Identification of Endoplasmic Reticulum Stress-Related Subtypes, Infiltration Analysis of Tumor Microenvironment, and Construction of a Prognostic Model in Colorectal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
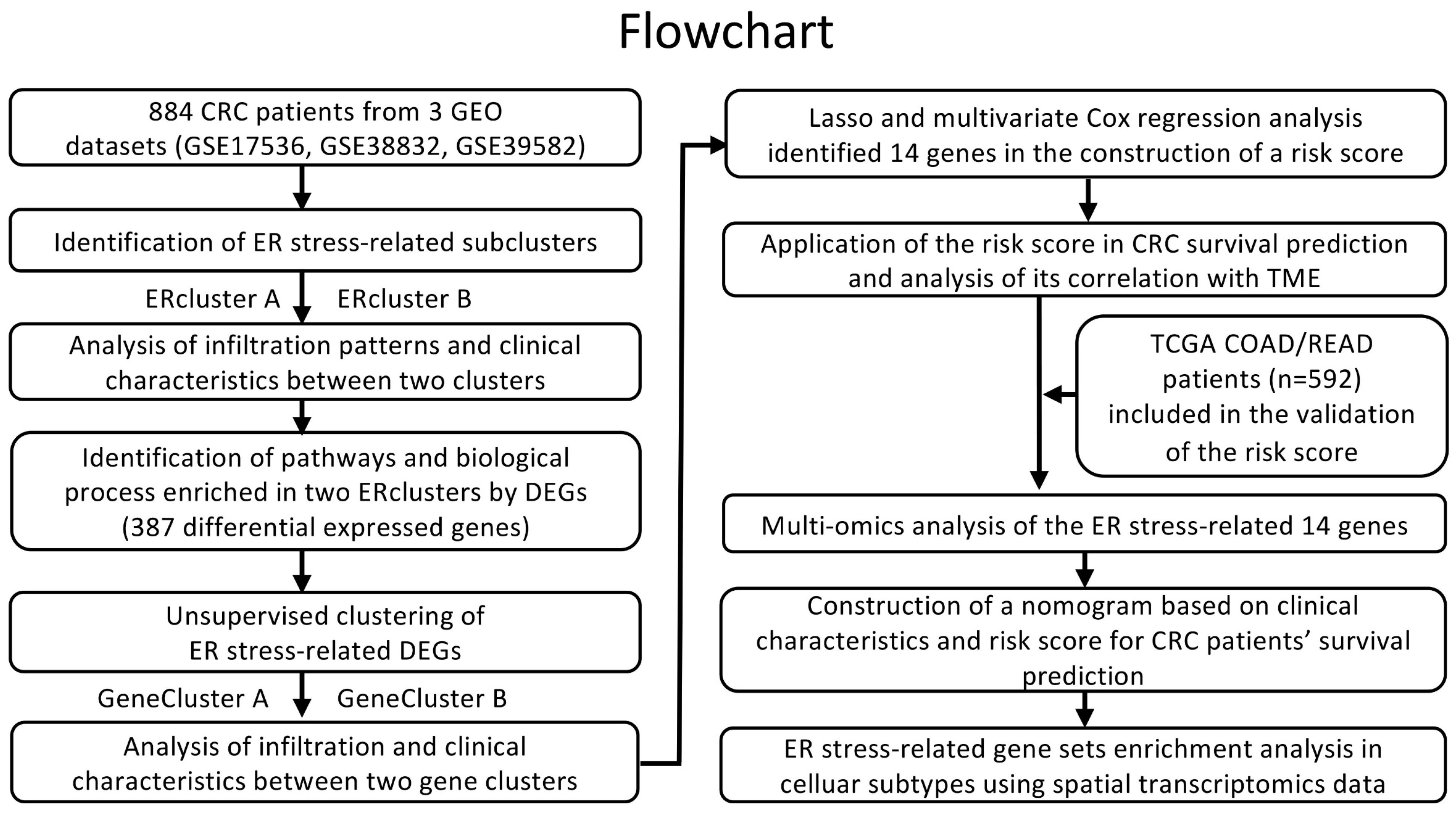
2. Materials and Methods
2.1. Data and Resources
2.2. Unsupervised Clustering Analysis of ER Stress-Related Genes
2.3. Estimation of Lymphocyte Infiltration in the Bulk Sequencing Data
2.4. Biological Functions and Pathways Enrichment Analysis
2.5. Construction of ER Stress-Related Risk Score
2.6. Mutation, Copy number Variation, and DNA Methylation Analysis
2.7. Real-Time Quantitative PCR Analysis
2.8. Construction of a Nomogram in Predicting Survival
2.9. Spatial Transcriptomics Data Analysis
2.10. Statistical Analysis
3. Results
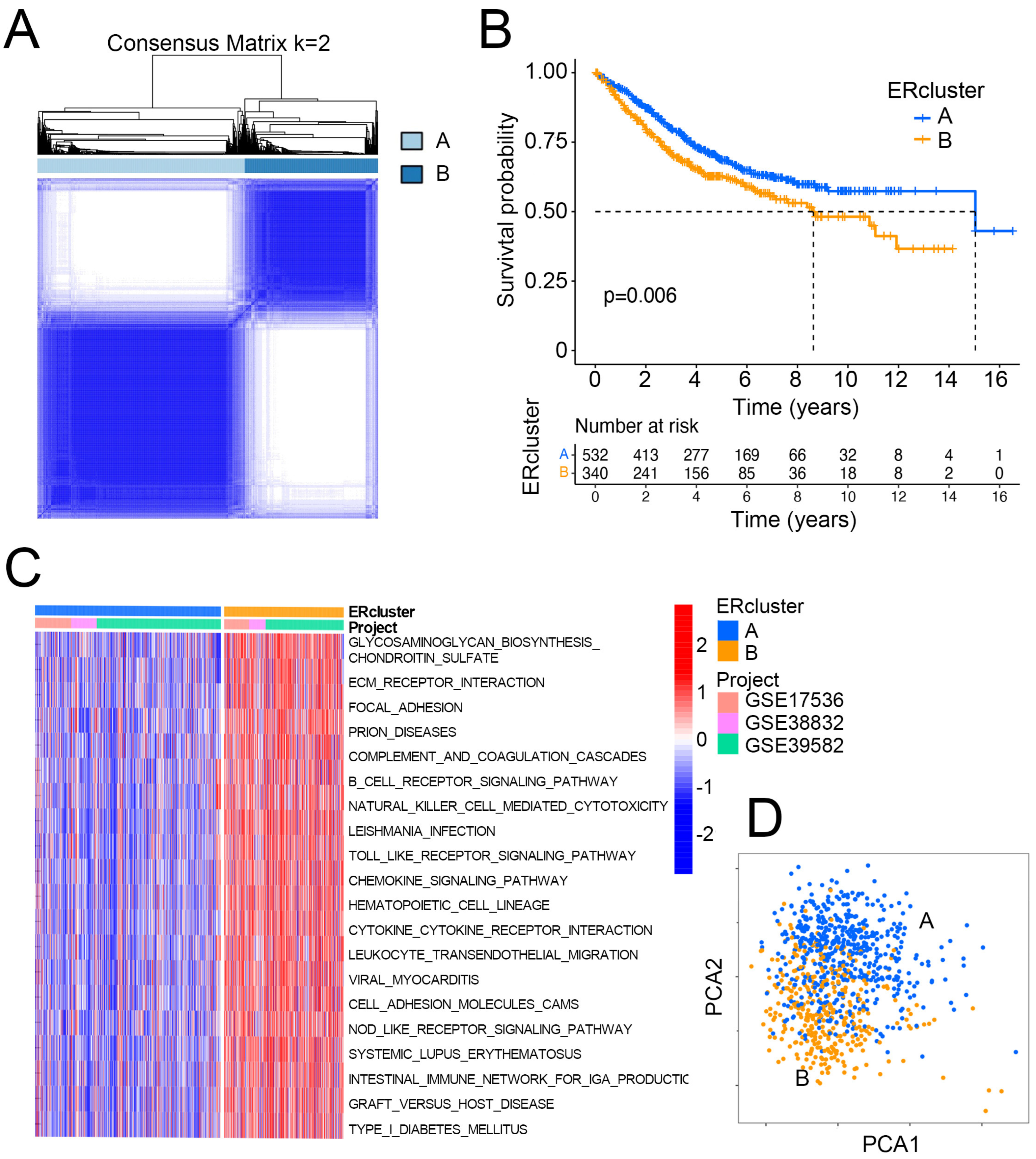
3.1. Identification of ER Stress Related Sub-Clusters in CRC
3.2. ERcluster A and B Had Distinct Cell-Infiltrating Patterns in the TME
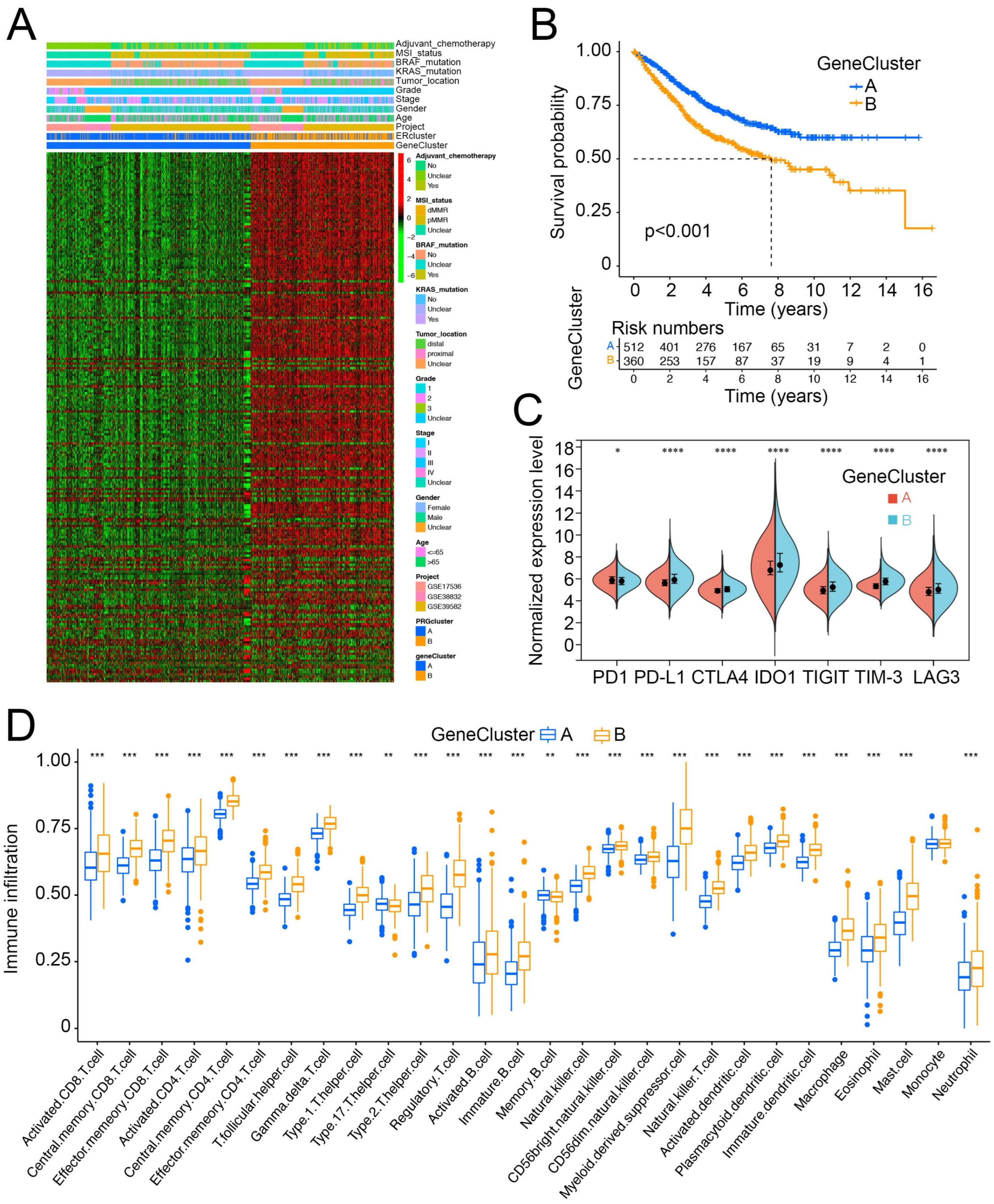
3.3. ER Stress-Related Gene Clusters Had Distinct Clinical Characteristics and Cell-Infiltration Patterns in the TME
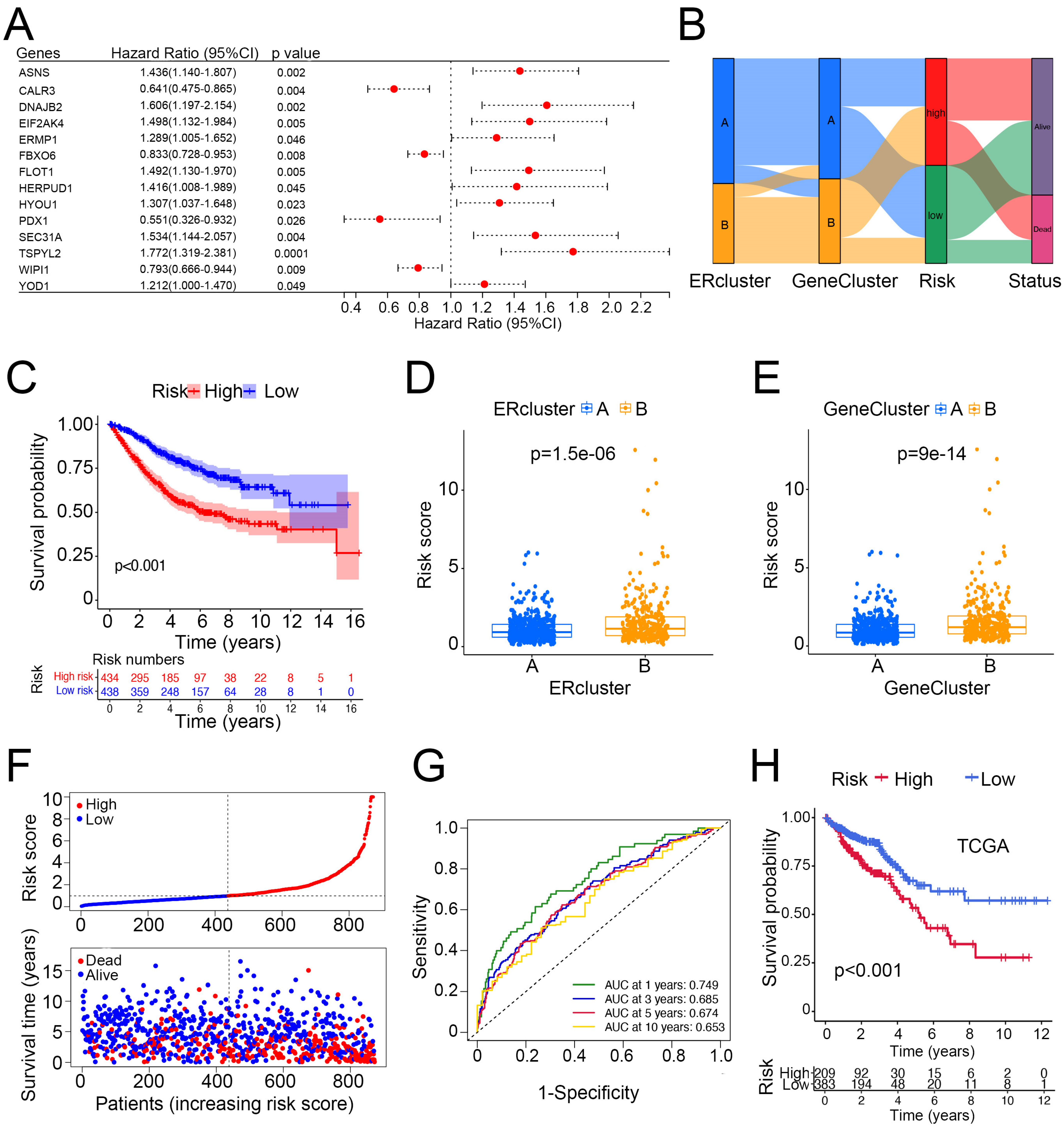
3.4. Construction and Validation of Risk Score and Its Clinical Significance
3.5. Mutation, CNV, Transcription, and Methylation Level of the 14 Risk Score-Building Genes
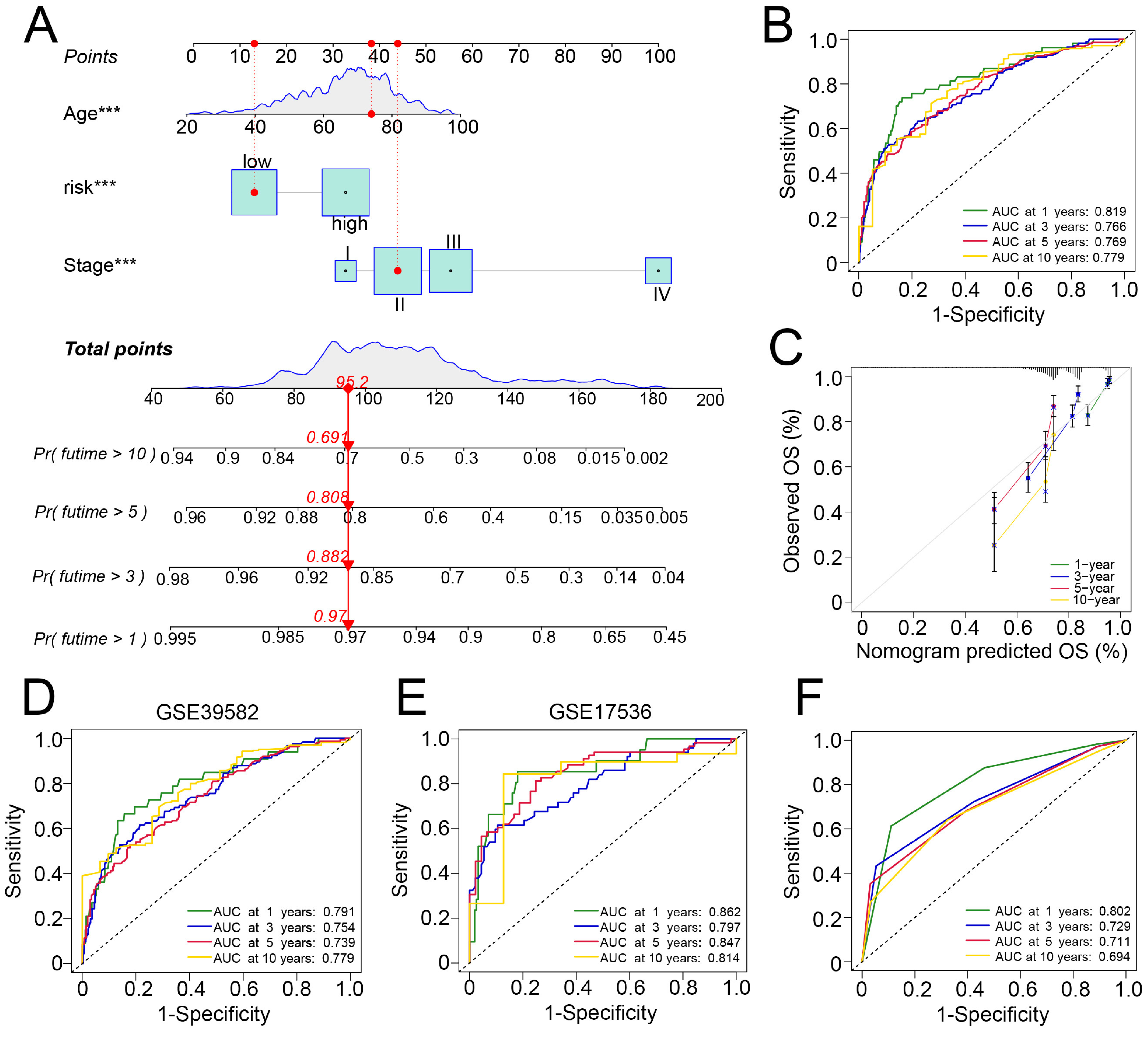
3.6. Development and Verification of a Nomogram in Predicting Survival of Patients with CRC
3.7. Insights into ER Stress-Related Gene Signatures at the ST Level
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef]
- Ganesh, K.; Stadler, Z.K.; Cercek, A.; Mendelsohn, R.B.; Shia, J.; Segal, N.H.; Diaz, L.A. Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, L.; Rodríguez-Abreu, D.; Gadgeel, S.; Esteban, E.; Felip, E.; De Angelis, F.; Domine, M.; Clingan, P.; Hochmair, M.J.; Powell, S.F.; et al. Pembrolizumab plus Chemotherapy in Metastatic Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 2078–2092. [Google Scholar] [CrossRef] [PubMed]
- Hodi, F.S.; O′Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.R.; Chlon, L.; Pharoah, P.D.; Markowetz, F.; Caldas, C. Patterns of Immune Infiltration in Breast Cancer and Their Clinical Implications: A Gene-Expression-Based Retrospective Study. PLoS Med. 2016, 13, e1002194. [Google Scholar] [CrossRef]
- Wei, S.C.; Duffy, C.R.; Allison, J.P. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov. 2018, 8, 1069–1086. [Google Scholar] [CrossRef] [Green Version]
- Hetz, C.; Zhang, K.; Kaufman, R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438. [Google Scholar] [CrossRef]
- Hetz, C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012, 13, 89–102. [Google Scholar] [CrossRef]
- Taylor, R.C.; Dillin, A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell 2013, 153, 1435–1447. [Google Scholar] [CrossRef] [Green Version]
- Hetz, C.; Saxena, S. ER stress and the unfolded protein response in neurodegeneration. Nat. Rev. Neurol. 2017, 13, 477–491. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, U.; Cao, Q.; Yilmaz, E.; Lee, A.H.; Iwakoshi, N.N.; Ozdelen, E.; Tuncman, G.; Görgün, C.; Glimcher, L.H.; Hotamisligil, G.S. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 2004, 306, 457–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Cubillos-Ruiz, J.R. Endoplasmic reticulum stress signals in the tumour and its microenvironment. Nat. Rev. Cancer 2021, 21, 71–88. [Google Scholar] [CrossRef]
- Chevet, E.; Hetz, C.; Samali, A. Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer Discov. 2015, 5, 586–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grootjans, J.; Kaser, A.; Kaufman, R.J.; Blumberg, R.S. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016, 16, 469–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniel; Mellman, I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Adams, N.M.; Xu, Y.; Cao, J.; Allan, D.S.J.; Carlyle, J.R.; Chen, X.; Sun, J.C.; Glimcher, L.H. The IRE1 endoplasmic reticulum stress sensor activates natural killer cell immunity in part by regulating c-Myc. Nat. Immunol. 2019, 20, 865–878. [Google Scholar] [CrossRef]
- Osorio, F.; Tavernier, S.J.; Hoffmann, E.; Saeys, Y.; Martens, L.; Vetters, J.; Delrue, I.; De Rycke, R.; Parthoens, E.; Pouliot, P.; et al. The unfolded-protein-response sensor IRE-1α regulates the function of CD8α+ dendritic cells. Nat. Immunol. 2014, 15, 248–257. [Google Scholar] [CrossRef]
- Cubillos-Ruiz, J.R.; Silberman, P.C.; Rutkowski, M.R.; Chopra, S.; Perales-Puchalt, A.; Song, M.; Zhang, S.; Bettigole, S.E.; Gupta, D.; Holcomb, K.; et al. ER Stress Sensor XBP1 Controls Anti-tumor Immunity by Disrupting Dendritic Cell Homeostasis. Cell 2015, 161, 1527–1538. [Google Scholar] [CrossRef] [Green Version]
- Condamine, T.; Dominguez, G.A.; Youn, J.I.; Kossenkov, A.V.; Mony, S.; Alicea-Torres, K.; Tcyganov, E.; Hashimoto, A.; Nefedova, Y.; Lin, C.; et al. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci. Immunol. 2016, 1, aaf8943. [Google Scholar] [CrossRef] [Green Version]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautès-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [Green Version]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Li, Y.; Ge, D.; Lu, C. The SMART App: An interactive web application for comprehensive DNA methylation analysis and visualization. Epigenetics Chromatin 2019, 12, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iasonos, A.; Schrag, D.; Raj, G.V.; Panageas, K.S. How to build and interpret a nomogram for cancer prognosis. J. Clin. Oncol. 2008, 26, 1364–1370. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yang, S.; Ma, J.; Chen, Z.; Song, G.; Rao, D.; Cheng, Y.; Huang, S.; Liu, Y.; Jiang, S.; et al. Spatiotemporal Immune Landscape of Colorectal Cancer Liver Metastasis at Single-Cell Level. Cancer Discov. 2022, 12, 134–153. [Google Scholar] [CrossRef] [PubMed]
- Racle, J.; De Jonge, K.; Baumgaertner, P.; Speiser, D.E.; Gfeller, D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife 2017, 6, e26476. [Google Scholar] [CrossRef]
- Richards, K.E.; Zeleniak, A.E.; Fishel, M.L.; Wu, J.; Littlepage, L.E.; Hill, R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene 2017, 36, 1770–1778. [Google Scholar] [CrossRef] [Green Version]
- Tsujino, T.; Seshimo, I.; Yamamoto, H.; Ngan, C.Y.; Ezumi, K.; Takemasa, I.; Ikeda, M.; Sekimoto, M.; Matsuura, N.; Monden, M. Stromal myofibroblasts predict disease recurrence for colorectal cancer. Clin. Cancer Res. 2007, 13, 2082–2090. [Google Scholar] [CrossRef] [Green Version]
- Al Hrout, A.A.; Cervantes-Gracia, K.; Chahwan, R.; Amin, A. Modelling liver cancer microenvironment using a novel 3D culture system. Sci. Rep. 2022, 12, 8003. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; De Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Ma, Y.; Hendershot, L.M. The role of the unfolded protein response in tumour development: Friend or foe? Nat. Rev. Cancer 2004, 4, 966–977. [Google Scholar] [CrossRef]
- Chen, X.; Iliopoulos, D.; Zhang, Q.; Tang, Q.; Greenblatt, M.B.; Hatziapostolou, M.; Lim, E.; Tam, W.L.; Ni, M.; Chen, Y.; et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature 2014, 508, 103–107. [Google Scholar] [CrossRef]
- Romero-Ramirez, L.; Cao, H.; Nelson, D.; Hammond, E.; Lee, A.H.; Yoshida, H.; Mori, K.; Glimcher, L.H.; Denko, N.C.; Giaccia, A.J.; et al. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 2004, 64, 5943–5947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Back, S.H.; Hur, J.; Lin, Y.-H.; Gildersleeve, R.; Shan, J.; Yuan, C.L.; Krokowski, D.; Wang, S.; Hatzoglou, M.; et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 2013, 15, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Zinszner, H.; Kuroda, M.; Wang, X.; Batchvarova, N.; Lightfoot, R.T.; Remotti, H.; Stevens, J.L.; Ron, D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998, 12, 982–995. [Google Scholar] [CrossRef] [PubMed]
- Mahadevan, N.R.; Rodvold, J.; Sepulveda, H.; Rossi, S.; Drew, A.F.; Zanetti, M. Transmission of endoplasmic reticulum stress and pro-inflammation from tumor cells to myeloid cells. Proc. Natl. Acad. Sci. USA 2011, 108, 6561–6566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, E.; Sierra, R.A.; Trillo-Tinoco, J.; Cao, Y.; Innamarato, P.; Payne, K.K.; De Mingo Pulido, A.; Mandula, J.; Zhang, S.; Thevenot, P.; et al. The Unfolded Protein Response Mediator PERK Governs Myeloid Cell-Driven Immunosuppression in Tumors through Inhibition of STING Signaling. Immunity 2020, 52, 668–682.e667. [Google Scholar] [CrossRef]
- Vanacker, H.; Vetters, J.; Moudombi, L.; Caux, C.; Janssens, S.; Michallet, M.C. Emerging Role of the Unfolded Protein Response in Tumor Immunosurveillance. Trends Cancer 2017, 3, 491–505. [Google Scholar] [CrossRef]
- Li, X.; Wen, D.; Li, X.; Yao, C.; Chong, W.; Chen, H. Identification of an Immune Signature Predicting Prognosis Risk and Lymphocyte Infiltration in Colon Cancer. Front. Immunol. 2020, 11, 1678. [Google Scholar] [CrossRef]
- Tanaka, A.; Sakaguchi, S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017, 27, 109–118. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Zhao, Y.; Dai, Y.; Cheng, J.N.; Gong, Z.; Feng, Y.; Sun, C.; Jia, Q.; Zhu, B. Immune Landscape of Colorectal Cancer Tumor Microenvironment from Different Primary Tumor Location. Front. Immunol. 2018, 9, 1578. [Google Scholar] [CrossRef] [Green Version]
- Mezheyeuski, A.; Micke, P.; Martín-Bernabé, A.; Backman, M.; Hrynchyk, I.; Hammarström, K.; Ström, S.; Ekström, J.; Edqvist, P.-H.; Sundström, M.; et al. The Immune Landscape of Colorectal Cancer. Cancers 2021, 13, 5545. [Google Scholar] [CrossRef]
- Xia, A.; Zhang, Y.; Xu, J.; Yin, T.; Lu, X.J. T Cell Dysfunction in Cancer Immunity and Immunotherapy. Front. Immunol. 2019, 10, 1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gajewski, T.F.; Woo, S.R.; Zha, Y.; Spaapen, R.; Zheng, Y.; Corrales, L.; Spranger, S. Cancer immunotherapy strategies based on overcoming barriers within the tumor microenvironment. Curr. Opin. Immunol. 2013, 25, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.; Farrukh, A.; Murali, C.; Soleimani, A.; Praz, F.; Graziani, G.; Brim, H.; Ashktorab, H. Saffron and Its Major Ingredients’ Effect on Colon Cancer Cells with Mismatch Repair Deficiency and Microsatellite Instability. Molecules 2021, 26, 3855. [Google Scholar] [CrossRef] [PubMed]
- Hurst, K.E.; Lawrence, K.A.; Essman, M.T.; Walton, Z.J.; Leddy, L.R.; Thaxton, J.E. Endoplasmic Reticulum Stress Contributes to Mitochondrial Exhaustion of CD8(+) T Cells. Cancer Immunol. Res. 2019, 7, 476–486. [Google Scholar] [CrossRef]
- Deng, L.; Yao, P.; Li, L.; Ji, F.; Zhao, S.; Xu, C.; Lan, X.; Jiang, P. p53-mediated control of aspartate-asparagine homeostasis dictates LKB1 activity and modulates cell survival. Nat. Commun. 2020, 11, 1755. [Google Scholar] [CrossRef]
- Schmidt, S.; Gay, D.; Uthe, F.W.; Denk, S.; Paauwe, M.; Matthes, N.; Diefenbacher, M.E.; Bryson, S.; Warrander, F.C.; Erhard, F.; et al. A MYC–GCN2–eIF2α negative feedback loop limits protein synthesis to prevent MYC-dependent apoptosis in colorectal cancer. Nat. Cell Biol. 2019, 21, 1413–1424. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Wang, S.; Tong, H.; Tang, Z.; Wang, J.; Zhang, Y.; Ou, J.; Quan, Z. Long Non-coding RNA FIRRE Acts as a miR-520a-3p Sponge to Promote Gallbladder Cancer Progression via Mediating YOD1 Expression. Front. Genet. 2021, 12, 674653. [Google Scholar] [CrossRef]
- Biffi, G.; Tuveson, D.A. Diversity and Biology of Cancer-Associated Fibroblasts. Physiol. Rev. 2021, 101, 147–176. [Google Scholar] [CrossRef]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer 2019, 18, 91. [Google Scholar] [CrossRef] [Green Version]
- Turley, S.J.; Cremasco, V.; Astarita, J.L. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat. Rev. Immunol. 2015, 15, 669–682. [Google Scholar] [CrossRef]



| Gene | Coef |
|---|---|
| ASNS | 0.688343247 |
| CALR3 | −0.572927078 |
| DNAJB2 | 0.455677169 |
| EIF2AK4 | 0.831306602 |
| ERMP1 | 0.764514094 |
| FBXO6 | −0.388996026 |
| FLOT1 | 0.463952998 |
| HERPUD1 | 0.803594073 |
| HYOU1 | 0.233149469 |
| PDX1 | −0.60035933 |
| SEC31A | 0.493525152 |
| TSPYL2 | 0.646208663 |
| WIPI1 | −0.244404952 |
| YOD1 | 0.292412602 |
| Characteristics | ERcluster A (n = 538) | ERcluster B (n = 346) | Total (n = 884) | p Value |
|---|---|---|---|---|
| Age | 0.84 | |||
| <65 | 183 (34.01%) | 111 (32.08%) | 294 (33.26%) | |
| ≥65 | 281 (52.23%) | 186 (53.76%) | 467 (52.83%) | |
| Unclear | 74 (13.76%) | 49 (14.16%) | 123 (13.91%) | |
| Gender | 0.63 | |||
| Female | 203 (37.73%) | 141 (40.75%) | 344 (38.91%) | |
| Male | 261 (48.51%) | 157 (45.38%) | 418 (47.29%) | |
| Unclear | 74 (13.76%) | 48 (13.87%) | 122 (13.80%) | |
| Stage | 0.17 | |||
| I | 61 (11.34%) | 23 (6.65%) | 84 (9.50%) | |
| II | 221 (41.08%) | 142 (41.04%) | 363 (41.06%) | |
| III | 176 (32.71%) | 130 (37.57%) | 306 (34.62%) | |
| IV | 79 (14.68%) | 50 (14.45%) | 129 (14.59%) | |
| Unclear | 1 (0.19%) | 1 (0.29%) | 2 (0.23%) | |
| Grade | 0.18 | |||
| 1 | 10 (1.86%) | 6 (1.73%) | 16 (1.81%) | |
| 2 | 84 (15.61%) | 50 (14.45%) | 134 (15.16%) | |
| 3 | 11 (2.04%) | 16 (4.62%) | 27 (3.05%) | |
| Unclear | 433 (80.49%) | 274 (79.20%) | 707 (79.98%) | |
| Tumor_location | 6.20 × 10−10 | |||
| Distal | 253 (47.03%) | 98 (28.32%) | 351 (39.71%) | |
| Proximal | 105 (19.52%) | 127 (36.71%) | 232 (26.24%) | |
| Unclear | 180 (33.45%) | 121 (34.97%) | 301 (34.05%) | |
| KRAS_mutation | 0.13 | |||
| No | 188 (34.94%) | 140 (40.46%) | 328 (37.10%) | |
| Yes | 143 (26.58%) | 74 (21.39%) | 217 (24.55%) | |
| Unclear | 207 (38.48%) | 132 (38.15%) | 339 (38.35%) | |
| BRAF_mutation | 3.20 × 10−14 | |||
| No | 307 (57.06%) | 154 (44.51%) | 461 (52.15%) | |
| Yes | 5 (0.93%) | 46 (13.29%) | 51 (5.77%) | |
| Unclear | 226 (42.01%) | 146 (42.20%) | 372 (42.08%) | |
| MSI_status | 2.20 × 10−17 | |||
| dMMR | 14 (2.60%) | 63 (18.20%) | 77 (8.71%) | |
| pMMR | 322 (59.85%) | 137 (39.60%) | 459 (51.92%) | |
| Unclear | 202 (37.55%) | 146 (42.20%) | 348 (39.37%) | |
| Adjuvant_chemotherapy | 0.38 | |||
| No | 194 (36.06%) | 132 (38.15%) | 326 (36.88%) | |
| Yes | 155 (28.81%) | 85 (24.57%) | 240 (27.15%) | |
| Unclear | 189 (35.13%) | 129 (37.28%) | 318 (35.97%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Yin, X.; Jiang, G.; Li, Y.; Jiang, Z.; Qiang, L.; Chen, N.; Fan, Y.; Shen, C.; Dai, L.; et al. Identification of Endoplasmic Reticulum Stress-Related Subtypes, Infiltration Analysis of Tumor Microenvironment, and Construction of a Prognostic Model in Colorectal Cancer. Cancers 2022, 14, 3326. https://doi.org/10.3390/cancers14143326
Liu B, Yin X, Jiang G, Li Y, Jiang Z, Qiang L, Chen N, Fan Y, Shen C, Dai L, et al. Identification of Endoplasmic Reticulum Stress-Related Subtypes, Infiltration Analysis of Tumor Microenvironment, and Construction of a Prognostic Model in Colorectal Cancer. Cancers. 2022; 14(14):3326. https://doi.org/10.3390/cancers14143326
Chicago/Turabian StyleLiu, Baike, Xiaonan Yin, Guangfu Jiang, Yang Li, Zhiyuan Jiang, Liming Qiang, Na Chen, Yating Fan, Chaoyong Shen, Lei Dai, and et al. 2022. "Identification of Endoplasmic Reticulum Stress-Related Subtypes, Infiltration Analysis of Tumor Microenvironment, and Construction of a Prognostic Model in Colorectal Cancer" Cancers 14, no. 14: 3326. https://doi.org/10.3390/cancers14143326
APA StyleLiu, B., Yin, X., Jiang, G., Li, Y., Jiang, Z., Qiang, L., Chen, N., Fan, Y., Shen, C., Dai, L., Yin, Y., & Zhang, B. (2022). Identification of Endoplasmic Reticulum Stress-Related Subtypes, Infiltration Analysis of Tumor Microenvironment, and Construction of a Prognostic Model in Colorectal Cancer. Cancers, 14(14), 3326. https://doi.org/10.3390/cancers14143326






