An Update on the Pathology and Molecular Features of Hodgkin Lymphoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Classic HL
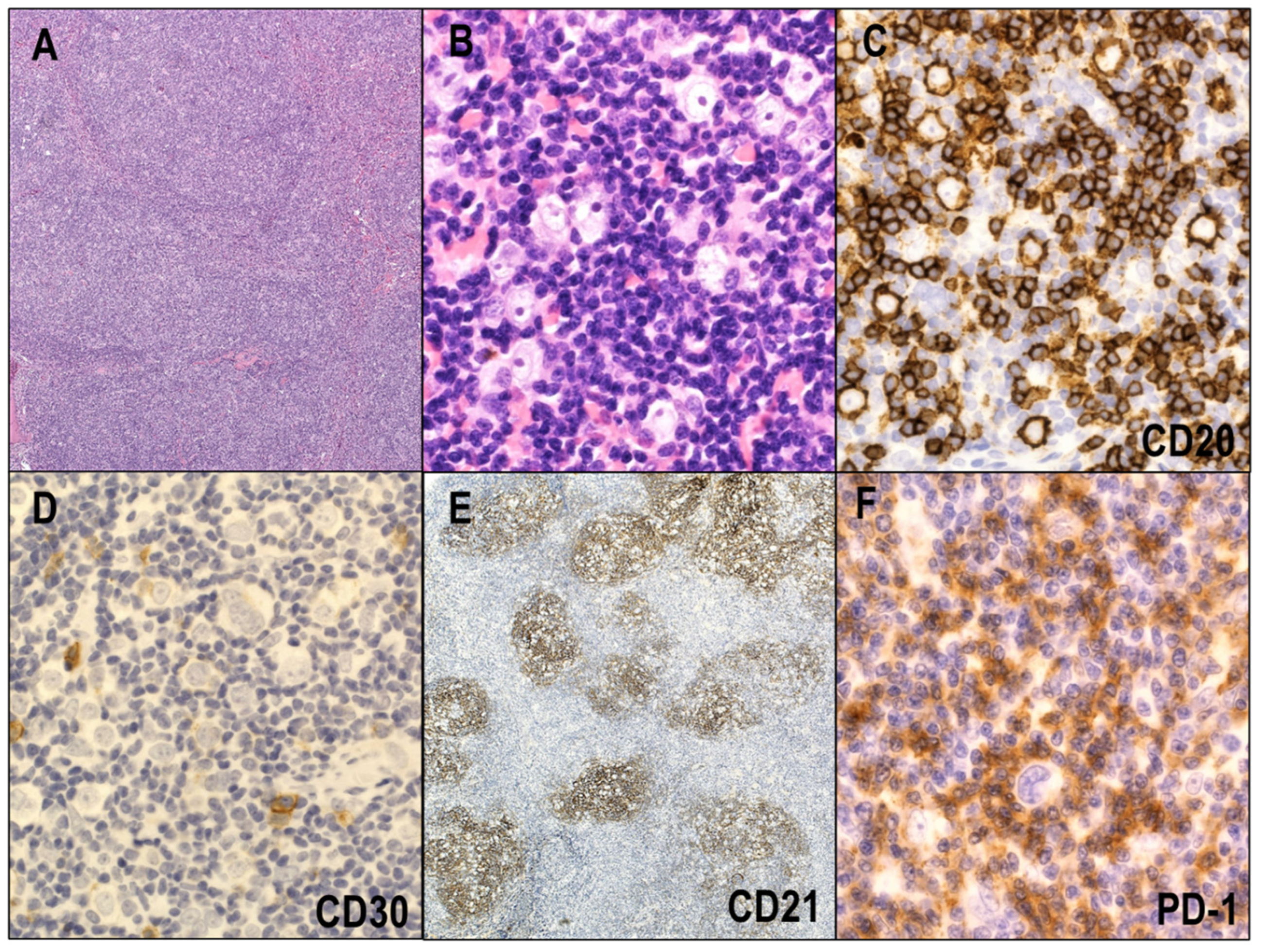
2.1. Clinical and Pathological Features
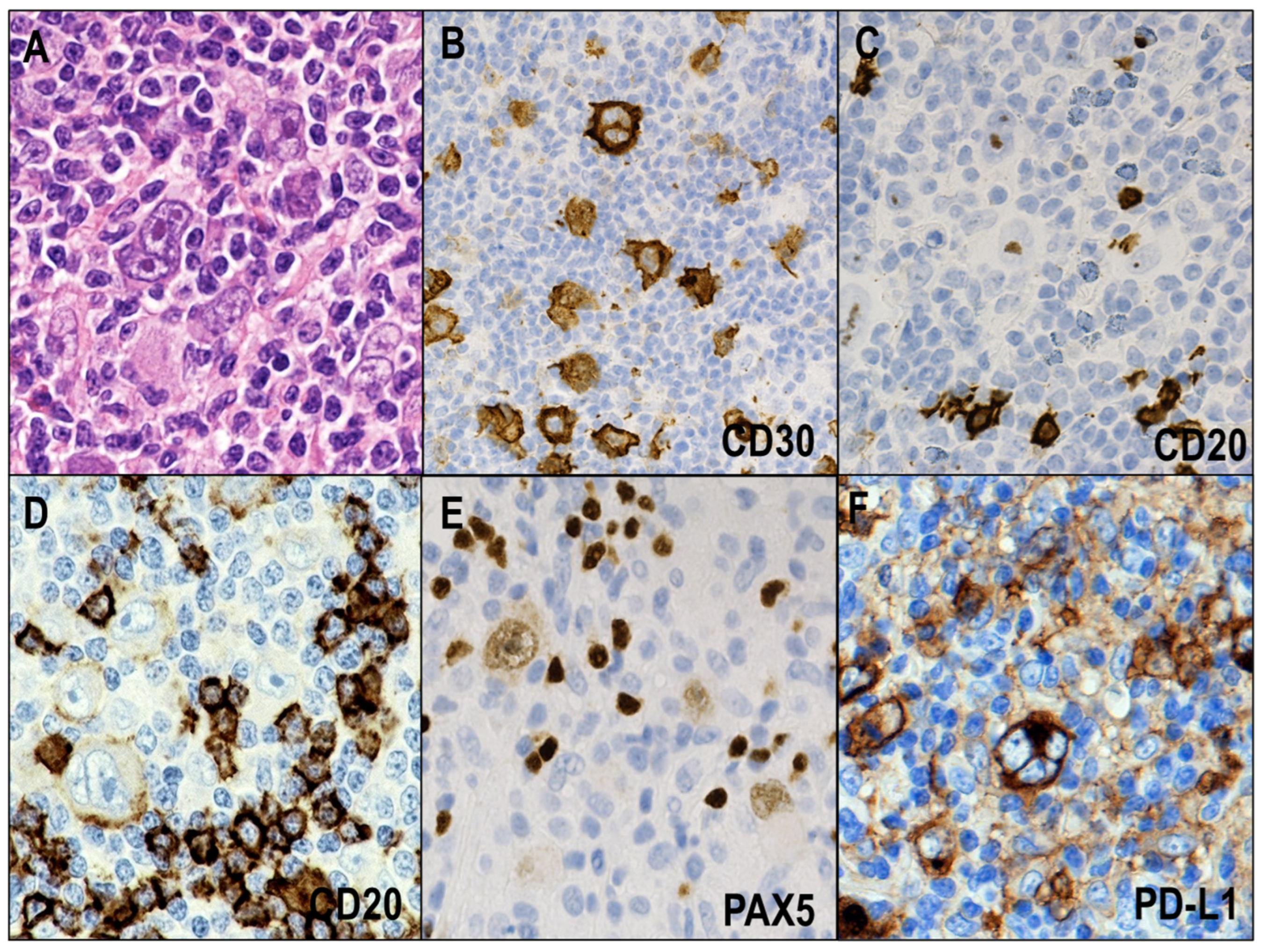
2.1.1. NSCHL
2.1.2. MCCHL
2.1.3. LRCHL
2.1.4. LDCHL
2.2. Key Pathways and Genetic Lesions in HRS Cells
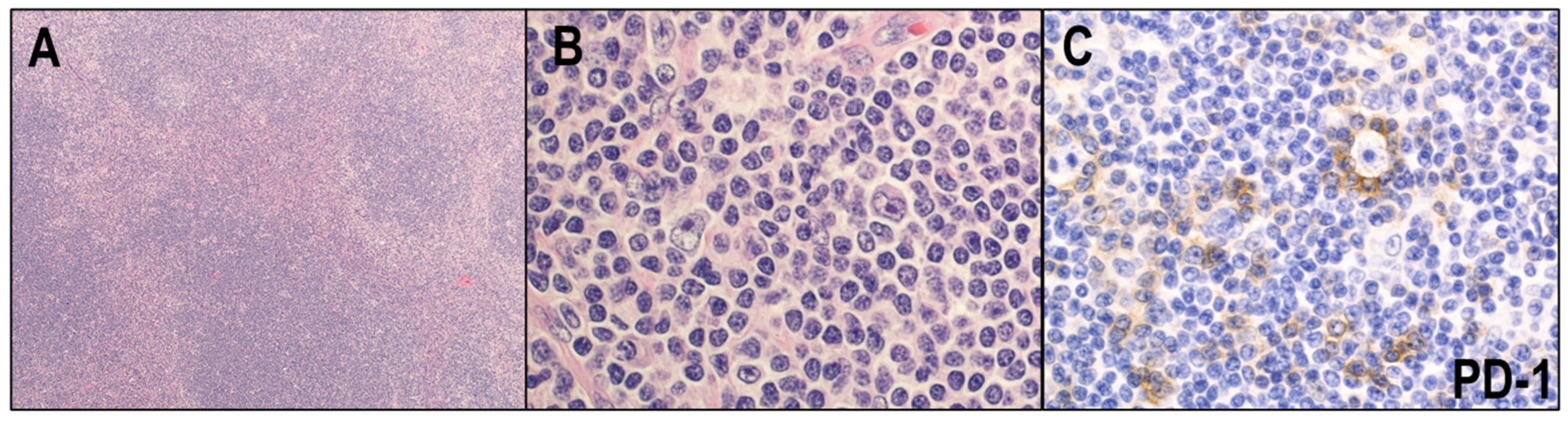
2.3. Immune Evasion Mechanisms in CHL
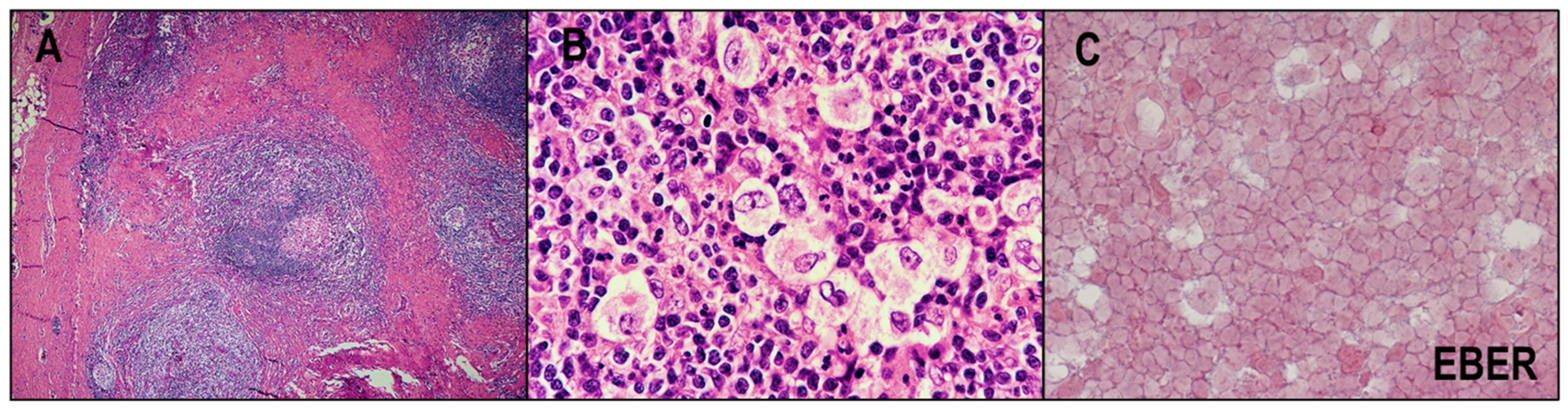
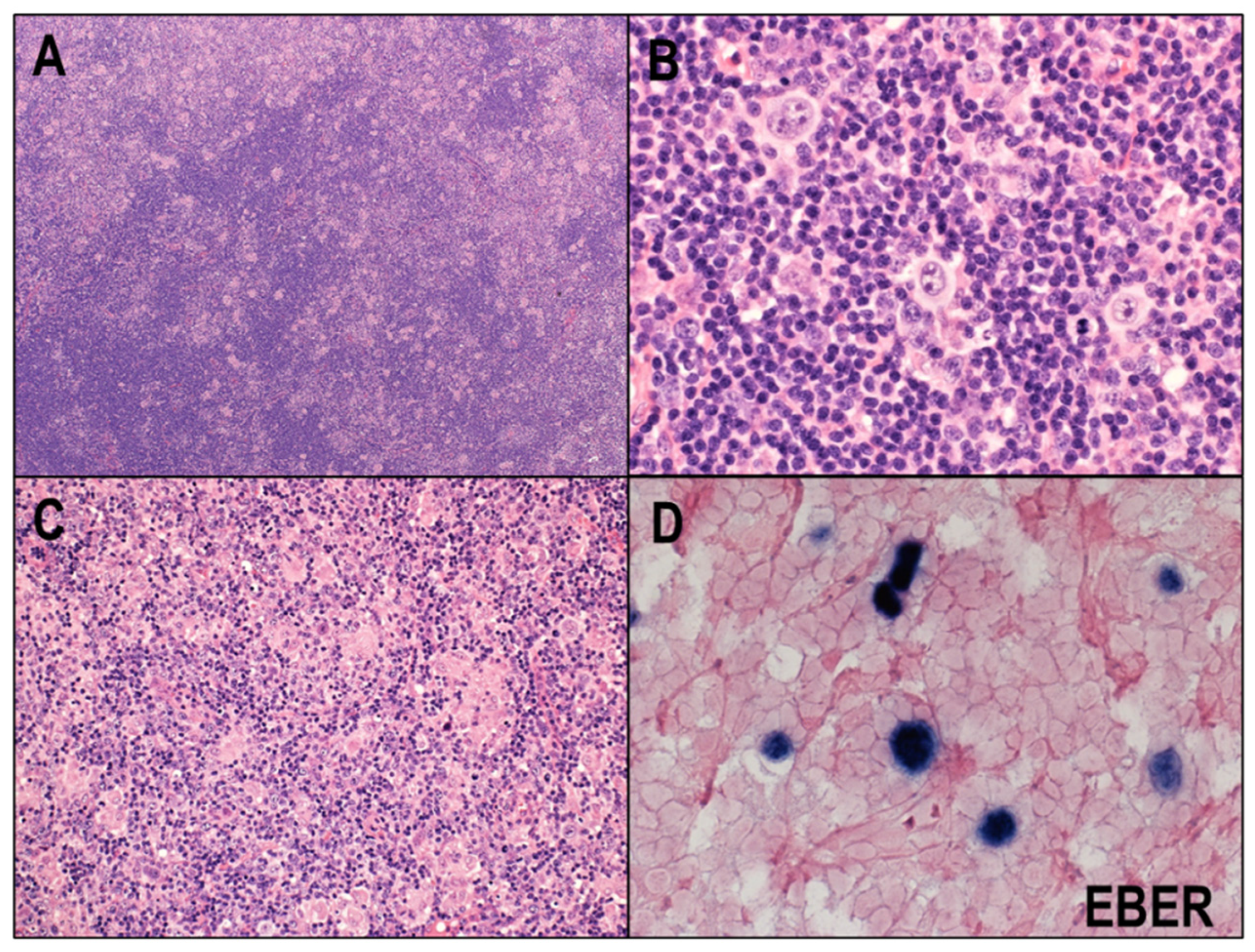
2.4. EBV and CHL
3. NLPHL
3.1. Clinical and Pathological Features
3.2. Key Pathways and Genetic Lesions in LP Cells
3.3. Relationship between NLPHL and THRLBCL
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Foss, H.D.; Reusch, R.; Demel, G.; Lenz, G.; Anagnostopoulos, I.; Hummel, M.; Stein, H. Frequent expression of the B-cell-specific activator protein in Reed-Sternberg cells of classical Hodgkin’s disease provides further evidence for its B-cell origin. Blood 1999, 94, 3108–3113. [Google Scholar] [CrossRef] [PubMed]
- Schwering, I.; Brauninger, A.; Klein, U.; Jungnickel, B.; Tinguely, M.; Diehl, V.; Hansmann, M.L.; Dalla-Favera, R.; Rajewsky, K.; Kuppers, R. Loss of the B-lineage-specific gene expression program in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 2003, 101, 1505–1512. [Google Scholar] [CrossRef]
- Tiacci, E.; Doring, C.; Brune, V.; van Noesel, C.J.; Klapper, W.; Mechtersheimer, G.; Falini, B.; Kuppers, R.; Hansmann, M.L. Analyzing primary Hodgkin and Reed-Sternberg cells to capture the molecular and cellular pathogenesis of classical Hodgkin lymphoma. Blood 2012, 120, 4609–4620. [Google Scholar] [CrossRef] [PubMed]
- Kanzler, H.; Kuppers, R.; Hansmann, M.L.; Rajewsky, K. Hodgkin and Reed-Sternberg cells in Hodgkin’s disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J. Exp. Med. 1996, 184, 1495–1505. [Google Scholar] [CrossRef] [PubMed]
- Kuppers, R.; Rajewsky, K.; Zhao, M.; Simons, G.; Laumann, R.; Fischer, R.; Hansmann, M.L. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc. Natl. Acad. Sci. USA 1994, 91, 10962–10966. [Google Scholar] [CrossRef] [PubMed]
- Marafioti, T.; Hummel, M.; Foss, H.D.; Laumen, H.; Korbjuhn, P.; Anagnostopoulos, I.; Lammert, H.; Demel, G.; Theil, J.; Wirth, T.; et al. Hodgkin and reed-sternberg cells represent an expansion of a single clone originating from a germinal center B-cell with functional immunoglobulin gene rearrangements but defective immunoglobulin transcription. Blood 2000, 95, 1443–1450. [Google Scholar] [CrossRef] [PubMed]
- Diehl, V.; Thomas, R.K.; Re, D. Part II: Hodgkin’s lymphoma—Diagnosis and treatment. Lancet Oncol. 2004, 5, 19–26. [Google Scholar] [CrossRef]
- Swerdlow, S.; Campo, E.; Harris, N.; Jaffe, E.; Pileri, S.; Stein, H.; Thiele, J. (Eds.) WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th ed.; International Agency for Research on Cancer: Lyon, France, 2017. [Google Scholar]
- Chang, K.C.; Huang, G.C.; Jones, D.; Tsao, C.J.; Lee, J.Y.; Su, I.J. Distribution and prognosis of WHO lymphoma subtypes in Taiwan reveals a low incidence of germinal-center derived tumors. Leuk. Lymphoma 2004, 45, 1375–1384. [Google Scholar] [CrossRef]
- Makita, S.; Maruyama, D.; Maeshima, A.M.; Taniguchi, H.; Miyamoto, K.; Kitahara, H.; Fukuhara, S.; Munakata, W.; Kobayashi, Y.; Itami, J.; et al. Clinical features and outcomes of 139 Japanese patients with Hodgkin lymphoma. Int. J. Hematol. 2016, 104, 236–244. [Google Scholar] [CrossRef]
- Meng, J.; Chang, C.; Pan, H.; Zhu, F.; Xiao, Y.; Liu, T.; Nie, X.; Wu, G.; Zhang, L. Epidemiologic characteristics of malignant lymphoma in Hubei, China: A single-center 5-year retrospective study. Medicine 2018, 97, e12120. [Google Scholar] [CrossRef]
- Medeiros, L.J.; Greiner, T.C. Hodgkin’s disease. Cancer 1995, 75, 357–369. [Google Scholar] [CrossRef]
- Connors, J.M.; Jurczak, W.; Straus, D.J.; Ansell, S.M.; Kim, W.S.; Gallamini, A.; Younes, A.; Alekseev, S.; Illes, A.; Picardi, M.; et al. Brentuximab Vedotin with Chemotherapy for Stage III or IV Hodgkin’s Lymphoma. N. Engl. J. Med. 2018, 378, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Karube, K.; Kakimoto, Y.; Tonozuka, Y.; Ohshima, K. The expression of CD30 and its clinico-pathologic significance in peripheral T-cell lymphomas. Expert Rev. Hematol. 2021, 14, 777–787. [Google Scholar] [CrossRef]
- van der Weyden, C.A.; Pileri, S.A.; Feldman, A.L.; Whisstock, J.; Prince, H.M. Understanding CD30 biology and therapeutic targeting: A historical perspective providing insight into future directions. Blood Cancer J. 2017, 7, e603. [Google Scholar] [CrossRef] [PubMed]
- Ansell, S.M.; Lesokhin, A.M.; Borrello, I.; Halwani, A.; Scott, E.C.; Gutierrez, M.; Schuster, S.J.; Millenson, M.M.; Cattry, D.; Freeman, G.J.; et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N. Engl. J. Med. 2015, 372, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Rodig, S.; Juszczynski, P.; Ouyang, J.; Sinha, P.; O’Donnell, E.; Neuberg, D.; Shipp, M.A. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: Implications for targeted therapy. Clin. Cancer Res. 2012, 18, 1611–1618. [Google Scholar] [CrossRef]
- Roemer, M.G.; Advani, R.H.; Ligon, A.H.; Natkunam, Y.; Redd, R.A.; Homer, H.; Connelly, C.F.; Sun, H.H.; Daadi, S.E.; Freeman, G.J.; et al. PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome. J. Clin. Oncol. 2016, 34, 2690–2697. [Google Scholar] [CrossRef]
- Sakakibara, A.; Kohno, K.; Eladl, A.E.; Klaisuwan, T.; Ishikawa, E.; Suzuki, Y.; Shimada, S.; Nakaguro, M.; Shimoyama, Y.; Takahara, T.; et al. Immunohistochemical assessment of the diagnostic utility of PD-L1: A preliminary analysis of anti-PD-L1 antibody (SP142) for lymphoproliferative diseases with tumour and non-malignant Hodgkin-Reed-Sternberg (HRS)-like cells. Histopathology 2018, 72, 1156–1163. [Google Scholar] [CrossRef]
- Aoki, T.; Chong, L.C.; Takata, K.; Milne, K.; Hav, M.; Colombo, A.; Chavez, E.A.; Nissen, M.; Wang, X.; Miyata-Takata, T.; et al. Single-Cell Transcriptome Analysis Reveals Disease-Defining T-cell Subsets in the Tumor Microenvironment of Classic Hodgkin Lymphoma. Cancer Discov. 2020, 10, 406–421. [Google Scholar] [CrossRef]
- Greaves, P.; Clear, A.; Owen, A.; Iqbal, S.; Lee, A.; Matthews, J.; Wilson, A.; Calaminici, M.; Gribben, J.G. Defining characteristics of classical Hodgkin lymphoma microenvironment T-helper cells. Blood 2013, 122, 2856–2863. [Google Scholar] [CrossRef]
- Carey, C.D.; Gusenleitner, D.; Lipschitz, M.; Roemer, M.G.M.; Stack, E.C.; Gjini, E.; Hu, X.; Redd, R.; Freeman, G.J.; Neuberg, D.; et al. Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood 2017, 130, 2420–2430. [Google Scholar] [CrossRef] [PubMed]
- Cader, F.Z.; Schackmann, R.C.J.; Hu, X.; Wienand, K.; Redd, R.; Chapuy, B.; Ouyang, J.; Paul, N.; Gjini, E.; Lipschitz, M.; et al. Mass cytometry of Hodgkin lymphoma reveals a CD4(+) regulatory T-cell-rich and exhausted T-effector microenvironment. Blood 2018, 132, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Connors, J.M.; Cozen, W.; Steidl, C.; Carbone, A.; Hoppe, R.T.; Flechtner, H.H.; Bartlett, N.L. Hodgkin lymphoma. Nat. Rev. Dis. Primers 2020, 6, 61. [Google Scholar] [CrossRef] [PubMed]
- Asano, N.; Oshiro, A.; Matsuo, K.; Kagami, Y.; Ishida, F.; Suzuki, R.; Kinoshita, T.; Shimoyama, Y.; Tamaru, J.; Yoshino, T.; et al. Prognostic significance of T-cell or cytotoxic molecules phenotype in classical Hodgkin’s lymphoma: A clinicopathologic study. J. Clin. Oncol. 2006, 24, 4626–4633. [Google Scholar] [CrossRef] [PubMed]
- Asano, N.; Kinoshita, T.; Tamaru, J.; Ohshima, K.; Yoshino, T.; Niitsu, N.; Tsukamoto, N.; Hirabayashi, K.; Izutsu, K.; Taniwaki, M.; et al. Cytotoxic molecule-positive classical Hodgkin’s lymphoma: A clinicopathological comparison with cytotoxic molecule-positive peripheral T-cell lymphoma of not otherwise specified type. Haematologica 2011, 96, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Piris, M.A.; Medeiros, L.J.; Chang, K.C. Hodgkin lymphoma: A review of pathological features and recent advances in pathogenesis. Pathology 2020, 52, 154–165. [Google Scholar] [CrossRef]
- Clarke, C.A.; Glaser, S.L.; Keegan, T.H.; Stroup, A. Neighborhood socioeconomic status and Hodgkin’s lymphoma incidence in California. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1441–1447. [Google Scholar] [CrossRef]
- Au, W.Y.; Gascoyne, R.D.; Gallagher, R.E.; Le, N.; Klasa, R.D.; Liang, R.H.; Choy, C.; Foo, W.; Connors, J.M. Hodgkin’s lymphoma in Chinese migrants to British Columbia: A 25-year survey. Ann. Oncol. 2004, 15, 626–630. [Google Scholar] [CrossRef]
- Morton, L.M.; Wang, S.S.; Devesa, S.S.; Hartge, P.; Weisenburger, D.D.; Linet, M.S. Lymphoma incidence patterns by WHO subtype in the United States, 1992–2001. Blood 2006, 107, 265–276. [Google Scholar] [CrossRef]
- Colby, T.V.; Hoppe, R.T.; Warnke, R.A. Hodgkin’s disease: A clinicopathologic study of 659 cases. Cancer 1982, 49, 1848–1858. [Google Scholar] [CrossRef]
- Herbst, H.; Dallenbach, F.; Hummel, M.; Niedobitek, G.; Pileri, S.; Muller-Lantzsch, N.; Stein, H. Epstein-Barr virus latent membrane protein expression in Hodgkin and Reed-Sternberg cells. Proc. Natl. Acad. Sci. USA 1991, 88, 4766–4770. [Google Scholar] [CrossRef] [PubMed]
- Herbst, H.; Niedobitek, G.; Kneba, M.; Hummel, M.; Finn, T.; Anagnostopoulos, I.; Bergholz, M.; Krieger, G.; Stein, H. High incidence of Epstein-Barr virus genomes in Hodgkin’s disease. Am. J. Pathol. 1990, 137, 13–18. [Google Scholar] [PubMed]
- Weiss, L.M. Epstein-Barr virus and Hodgkin’s disease. Curr. Oncol. Rep. 2000, 2, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Allemani, C.; Sant, M.; De Angelis, R.; Marcos-Gragera, R.; Coebergh, J.W.; Group, E.W. Hodgkin disease survival in Europe and the U.S.: Prognostic significance of morphologic groups. Cancer 2006, 107, 352–360. [Google Scholar] [CrossRef]
- Biggar, R.J.; Jaffe, E.S.; Goedert, J.J.; Chaturvedi, A.; Pfeiffer, R.; Engels, E.A. Hodgkin lymphoma and immunodeficiency in persons with HIV/AIDS. Blood 2006, 108, 3786–3791. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.G.; Sandvej, K.; Li, P.J.; Ji, X.L.; Yan, Q.H.; Zhang, X.P.; Da, J.P.; Hamilton-Dutoit, S.J. Epstein-Barr virus (EBV) in Chinese pediatric Hodgkin disease: Hodgkin disease in young children is an EBV-related lymphoma. Cancer 2001, 92, 1621–1631. [Google Scholar] [CrossRef]
- Anagnostopoulos, I.; Hansmann, M.L.; Franssila, K.; Harris, M.; Harris, N.L.; Jaffe, E.S.; Han, J.; van Krieken, J.M.; Poppema, S.; Marafioti, T.; et al. European Task Force on Lymphoma project on lymphocyte predominance Hodgkin disease: Histologic and immunohistologic analysis of submitted cases reveals 2 types of Hodgkin disease with a nodular growth pattern and abundant lymphocytes. Blood 2000, 96, 1889–1899. [Google Scholar]
- Shimabukuro-Vornhagen, A.; Haverkamp, H.; Engert, A.; Balleisen, L.; Majunke, P.; Heil, G.; Eich, H.T.; Stein, H.; Diehl, V.; Josting, A. Lymphocyte-rich classical Hodgkin’s lymphoma: Clinical presentation and treatment outcome in 100 patients treated within German Hodgkin’s Study Group trials. J. Clin. Oncol. 2005, 23, 5739–5745. [Google Scholar] [CrossRef]
- Diehl, V.; Sextro, M.; Franklin, J.; Hansmann, M.L.; Harris, N.; Jaffe, E.; Poppema, S.; Harris, M.; Franssila, K.; van Krieken, J.; et al. Clinical presentation, course, and prognostic factors in lymphocyte-predominant Hodgkin’s disease and lymphocyte-rich classical Hodgkin’s disease: Report from the European Task Force on Lymphoma Project on Lymphocyte-Predominant Hodgkin’s Disease. J. Clin. Oncol. 1999, 17, 776–783. [Google Scholar] [CrossRef]
- Shiels, M.S.; Koritzinsky, E.H.; Clarke, C.A.; Suneja, G.; Morton, L.M.; Engels, E.A. Prevalence of HIV Infection among U.S. Hodgkin lymphoma cases. Cancer Epidemiol. Biomark. Prev. 2014, 23, 274–281. [Google Scholar] [CrossRef]
- Benharroch, D.; Levy, A.; Gopas, J.; Sacks, M. Lymphocyte-depleted classic Hodgkin lymphoma-a neglected entity? Virchows Arch. 2008, 453, 611–616. [Google Scholar] [CrossRef]
- Slack, G.W.; Ferry, J.A.; Hasserjian, R.P.; Sohani, A.R.; Longtine, J.A.; Harris, N.L.; Zukerberg, L.R. Lymphocyte depleted Hodgkin lymphoma: An evaluation with immunophenotyping and genetic analysis. Leuk. Lymphoma 2009, 50, 937–943. [Google Scholar] [CrossRef]
- Klimm, B.; Franklin, J.; Stein, H.; Eichenauer, D.A.; Haverkamp, H.; Diehl, V.; Fuchs, M.; Borchmann, P.; Engert, A. Lymphocyte-depleted classical Hodgkin’s lymphoma: A comprehensive analysis from the German Hodgkin study group. J. Clin. Oncol. 2011, 29, 3914–3920. [Google Scholar] [CrossRef] [PubMed]
- Schlegelberger, B.; Weber-Matthiesen, K.; Himmler, A.; Bartels, H.; Sonnen, R.; Kuse, R.; Feller, A.C.; Grote, W. Cytogenetic findings and results of combined immunophenotyping and karyotyping in Hodgkin’s disease. Leukemia 1994, 8, 72–80. [Google Scholar] [PubMed]
- Joos, S.; Kupper, M.; Ohl, S.; von Bonin, F.; Mechtersheimer, G.; Bentz, M.; Marynen, P.; Moller, P.; Pfreundschuh, M.; Trumper, L.; et al. Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res. 2000, 60, 549–552. [Google Scholar] [PubMed]
- Bargou, R.C.; Emmerich, F.; Krappmann, D.; Bommert, K.; Mapara, M.Y.; Arnold, W.; Royer, H.D.; Grinstein, E.; Greiner, A.; Scheidereit, C.; et al. Constitutive nuclear factor-kappaB-RelA activation is required for proliferation and survival of Hodgkin’s disease tumor cells. J. Clin. Investig. 1997, 100, 2961–2969. [Google Scholar] [CrossRef]
- Krappmann, D.; Emmerich, F.; Kordes, U.; Scharschmidt, E.; Dorken, B.; Scheidereit, C. Molecular mechanisms of constitutive NF-kappaB/Rel activation in Hodgkin/Reed-Sternberg cells. Oncogene 1999, 18, 943–953. [Google Scholar] [CrossRef]
- Barth, T.F.; Martin-Subero, J.I.; Joos, S.; Menz, C.K.; Hasel, C.; Mechtersheimer, G.; Parwaresch, R.M.; Lichter, P.; Siebert, R.; Mooller, P. Gains of 2p involving the REL locus correlate with nuclear c-Rel protein accumulation in neoplastic cells of classical Hodgkin lymphoma. Blood 2003, 101, 3681–3686. [Google Scholar] [CrossRef]
- Ranuncolo, S.M.; Pittaluga, S.; Evbuomwan, M.O.; Jaffe, E.S.; Lewis, B.A. Hodgkin lymphoma requires stabilized NIK and constitutive RelB expression for survival. Blood 2012, 120, 3756–3763. [Google Scholar] [CrossRef]
- Joos, S.; Menz, C.K.; Wrobel, G.; Siebert, R.; Gesk, S.; Ohl, S.; Mechtersheimer, G.; Trumper, L.; Moller, P.; Lichter, P.; et al. Classical Hodgkin lymphoma is characterized by recurrent copy number gains of the short arm of chromosome 2. Blood 2002, 99, 1381–1387. [Google Scholar] [CrossRef]
- Martin-Subero, J.I.; Gesk, S.; Harder, L.; Sonoki, T.; Tucker, P.W.; Schlegelberger, B.; Grote, W.; Novo, F.J.; Calasanz, M.J.; Hansmann, M.L.; et al. Recurrent involvement of the REL and BCL11A loci in classical Hodgkin lymphoma. Blood 2002, 99, 1474–1477. [Google Scholar] [CrossRef]
- Martin-Subero, J.I.; Wlodarska, I.; Bastard, C.; Picquenot, J.M.; Hoppner, J.; Giefing, M.; Klapper, W.; Siebert, R. Chromosomal rearrangements involving the BCL3 locus are recurrent in classical Hodgkin and peripheral T-cell lymphoma. Blood 2006, 108, 401–402; author reply 402–403. [Google Scholar] [CrossRef] [PubMed]
- Steidl, C.; Telenius, A.; Shah, S.P.; Farinha, P.; Barclay, L.; Boyle, M.; Connors, J.M.; Horsman, D.E.; Gascoyne, R.D. Genome-wide copy number analysis of Hodgkin Reed-Sternberg cells identifies recurrent imbalances with correlations to treatment outcome. Blood 2010, 116, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Weniger, M.A.; Kuppers, R. NF-kappaB deregulation in Hodgkin lymphoma. Semin. Cancer Biol. 2016, 39, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Sanada, M.; Kato, I.; Sato, Y.; Takita, J.; Takeuchi, K.; Niwa, A.; Chen, Y.; Nakazaki, K.; Nomoto, J.; et al. Frequent inactivation of A20 in B-cell lymphomas. Nature 2009, 459, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.; Hansmann, M.L.; Bohle, V.; Martin-Subero, J.I.; Hartmann, S.; Mechtersheimer, G.; Klapper, W.; Vater, I.; Giefing, M.; Gesk, S.; et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J. Exp. Med. 2009, 206, 981–989. [Google Scholar] [CrossRef]
- Emmerich, F.; Theurich, S.; Hummel, M.; Haeffker, A.; Vry, M.S.; Dohner, K.; Bommert, K.; Stein, H.; Dorken, B. Inactivating I kappa B epsilon mutations in Hodgkin/Reed-Sternberg cells. J. Pathol. 2003, 201, 413–420. [Google Scholar] [CrossRef]
- Emmerich, F.; Meiser, M.; Hummel, M.; Demel, G.; Foss, H.D.; Jundt, F.; Mathas, S.; Krappmann, D.; Scheidereit, C.; Stein, H.; et al. Overexpression of I kappa B alpha without inhibition of NF-kappaB activity and mutations in the I kappa B alpha gene in Reed-Sternberg cells. Blood 1999, 94, 3129–3134. [Google Scholar] [CrossRef]
- Otto, C.; Giefing, M.; Massow, A.; Vater, I.; Gesk, S.; Schlesner, M.; Richter, J.; Klapper, W.; Hansmann, M.L.; Siebert, R.; et al. Genetic lesions of the TRAF3 and MAP3K14 genes in classical Hodgkin lymphoma. Br. J. Haematol. 2012, 157, 702–708. [Google Scholar] [CrossRef]
- Schmidt, A.; Schmitz, R.; Giefing, M.; Martin-Subero, J.I.; Gesk, S.; Vater, I.; Massow, A.; Maggio, E.; Schneider, M.; Hansmann, M.L.; et al. Rare occurrence of biallelic CYLD gene mutations in classical Hodgkin lymphoma. Genes Chromosomes Cancer 2010, 49, 803–809. [Google Scholar] [CrossRef]
- Hinz, M.; Lemke, P.; Anagnostopoulos, I.; Hacker, C.; Krappmann, D.; Mathas, S.; Dorken, B.; Zenke, M.; Stein, H.; Scheidereit, C. Nuclear factor kappaB-dependent gene expression profiling of Hodgkin’s disease tumor cells, pathogenetic significance, and link to constitutive signal transducer and activator of transcription 5a activity. J. Exp. Med. 2002, 196, 605–617. [Google Scholar] [CrossRef]
- Skinnider, B.F.; Elia, A.J.; Gascoyne, R.D.; Patterson, B.; Trumper, L.; Kapp, U.; Mak, T.W. Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 2002, 99, 618–626. [Google Scholar] [CrossRef]
- Meier, C.; Hoeller, S.; Bourgau, C.; Hirschmann, P.; Schwaller, J.; Went, P.; Pileri, S.A.; Reiter, A.; Dirnhofer, S.; Tzankov, A. Recurrent numerical aberrations of JAK2 and deregulation of the JAK2-STAT cascade in lymphomas. Mod. Pathol. 2009, 22, 476–487. [Google Scholar] [CrossRef] [PubMed]
- Van Roosbroeck, K.; Cox, L.; Tousseyn, T.; Lahortiga, I.; Gielen, O.; Cauwelier, B.; De Paepe, P.; Verhoef, G.; Marynen, P.; Vandenberghe, P.; et al. JAK2 rearrangements, including the novel SEC31A-JAK2 fusion, are recurrent in classical Hodgkin lymphoma. Blood 2011, 117, 4056–4064. [Google Scholar] [CrossRef] [PubMed]
- Tiacci, E.; Ladewig, E.; Schiavoni, G.; Penson, A.; Fortini, E.; Pettirossi, V.; Wang, Y.; Rosseto, A.; Venanzi, A.; Vlasevska, S.; et al. Pervasive mutations of JAK-STAT pathway genes in classical Hodgkin lymphoma. Blood 2018, 131, 2454–2465. [Google Scholar] [CrossRef] [PubMed]
- Wienand, K.; Chapuy, B.; Stewart, C.; Dunford, A.J.; Wu, D.; Kim, J.; Kamburov, A.; Wood, T.R.; Cader, F.Z.; Ducar, M.D.; et al. Genomic analyses of flow-sorted Hodgkin Reed-Sternberg cells reveal complementary mechanisms of immune evasion. Blood Adv. 2019, 3, 4065–4080. [Google Scholar] [CrossRef]
- Gunawardana, J.; Chan, F.C.; Telenius, A.; Woolcock, B.; Kridel, R.; Tan, K.L.; Ben-Neriah, S.; Mottok, A.; Lim, R.S.; Boyle, M.; et al. Recurrent somatic mutations of PTPN1 in primary mediastinal B cell lymphoma and Hodgkin lymphoma. Nat. Genet. 2014, 46, 329–335. [Google Scholar] [CrossRef]
- Weniger, M.A.; Melzner, I.; Menz, C.K.; Wegener, S.; Bucur, A.J.; Dorsch, K.; Mattfeldt, T.; Barth, T.F.; Moller, P. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene 2006, 25, 2679–2684. [Google Scholar] [CrossRef] [PubMed]
- Kapp, U.; Yeh, W.C.; Patterson, B.; Elia, A.J.; Kagi, D.; Ho, A.; Hessel, A.; Tipsword, M.; Williams, A.; Mirtsos, C.; et al. Interleukin 13 is secreted by and stimulates the growth of Hodgkin and Reed-Sternberg cells. J. Exp. Med. 1999, 189, 1939–1946. [Google Scholar] [CrossRef]
- Lamprecht, B.; Kreher, S.; Anagnostopoulos, I.; Johrens, K.; Monteleone, G.; Jundt, F.; Stein, H.; Janz, M.; Dorken, B.; Mathas, S. Aberrant expression of the Th2 cytokine IL-21 in Hodgkin lymphoma cells regulates STAT3 signaling and attracts Treg cells via regulation of MIP-3alpha. Blood 2008, 112, 3339–3347. [Google Scholar] [CrossRef] [PubMed]
- Scheeren, F.A.; Diehl, S.A.; Smit, L.A.; Beaumont, T.; Naspetti, M.; Bende, R.J.; Blom, B.; Karube, K.; Ohshima, K.; van Noesel, C.J.; et al. IL-21 is expressed in Hodgkin lymphoma and activates STAT5: Evidence that activated STAT5 is required for Hodgkin lymphomagenesis. Blood 2008, 111, 4706–4715. [Google Scholar] [CrossRef] [PubMed]
- Cattaruzza, L.; Gloghini, A.; Olivo, K.; Di Francia, R.; Lorenzon, D.; De Filippi, R.; Carbone, A.; Colombatti, A.; Pinto, A.; Aldinucci, D. Functional coexpression of Interleukin (IL)-7 and its receptor (IL-7R) on Hodgkin and Reed-Sternberg cells: Involvement of IL-7 in tumor cell growth and microenvironmental interactions of Hodgkin’s lymphoma. Int. J. Cancer 2009, 125, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Mathas, S.; Hinz, M.; Anagnostopoulos, I.; Krappmann, D.; Lietz, A.; Jundt, F.; Bommert, K.; Mechta-Grigoriou, F.; Stein, H.; Dorken, B.; et al. Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-kappa B. EMBO J. 2002, 21, 4104–4113. [Google Scholar] [CrossRef]
- Lollies, A.; Hartmann, S.; Schneider, M.; Bracht, T.; Weiss, A.L.; Arnolds, J.; Klein-Hitpass, L.; Sitek, B.; Hansmann, M.L.; Kuppers, R.; et al. An oncogenic axis of STAT-mediated BATF3 upregulation causing MYC activity in classical Hodgkin lymphoma and anaplastic large cell lymphoma. Leukemia 2018, 32, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Vrzalikova, K.; Ibrahim, M.; Vockerodt, M.; Perry, T.; Margielewska, S.; Lupino, L.; Nagy, E.; Soilleux, E.; Liebelt, D.; Hollows, R.; et al. S1PR1 drives a feedforward signalling loop to regulate BATF3 and the transcriptional programme of Hodgkin lymphoma cells. Leukemia 2018, 32, 214–223. [Google Scholar] [CrossRef]
- Schwarzer, R.; Dorken, B.; Jundt, F. Notch is an essential upstream regulator of NF-kappaB and is relevant for survival of Hodgkin and Reed-Sternberg cells. Leukemia 2012, 26, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef]
- Satou, A.; Nakamura, S. EBV-positive B-cell lymphomas and lymphoproliferative disorders: Review from the perspective of immune escape and immunodeficiency. Cancer Med. 2021, 10, 6777–6785. [Google Scholar] [CrossRef]
- Topalian, S.L.; Drake, C.G.; Pardoll, D.M. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 2015, 27, 450–461. [Google Scholar] [CrossRef]
- Topalian, S.L.; Taube, J.M.; Anders, R.A.; Pardoll, D.M. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer 2016, 16, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Parsa, A.T.; Waldron, J.S.; Panner, A.; Crane, C.A.; Parney, I.F.; Barry, J.J.; Cachola, K.E.; Murray, J.C.; Tihan, T.; Jensen, M.C.; et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat. Med. 2007, 13, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Sumimoto, H.; Imabayashi, F.; Iwata, T.; Kawakami, Y. The BRAF-MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells. J. Exp. Med. 2006, 203, 1651–1656. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Zhang, Q.; Goradia, A.; Raghunath, P.N.; Liu, X.; Paessler, M.; Wang, H.Y.; Wysocka, M.; Cheng, M.; Ruggeri, B.A.; et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1). Proc. Natl. Acad. Sci. USA 2008, 105, 20852–20857. [Google Scholar] [CrossRef]
- Akbay, E.A.; Koyama, S.; Carretero, J.; Altabef, A.; Tchaicha, J.H.; Christensen, C.L.; Mikse, O.R.; Cherniack, A.D.; Beauchamp, E.M.; Pugh, T.J.; et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013, 3, 1355–1363. [Google Scholar] [CrossRef]
- Gulley, M.L. Genomic assays for Epstein-Barr virus-positive gastric adenocarcinoma. Exp. Mol. Med. 2015, 47, e134. [Google Scholar] [CrossRef]
- Kataoka, K.; Miyoshi, H.; Sakata, S.; Dobashi, A.; Couronne, L.; Kogure, Y.; Sato, Y.; Nishida, K.; Gion, Y.; Shiraishi, Y.; et al. Frequent structural variations involving programmed death ligands in Epstein-Barr virus-associated lymphomas. Leukemia 2019, 33, 1687–1699. [Google Scholar] [CrossRef]
- Kataoka, K.; Shiraishi, Y.; Takeda, Y.; Sakata, S.; Matsumoto, M.; Nagano, S.; Maeda, T.; Nagata, Y.; Kitanaka, A.; Mizuno, S.; et al. Aberrant PD-L1 expression through 3’-UTR disruption in multiple cancers. Nature 2016, 534, 402–406. [Google Scholar] [CrossRef]
- Steidl, C.; Shah, S.P.; Woolcock, B.W.; Rui, L.; Kawahara, M.; Farinha, P.; Johnson, N.A.; Zhao, Y.; Telenius, A.; Neriah, S.B.; et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature 2011, 471, 377–381. [Google Scholar] [CrossRef]
- Rosenwald, A.; Wright, G.; Leroy, K.; Yu, X.; Gaulard, P.; Gascoyne, R.D.; Chan, W.C.; Zhao, T.; Haioun, C.; Greiner, T.C.; et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J. Exp. Med. 2003, 198, 851–862. [Google Scholar] [CrossRef]
- Green, M.R.; Monti, S.; Rodig, S.J.; Juszczynski, P.; Currie, T.; O’Donnell, E.; Chapuy, B.; Takeyama, K.; Neuberg, D.; Golub, T.R.; et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 2010, 116, 3268–3277. [Google Scholar] [CrossRef] [PubMed]
- Rui, L.; Emre, N.C.; Kruhlak, M.J.; Chung, H.J.; Steidl, C.; Slack, G.; Wright, G.W.; Lenz, G.; Ngo, V.N.; Shaffer, A.L.; et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell 2010, 18, 590–605. [Google Scholar] [CrossRef] [PubMed]
- Taube, J.M.; Anders, R.A.; Young, G.D.; Xu, H.; Sharma, R.; McMiller, T.L.; Chen, S.; Klein, A.P.; Pardoll, D.M.; Topalian, S.L.; et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci. Transl. Med. 2012, 4, 127ra137. [Google Scholar] [CrossRef]
- Volaric, A.; Bacchi, C.E.; Gru, A.A. PD-1 and PD-L1 Immunohistochemistry as a Diagnostic Tool for Classic Hodgkin Lymphoma in Small-volume Biopsies. Am. J. Surg. Pathol. 2020, 44, 1353–1366. [Google Scholar] [CrossRef] [PubMed]
- Uccella, S.; Magnoli, F.; Vivian, L.F.; Marchiori, D.; Leoni, E.; Tibiletti, M.G.; Sessa, F. PD-1 and PD-L1 Immunohistochemistry as a Diagnostic Tool for Classic Hodgkin Lymphoma in Small-volume Biopsies. Am. J. Surg. Pathol. 2021, 45, 1293–1296. [Google Scholar] [CrossRef] [PubMed]
- Eladl, A.E.; Satou, A.; Elsayed, A.A.; Suzuki, Y.; Kato, S.; Asano, N.; Nakamura, S. Clinicopathological Study of 30 Cases of Peripheral T-cell Lymphoma with Hodgkin and Reed-Sternberg-like B-cells from Japan. Am. J. Surg. Pathol. 2017, 41, 506–516. [Google Scholar] [CrossRef]
- Kawashima, M.; Carreras, J.; Higuchi, H.; Kotaki, R.; Hoshina, T.; Okuyama, K.; Suzuki, N.; Kakizaki, M.; Miyatake, Y.; Ando, K.; et al. PD-L1/L2 protein levels rapidly increase on monocytes via trogocytosis from tumor cells in classical Hodgkin lymphoma. Leukemia 2020, 34, 2405–2417. [Google Scholar] [CrossRef]
- Reichel, J.; Chadburn, A.; Rubinstein, P.G.; Giulino-Roth, L.; Tam, W.; Liu, Y.; Gaiolla, R.; Eng, K.; Brody, J.; Inghirami, G.; et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood 2015, 125, 1061–1072. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A.; Gross, A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 2004, 350, 1328–1337. [Google Scholar] [CrossRef]
- Taylor, G.S.; Long, H.M.; Brooks, J.M.; Rickinson, A.B.; Hislop, A.D. The immunology of Epstein-Barr virus-induced disease. Annu. Rev. Immunol. 2015, 33, 787–821. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A. Epstein-Barr virus: Exploiting the immune system. Nat. Rev. Immunol. 2001, 1, 75–82. [Google Scholar] [CrossRef]
- Satou, A.; Asano, N.; Nakazawa, A.; Osumi, T.; Tsurusawa, M.; Ishiguro, A.; Elsayed, A.A.; Nakamura, N.; Ohshima, K.; Kinoshita, T.; et al. Epstein-Barr virus (EBV)-positive sporadic burkitt lymphoma: An age-related lymphoproliferative disorder? Am. J. Surg. Pathol. 2015, 39, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Glaser, S.L.; Lin, R.J.; Stewart, S.L.; Ambinder, R.F.; Jarrett, R.F.; Brousset, P.; Pallesen, G.; Gulley, M.L.; Khan, G.; O’Grady, J.; et al. Epstein-Barr virus-associated Hodgkin’s disease: Epidemiologic characteristics in international data. Int. J. Cancer 1997, 70, 375–382. [Google Scholar] [CrossRef]
- Flavell, K.J.; Biddulph, J.P.; Constandinou, C.M.; Lowe, D.; Scott, K.; Crocker, J.; Young, L.S.; Murray, P.G. Variation in the frequency of Epstein-Barr virus-associated Hodgkin’s disease with age. Leukemia 2000, 14, 748–753. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Young, L.S.; Rickinson, A.B. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 2004, 4, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Young, L.S.; Yap, L.F.; Murray, P.G. Epstein-Barr virus: More than 50 years old and still providing surprises. Nat. Rev. Cancer 2016, 16, 789–802. [Google Scholar] [CrossRef]
- Hochberg, D.; Souza, T.; Catalina, M.; Sullivan, J.L.; Luzuriaga, K.; Thorley-Lawson, D.A. Acute infection with Epstein-Barr virus targets and overwhelms the peripheral memory B-cell compartment with resting, latently infected cells. J. Virol. 2004, 78, 5194–5204. [Google Scholar] [CrossRef]
- Chang, K.C.; Chen, P.C.; Chen, Y.P.; Chang, Y.; Su, I.J. Dominant expression of survival signals of endoplasmic reticulum stress response in Hodgkin lymphoma. Cancer Sci. 2011, 102, 275–281. [Google Scholar] [CrossRef]
- Kilger, E.; Kieser, A.; Baumann, M.; Hammerschmidt, W. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J. 1998, 17, 1700–1709. [Google Scholar] [CrossRef]
- Zimber-Strobl, U.; Kempkes, B.; Marschall, G.; Zeidler, R.; Van Kooten, C.; Banchereau, J.; Bornkamm, G.W.; Hammerschmidt, W. Epstein-Barr virus latent membrane protein (LMP1) is not sufficient to maintain proliferation of B cells but both it and activated CD40 can prolong their survival. EMBO J. 1996, 15, 7070–7078. [Google Scholar] [CrossRef]
- Beaufils, P.; Choquet, D.; Mamoun, R.Z.; Malissen, B. The (YXXL/I)2 signalling motif found in the cytoplasmic segments of the bovine leukaemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO J. 1993, 12, 5105–5112. [Google Scholar] [CrossRef] [PubMed]
- Mancao, C.; Altmann, M.; Jungnickel, B.; Hammerschmidt, W. Rescue of "crippled" germinal center B cells from apoptosis by Epstein-Barr virus. Blood 2005, 106, 4339–4344. [Google Scholar] [CrossRef] [PubMed]
- Dirmeier, U.; Hoffmann, R.; Kilger, E.; Schultheiss, U.; Briseno, C.; Gires, O.; Kieser, A.; Eick, D.; Sugden, B.; Hammerschmidt, W. Latent membrane protein 1 of Epstein-Barr virus coordinately regulates proliferation with control of apoptosis. Oncogene 2005, 24, 1711–1717. [Google Scholar] [CrossRef] [PubMed]
- Dirmeier, U.; Neuhierl, B.; Kilger, E.; Reisbach, G.; Sandberg, M.L.; Hammerschmidt, W. Latent membrane protein 1 is critical for efficient growth transformation of human B cells by epstein-barr virus. Cancer Res. 2003, 63, 2982–2989. [Google Scholar]
- Gutensohn, N.M. Social class and age at diagnosis of Hodgkin’s disease: New epidemiologic evidence for the “two-disease hypothesis”. Cancer Treat. Rep. 1982, 66, 689–695. [Google Scholar]
- Hjalgrim, H.; Askling, J.; Rostgaard, K.; Hamilton-Dutoit, S.; Frisch, M.; Zhang, J.S.; Madsen, M.; Rosdahl, N.; Konradsen, H.B.; Storm, H.H.; et al. Characteristics of Hodgkin’s lymphoma after infectious mononucleosis. N. Engl. J. Med. 2003, 349, 1324–1332. [Google Scholar] [CrossRef]
- Lake, A.; Shield, L.A.; Cordano, P.; Chui, D.T.; Osborne, J.; Crae, S.; Wilson, K.S.; Tosi, S.; Knight, S.J.; Gesk, S.; et al. Mutations of NFKBIA, encoding IkappaB alpha, are a recurrent finding in classical Hodgkin lymphoma but are not a unifying feature of non-EBV-associated cases. Int. J. Cancer 2009, 125, 1334–1342. [Google Scholar] [CrossRef]
- Younes, S.; Rojansky, R.B.; Menke, J.R.; Gratzinger, D.; Natkunam, Y. Pitfalls in the Diagnosis of Nodular Lymphocyte Predominant Hodgkin Lymphoma: Variant Patterns, Borderlines and Mimics. Cancers 2021, 13, 3021. [Google Scholar] [CrossRef]
- Hartmann, S.; Doring, C.; Vucic, E.; Chan, F.C.; Ennishi, D.; Tousseyn, T.; de Wolf-Peeters, C.; Perner, S.; Wlodarska, I.; Steidl, C.; et al. Array comparative genomic hybridization reveals similarities between nodular lymphocyte predominant Hodgkin lymphoma and T cell/histiocyte rich large B cell lymphoma. Br. J. Haematol. 2015, 169, 415–422. [Google Scholar] [CrossRef]
- Hartmann, S.; Eichenauer, D.A.; Plutschow, A.; Mottok, A.; Bob, R.; Koch, K.; Bernd, H.W.; Cogliatti, S.; Hummel, M.; Feller, A.C.; et al. The prognostic impact of variant histology in nodular lymphocyte-predominant Hodgkin lymphoma: A report from the German Hodgkin Study Group (GHSG). Blood 2013, 122, 4246–4252. [Google Scholar] [CrossRef]
- Shankar, A.G.; Kirkwood, A.A.; Hall, G.W.; Hayward, J.; O’Hare, P.; Ramsay, A.D. Childhood and Adolescent nodular lymphocyte predominant Hodgkin lymphoma-A review of clinical outcome based on the histological variants. Br. J. Haematol. 2015, 171, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Natkunam, Y.; Bair, E.; Tibshirani, R.; Warnke, R.A. Characterization of variant patterns of nodular lymphocyte predominant hodgkin lymphoma with immunohistologic and clinical correlation. Am. J. Surg. Pathol. 2003, 27, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Huppmann, A.R.; Nicolae, A.; Slack, G.W.; Pittaluga, S.; Davies-Hill, T.; Ferry, J.A.; Harris, N.L.; Jaffe, E.S.; Hasserjian, R.P. EBV may be expressed in the LP cells of nodular lymphocyte-predominant Hodgkin lymphoma (NLPHL) in both children and adults. Am. J. Surg. Pathol. 2014, 38, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Medeiros, L.J.; Xu-Monette, Z.Y.; Zhang, S.; O’Malley, D.P.; Orazi, A.; Zuo, Z.; Bueso-Ramos, C.E.; Yin, C.C.; Liu, Z.; et al. Epstein-Barr virus-positive nodular lymphocyte predominant Hodgkin lymphoma. Ann. Diagn. Pathol. 2014, 18, 203–209. [Google Scholar] [CrossRef]
- Boudova, L.; Torlakovic, E.; Delabie, J.; Reimer, P.; Pfistner, B.; Wiedenmann, S.; Diehl, V.; Muller-Hermelink, H.K.; Rudiger, T. Nodular lymphocyte-predominant Hodgkin lymphoma with nodules resembling T-cell/histiocyte-rich B-cell lymphoma: Differential diagnosis between nodular lymphocyte-predominant Hodgkin lymphoma and T-cell/histiocyte-rich B-cell lymphoma. Blood 2003, 102, 3753–3758. [Google Scholar] [CrossRef] [PubMed]
- El Weshi, A.; Akhtar, S.; Mourad, W.A.; Ajarim, D.; Abdelsalm, M.; Khafaga, Y.; Bazarbashi, S.; Maghfoor, I. T-cell/histiocyte-rich B-cell lymphoma: Clinical presentation, management and prognostic factors: Report on 61 patients and review of literature. Leuk. Lymphoma 2007, 48, 1764–1773. [Google Scholar] [CrossRef]
- Al-Mansour, M.; Connors, J.M.; Gascoyne, R.D.; Skinnider, B.; Savage, K.J. Transformation to aggressive lymphoma in nodular lymphocyte-predominant Hodgkin’s lymphoma. J. Clin. Oncol. 2010, 28, 793–799. [Google Scholar] [CrossRef]
- Kenderian, S.S.; Habermann, T.M.; Macon, W.R.; Ristow, K.M.; Ansell, S.M.; Colgan, J.P.; Johnston, P.B.; Inwards, D.J.; Markovic, S.N.; Micallef, I.N.; et al. Large B-cell transformation in nodular lymphocyte-predominant Hodgkin lymphoma: 40-year experience from a single institution. Blood 2016, 127, 1960–1966. [Google Scholar] [CrossRef]
- Mottok, A.; Renne, C.; Willenbrock, K.; Hansmann, M.L.; Brauninger, A. Somatic hypermutation of SOCS1 in lymphocyte-predominant Hodgkin lymphoma is accompanied by high JAK2 expression and activation of STAT6. Blood 2007, 110, 3387–3390. [Google Scholar] [CrossRef]
- Brune, V.; Tiacci, E.; Pfeil, I.; Doring, C.; Eckerle, S.; van Noesel, C.J.; Klapper, W.; Falini, B.; von Heydebreck, A.; Metzler, D.; et al. Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J. Exp. Med. 2008, 205, 2251–2268. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Schmitz, R.; Brune, V.; Tiacci, E.; Doring, C.; Hansmann, M.L.; Siebert, R.; Kuppers, R. Mutations in the genes coding for the NF-kappaB regulating factors IkappaBalpha and A20 are uncommon in nodular lymphocyte-predominant Hodgkin’s lymphoma. Haematologica 2010, 95, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Thurner, L.; Hartmann, S.; Fadle, N.; Regitz, E.; Kemele, M.; Kim, Y.J.; Bohle, R.M.; Nimmesgern, A.; von Muller, L.; Kempf, V.A.J.; et al. Lymphocyte predominant cells detect Moraxella catarrhalis-derived antigens in nodular lymphocyte-predominant Hodgkin lymphoma. Nat. Commun. 2020, 11, 2465. [Google Scholar] [CrossRef] [PubMed]
- Wlodarska, I.; Nooyen, P.; Maes, B.; Martin-Subero, J.I.; Siebert, R.; Pauwels, P.; De Wolf-Peeters, C.; Hagemeijer, A. Frequent occurrence of BCL6 rearrangements in nodular lymphocyte predominance Hodgkin lymphoma but not in classical Hodgkin lymphoma. Blood 2003, 101, 706–710. [Google Scholar] [CrossRef] [PubMed]
- Renne, C.; Martin-Subero, J.I.; Hansmann, M.L.; Siebert, R. Molecular cytogenetic analyses of immunoglobulin loci in nodular lymphocyte predominant Hodgkin’s lymphoma reveal a recurrent IGH-BCL6 juxtaposition. J. Mol. Diagn. 2005, 7, 352–356. [Google Scholar] [CrossRef]
- Liso, A.; Capello, D.; Marafioti, T.; Tiacci, E.; Cerri, M.; Distler, V.; Paulli, M.; Carbone, A.; Delsol, G.; Campo, E.; et al. Aberrant somatic hypermutation in tumor cells of nodular-lymphocyte-predominant and classic Hodgkin lymphoma. Blood 2006, 108, 1013–1020. [Google Scholar] [CrossRef][Green Version]
- Hartmann, S.; Schuhmacher, B.; Rausch, T.; Fuller, L.; Doring, C.; Weniger, M.; Lollies, A.; Weiser, C.; Thurner, L.; Rengstl, B.; et al. Highly recurrent mutations of SGK1, DUSP2 and JUNB in nodular lymphocyte predominant Hodgkin lymphoma. Leukemia 2016, 30, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Eladl, A.E.; Satou, A.; Elsayed, A.A.; Suzuki, Y.; Shimizu-Kohno, K.; Kato, S.; Tomita, A.; Kinoshita, T.; Nakamura, S.; Asano, N. Nodular lymphocyte predominant Hodgkin lymphoma: Clincopathological study of 25 cases from Japan with a reappraisal of tissue associated macrophages. Pathol. Int. 2015, 65, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Schuhmacher, B.; Bein, J.; Rausch, T.; Benes, V.; Tousseyn, T.; Vornanen, M.; Ponzoni, M.; Thurner, L.; Gascoyne, R.; Steidl, C.; et al. JUNB, DUSP2, SGK1, SOCS1 and CREBBP are frequently mutated in T-cell/histiocyte-rich large B-cell lymphoma. Haematologica 2019, 104, 330–337. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Satou, A.; Takahara, T.; Nakamura, S. An Update on the Pathology and Molecular Features of Hodgkin Lymphoma. Cancers 2022, 14, 2647. https://doi.org/10.3390/cancers14112647
Satou A, Takahara T, Nakamura S. An Update on the Pathology and Molecular Features of Hodgkin Lymphoma. Cancers. 2022; 14(11):2647. https://doi.org/10.3390/cancers14112647
Chicago/Turabian StyleSatou, Akira, Taishi Takahara, and Shigeo Nakamura. 2022. "An Update on the Pathology and Molecular Features of Hodgkin Lymphoma" Cancers 14, no. 11: 2647. https://doi.org/10.3390/cancers14112647
APA StyleSatou, A., Takahara, T., & Nakamura, S. (2022). An Update on the Pathology and Molecular Features of Hodgkin Lymphoma. Cancers, 14(11), 2647. https://doi.org/10.3390/cancers14112647




