Scholar] [CrossRef] [Green Version]
- Vaughan, K.R.; Stokes, L.; Prince, L.R.; Marriott, H.M.; Meis, S.; Kassack, M.U.; Bingle, C.D.; Sabroe, I.; Surprenant, A.; Whyte, M.K. Inhibition of neutrophil apoptosis by ATP is mediated by the P2Y11 receptor. J. Immunol. 2007, 179, 8544–8553. [Google Scholar] [CrossRef] [Green Version]
- Antonioli, L.; Pacher, P.; Vizi, E.S.; Haskó, G. CD39 and CD73 in immunity and inflammation. Trends. Mol. Med. 2013, 19, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Chen, D. Purinergic Regulation of Neutrophil Function. Front. Immunol. 2018, 9, 399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Corriden, R.; Inoue, Y.; Yip, L.; Hashiguchi, N.; Zinkernagel, A.; Nizet, V.; Insel, P.A.; Junger, W.G. ATP release guides neutrophil chemotaxis via P2Y2 and A3 receptors. Science 2006, 314, 1792–1795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barletta, K.E.; Ley, K.; Mehrad, B. Regulation of neutrophil function by adenosine. Arter. Thromb. Vasc. Biol. 2012, 32, 856–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yago, T.; Tsukamoto, H.; Liu, Z.; Wang, Y.; Thompson, L.F.; McEver, R.P. Multi-Inhibitory Effects of A2A Adenosine Receptor Signaling on Neutrophil Adhesion Under Flow. J. Immunol. 2015, 195, 3880–3889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sil, P.; Hayes, C.P.; Reaves, B.J.; Breen, P.; Quinn, S.; Sokolove, J.; Rada, B. P2Y6 Receptor Antagonist MRS2578 Inhibits Neutrophil Activation and Aggregated Neutrophil Extracellular Trap Formation Induced by Gout-Associated Monosodium Urate Crystals. J. Immunol. 2017, 198, 428–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barletta, K.E.; Cagnina, R.E.; Burdick, M.D.; Linden, J.; Mehrad, B. Adenosine A(2B) receptor deficiency promotes host defenses against gram-negative bacterial pneumonia. Am. J. Respir. Crit. Care Med. 2012, 186, 1044–1050. [Google Scholar] [CrossRef] [Green Version]
- Corriden, R.; Self, T.; Akong-Moore, K.; Nizet, V.; Kellam, B.; Briddon, S.J.; Hill, S.J. Adenosine-A3 receptors in neutrophil microdomains promote the formation of bacteria-tethering cytonemes. EMBO Rep. 2013, 14, 726–732. [Google Scholar] [CrossRef]
- Wu, L.; Saxena, S.; Goel, P.; Prajapati, D.R.; Wang, C.; Singh, R.K. Breast Cancer Cell-Neutrophil Interactions Enhance Neutrophil Survival and Pro-Tumorigenic Activities. Cancers 2020, 12, 2884. [Google Scholar] [CrossRef]
- Abrahamsson, A.; Rzepecka, A.; Dabrosin, C. Equal Pro-inflammatory Profiles of CCLs, CXCLs, and Matrix Metalloproteinases in the Extracellular Microenvironment In Vivo in Human Dense Breast Tissue and Breast Cancer. Front. Immunol. 2017, 8, 1994. [Google Scholar] [CrossRef] [Green Version]
- Queen, M.M.; Ryan, R.E.; Holzer, R.G.; Keller-Peck, C.R.; Jorcyk, C.L. Breast cancer cells stimulate neutrophils to produce oncostatin M: Potential implications for tumor progression. Cancer Res. 2005, 65, 8896–8904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coelho, B.A.; Belo, A.V.; Andrade, S.P.; Amorim, W.C.; Uemura, G.; da Silva Filho, A.L. N-acetylglucosaminidase, myeloperoxidase and vascular endothelial growth factor serum levels in breast cancer patients. Biomed. Pharmacother. 2014, 68, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.B.; Zhu, B.; Lin, W.J.; Gao, H.Y.; Dai, H.; Zheng, L.; Shi, W.H.; Chen, W.X. Identification of prognostic significance of BIRC5 in breast cancer using integrative bioinformatics analysis. Biosci. Rep. 2020, 40, BSR20193678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poudel, N.; Okusa, M.D. Pannexins in Acute Kidney Injury. Nephron Exp. Nephrol. 2019, 143, 158–161. [Google Scholar] [CrossRef]
- Buisseret, L.; Pommey, S.; Allard, B.; Garaud, S.; Bergeron, M.; Cousineau, I.; Ameye, L.; Bareche, Y.; Paesmans, M.; Crown, J.P.A.; et al. Clinical significance of CD73 in triple-negative breast cancer: Multiplex analysis of a phase III clinical trial. Ann. Oncol. 2018, 29, 1056–1062. [Google Scholar] [CrossRef] [Green Version]
- Allard, B.; Allard, D.; Buisseret, L.; Stagg, J. The adenosine pathway in immuno-oncology. Nat. Rev. Clin. Oncol. 2020, 17, 611–629. [Google Scholar] [CrossRef] [PubMed]
- Laird, D.W.; Penuela, S. Pannexin biology and emerging linkages to cancer. Trends Cancer 2021, 7, 1119–1131. [Google Scholar] [CrossRef]
| No. | Age | ER | PR | HER2 | Ki-67 | Subtype | WHO Grade | Stage | Stromal TILs% | Bulk RNA-Seq | PAM50 | Barcode | IHC | IF | TAN RNA-seq | Paired PBN RNA-seq | ATP/ADO Assay |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PT01 | 50 | Neg | Neg | Neg | 20% | TNBC | 3 | IIA | 5.0% | NA | Yes | ||||||
| PT02 | 37 | Neg | Neg | Neg | 70% | TNBC | 3 | IIA | 30.0% | NA | Yes | ||||||
| PT03 | 41 | Neg | Neg | Neg | 40% | TNBC | 3 | IIIC | 7.0% | NA | Yes | ||||||
| PT04 | 48 | Neg | Neg | 1+ | 50% | TNBC | 3 | IIB | 30.0% | Yes | Basal-like | TNBC08 | Yes | ||||
| PT05 | 62 | Neg | Neg | Neg | 40% | TNBC | 3 | IIA | 65.0% | Yes | Basal-like | TNBC02 | Yes | Yes | Yes | Yes | |
| PT06 | 61 | Neg | Neg | 1+ | 30% | TNBC | 3 | IIA | 10.0% | Yes | Basal-like | TNBC07 | Yes | Yes | Yes | ||
| PT07 | 44 | Neg | Neg | Neg | 70% | TNBC | 3 | IIA | 20.0% | NA | Yes | ||||||
| PT08 | 47 | Neg | Neg | Neg | 70% | TNBC | 3 | IIA | 7.0% | NA | Yes | ||||||
| PT09 | 54 | Neg | Neg | 1+ | 15% | TNBC | 2 | IIA | 3.0% | Yes | Basal-like | TNBC06 | Yes | ||||
| PT10 | 48 | Neg | Neg | Neg | 15% | TNBC | 3 | I | 60.0% | Yes | Basal-like | TNBC05 | Yes | Yes | |||
| PT11 | 59 | Neg | Neg | Neg | 30% | TNBC | 2 | I | 10.0% | Yes | Basal-like | TNBC09 | Yes | Yes | |||
| PT12 | 53 | Neg | Neg | 1+ | 40% | TNBC | 3 | IIA | 40.0% | Yes | Basal-like | TNBC03 | Yes | ||||
| PT13 | 61 | Pos | Pos | 1+ | 8% | Luminal A | 2 | IIA | 5.0% | NA | Yes | ||||||
| PT14 | 59 | Pos | Pos | 1+ | 5% | Luminal A | 2 | IIA | 5.0% | NA | Yes | ||||||
| PT15 | 87 | Pos | Pos | 1+ | 5% | Luminal A | 2 | IIA | 5.0% | NA | Yes | ||||||
| PT16 | 61 | Pos | Pos | 1+ | 10% | Luminal A | 3 | IIA | 5.0% | Yes | Luminal-A | LUM03 | |||||
| PT17 | 47 | Pos | Pos | 1+ | 10% | Luminal A | 3 | IIB | 4.0% | NA | Yes | ||||||
| PT18 | 51 | Pos | Pos | 1+ | 15% | Luminal A | 2 | IIA | 10.0% | Yes | Luminal-A | LUM01 | |||||
| PT19 | 66 | Pos | Pos | 1+ | 5% | Luminal A | 2 | I | 5.0% | NA | Yes | ||||||
| PT20 | 61 | Pos | Pos | Neg | 5% | Luminal A | 2 | IIA | 7.0% | NA | Yes | ||||||
| PT21 | 73 | Pos | Pos | 1+ | 15% | Luminal A | 3 | IIA | 5.0% | Yes | Luminal-A | LUM02 | |||||
| PT22 | 61 | Pos | Pos | Neg | 5% | Luminal A | 2 | IIA | 2.0% | NA | Yes | ||||||
| PT23 | 76 | Pos | Pos | Neg | 10% | Luminal A | 3 | IIA | 2.0% | NA | Yes | ||||||
| PT24 | 52 | Pos | Pos | Neg | 10% | Luminal A | 2 | IIA | 5.0% | NA | Yes | ||||||
| PT25 | 55 | Neg | Neg | Neg | 40% | TNBC | 2 | IIA | 55.0% | Yes | Basal-like | TNBC04 | Yes | Yes | Yes | Yes | |
| PT26 | 35 | Neg | Neg | 1+ | 10% | TNBC | 2 | IIB | 20.0% | Yes | Basal-like | TNBC10 | Yes | Yes | Yes | ||
| PT27 | 52 | Neg | Neg | Neg | 15% | TNBC | 2 | IIA | 12.0% | Yes | Basal-like | TNBC01 | Yes | Yes | Yes | Yes | |
| PT28 | 45 | Neg | Neg | Neg | 70% | TNBC | 3 | IIB | 25.0% | Yes | Basal-like | TNBC11 | |||||
| PT29 | 51 | Neg | Neg | Neg | 65% | TNBC | 3 | I | 45.0% | Yes | Basal-like | TNBC12 | |||||
| PT30 | 28 | Neg | Neg | Neg | 60% | TNBC | 3 | IIA | 50.0% | Yes | Basal-like | TNBC13 | |||||
| PT31 | 60 | Neg | Neg | Neg | 75% | TNBC | 3 | IIB | 30.0% | Yes | Basal-like | TNBC14 | |||||
| PT32 | 71 | Neg | Neg | Neg | 45% | TNBC | 3 | IIB | 10.0% | Yes | Basal-like | TNBC15 | |||||
| PT33 | 43 | Neg | Neg | Neg | 50% | TNBC | 2 | IIA | 7.0% | Yes | Basal-like | TNBC16 |
| No. | Age | Subtypes | Ki-67 | WHO Grade | Stage | Stromal TILs% | Bulk RNA-Seq | PAM50 | Barcode | IHC | IHC-PANX1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PT05 | 62 | TNBC | 40% | 3 | IIA | 65.0% | Yes | Basal-like | TNBC02 | Yes | High |
| PT25 | 55 | TNBC | 40% | 2 | IIA | 55.0% | Yes | Basal-like | TNBC04 | Yes | High |
| PT27 | 52 | TNBC | 15% | 2 | IIA | 12.0% | Yes | Basal-like | TNBC01 | Yes | High |
| PT09 | 54 | TNBC | 15% | 2 | IIA | 3.0% | Yes | Basal-like | TNBC06 | Yes | Low |
| PT12 | 53 | TNBC | 40% | 3 | IIA | 40.0% | Yes | Basal-like | TNBC03 | Yes | Low |
| PT26 | 35 | TNBC | 10% | 2 | IIB | 20.0% | Yes | Basal-like | TNBC10 | Yes | Low |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Li, B.; Jia, F.; Li, J.; Huang, H.; Ni, C.; Xia, W. High PANX1 Expression Leads to Neutrophil Recruitment and the Formation of a High Adenosine Immunosuppressive Tumor Microenvironment in Basal-like Breast Cancer. Cancers 2022, 14, 3369. https://doi.org/10.3390/cancers14143369
Chen W, Li B, Jia F, Li J, Huang H, Ni C, Xia W. High PANX1 Expression Leads to Neutrophil Recruitment and the Formation of a High Adenosine Immunosuppressive Tumor Microenvironment in Basal-like Breast Cancer. Cancers. 2022; 14(14):3369. https://doi.org/10.3390/cancers14143369
Chicago/Turabian StyleChen, Wuzhen, Baizhou Li, Fang Jia, Jiaxin Li, Huanhuan Huang, Chao Ni, and Wenjie Xia. 2022. "High PANX1 Expression Leads to Neutrophil Recruitment and the Formation of a High Adenosine Immunosuppressive Tumor Microenvironment in Basal-like Breast Cancer" Cancers 14, no. 14: 3369. https://doi.org/10.3390/cancers14143369
APA StyleChen, W., Li, B., Jia, F., Li, J., Huang, H., Ni, C., & Xia, W. (2022). High PANX1 Expression Leads to Neutrophil Recruitment and the Formation of a High Adenosine Immunosuppressive Tumor Microenvironment in Basal-like Breast Cancer. Cancers, 14(14), 3369. https://doi.org/10.3390/cancers14143369






