Latest Contributions of Genomics to T-Cell Acute Lymphoblastic Leukemia (T-ALL)
Abstract
Simple Summary
Abstract
1. Introduction
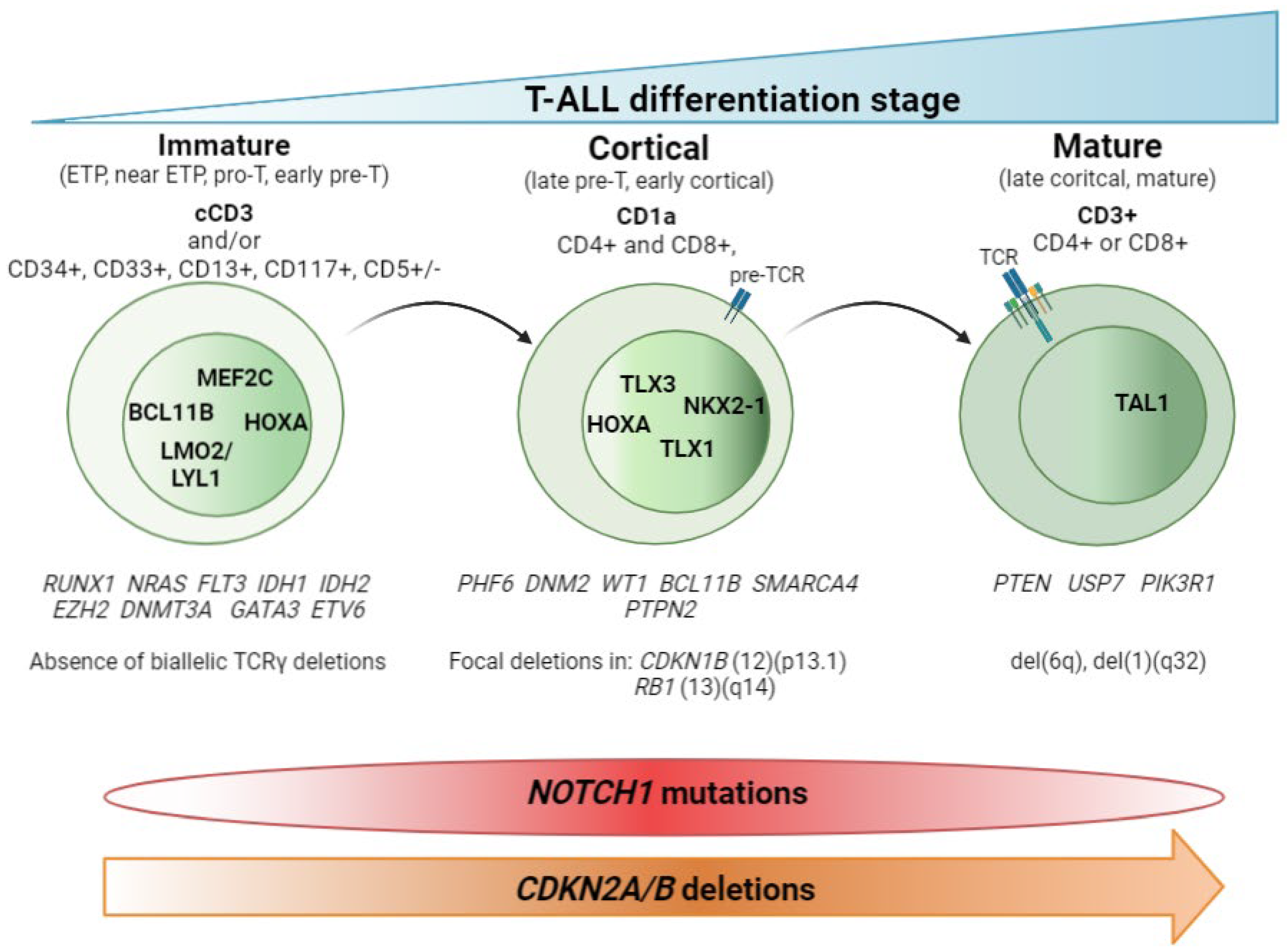
2. T-ALL Classification by Differentiation Stage
2.1. Non-Coding Mutations
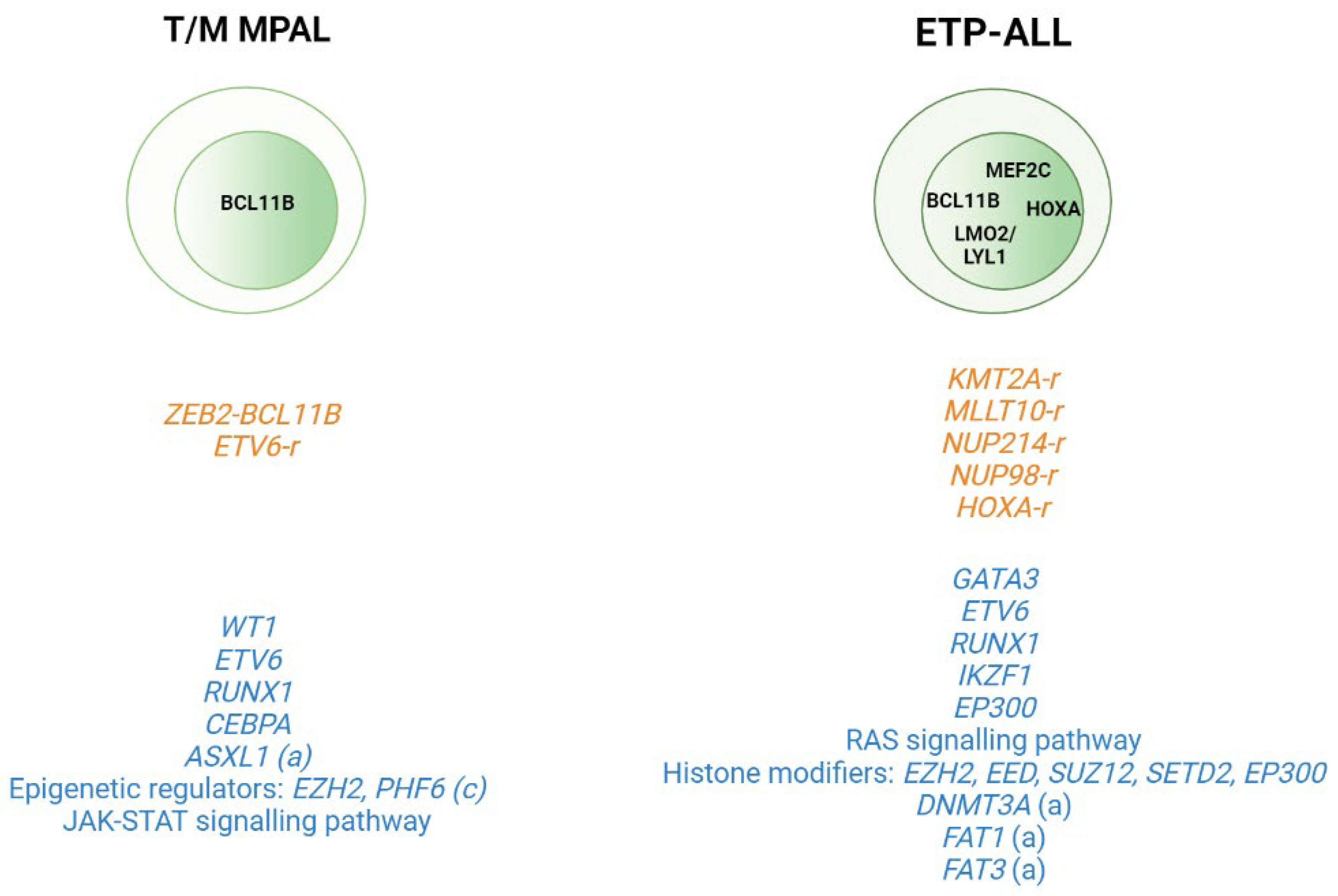
2.2. T-ALL Related Immature Subtypes
3. (Epi)genetic Modification
4. Germline Variants and Predisposition Alleles
4.1. Germline Predispostion Alterations Contributing to the Development of T-ALL
4.2. Predisposition Alleles Affecting Drug Response
5. Clonal Hematopoiesis and Aging
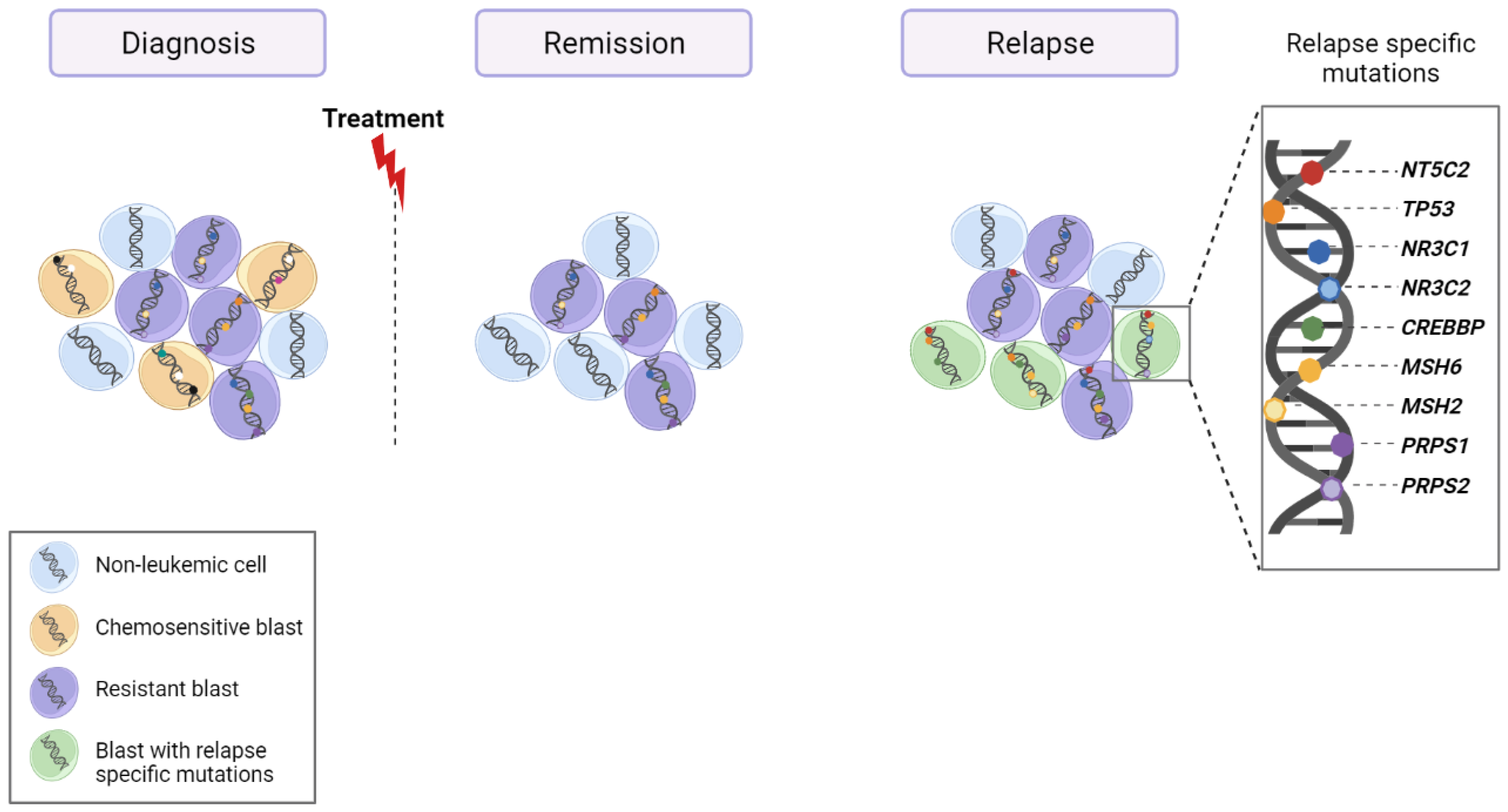
6. Relapse
7. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Yi, M.; Zhou, L.; Li, A.; Luo, S.; Wu, K. Global Burden and Trend of Acute Lymphoblastic Leukemia from 1990 to 2017. Aging 2020, 12, 22869–22891. [Google Scholar] [CrossRef] [PubMed]
- Belver, L.; Ferrando, A. The Genetics and Mechanisms of T Cell Acute Lymphoblastic Leukaemia. Nat. Rev. Cancer 2016, 16, 494–507. [Google Scholar] [CrossRef] [PubMed]
- Guru Murthy, G.S.; Pondaiah, S.K.; Abedin, S.; Atallah, E. Incidence and Survival of T-Cell Acute Lymphoblastic Leukemia in the United States. Leuk. Lymphoma 2019, 60, 1171–1178. [Google Scholar] [CrossRef]
- Bartram, J.; Veys, P.; Vora, A. Improvements in Outcome of Childhood Acute Lymphoblastic Leukaemia (ALL) in the UK—A Success Story of Modern Medicine through Successive UKALL Trials and International Collaboration. Br. J. Haematol. 2020, 191, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Ribera, J.-M.; Morgades, M.; Ciudad, J.; Montesinos, P.; Esteve, J.; Genescà, E.; Barba, P.; Ribera, J.; García-Cadenas, I.; Moreno, M.J.; et al. Chemotherapy or Allogeneic Transplantation in High-Risk Philadelphia Chromosome-Negative Adult Lymphoblastic Leukemia. Blood 2021, 137, 1879–1894. [Google Scholar] [CrossRef]
- Beldjord, K.; Chevret, S.; Asnafi, V.; Huguet, F.; Boulland, M.-L.; Leguay, T.; Thomas, X.; Cayuela, J.-M.; Grardel, N.; Chalandon, Y.; et al. Oncogenetics and Minimal Residual Disease Are Independent Outcome Predictors in Adult Patients with Acute Lymphoblastic Leukemia. Blood 2014, 123, 3739–3749. [Google Scholar] [CrossRef]
- Nguyen, K.; Devidas, M.; Cheng, S.-C.; La, M.; Raetz, E.A.; Carroll, W.L.; Winick, N.J.; Hunger, S.P.; Gaynon, P.S.; Loh, M.L. Factors Influencing Survival After Relapse From Acute Lymphoblastic Leukemia: A Children’s Oncology Group Study. Leukemia 2008, 22, 2142–2150. [Google Scholar] [CrossRef]
- Barba, P.; Morgades, M.; Montesinos, P.; Gil, C.; Fox, M.-L.; Ciudad, J.; Moreno, M.-J.; González-Campos, J.; Genescà, E.; Martínez-Carballeira, D.; et al. Increased Survival Due to Lower Toxicity for High-Risk T-Cell Acute Lymphoblastic Leukemia Patients in Two Consecutive Pediatric-Inspired PETHEMA Trials. Eur. J. Haematol. 2019, 102, 79–86. [Google Scholar] [CrossRef]
- Begley, C.G.; Aplan, P.D.; Davey, M.P.; Nakahara, K.; Tchorz, K.; Kurtzberg, J.; Hershfield, M.S.; Haynes, B.F.; Cohen, D.I.; Waldmann, T.A. Chromosomal Translocation in a Human Leukemic Stem-Cell Line Disrupts the T-Cell Antigen Receptor Delta-Chain Diversity Region and Results in a Previously Unreported Fusion Transcript. Proc. Natl. Acad. Sci. USA 1989, 86, 2031–2035. [Google Scholar] [CrossRef]
- Bernard, O.; Guglielmi, P.; Jonveaux, P.; Cherif, D.; Gisselbrecht, S.; Mauchauffe, M.; Berger, R.; Larsen, C.J.; Mathieu-Mahul, D. Two Distinct Mechanisms for the SCL Gene Activation in the t(1;14) Translocation of T-Cell Leukemias. Genes Chromosom. Cancer 1990, 1, 194–208. [Google Scholar] [CrossRef]
- Chen, Q.; Cheng, J.T.; Tasi, L.H.; Schneider, N.; Buchanan, G.; Carroll, A.; Crist, W.; Ozanne, B.; Siciliano, M.J.; Baer, R. The Tal Gene Undergoes Chromosome Translocation in T Cell Leukemia and Potentially Encodes a Helix-Loop-Helix Protein. EMBO J. 1990, 9, 415–424. [Google Scholar] [CrossRef]
- Xia, Y.; Brown, L.; Yang, C.Y.; Tsan, J.T.; Siciliano, M.J.; Espinosa, R.; Le Beau, M.M.; Baer, R.J. TAL2, a Helix-Loop-Helix Gene Activated by the (7;9)(Q34;Q32) Translocation in Human T-Cell Leukemia. Proc. Natl. Acad. Sci. USA 1991, 88, 11416–11420. [Google Scholar] [CrossRef]
- Mellentin, J.D.; Smith, S.D.; Cleary, M.L. Lyl-1, a Novel Gene Altered by Chromosomal Translocation in T Cell Leukemia, Codes for a Protein with a Helix-Loop-Helix DNA Binding Motif. Cell 1989, 58, 77–83. [Google Scholar] [CrossRef]
- McGuire, E.A.; Hockett, R.D.; Pollock, K.M.; Bartholdi, M.F.; O’Brien, S.J.; Korsmeyer, S.J. The t(11;14)(P15;Q11) in a T-Cell Acute Lymphoblastic Leukemia Cell Line Activates Multiple Transcripts, Including Ttg-1, a Gene Encoding a Potential Zinc Finger Protein. Mol. Cell Biol. 1989, 9, 2124–2132. [Google Scholar] [PubMed]
- Boehm, T.; Foroni, L.; Kaneko, Y.; Perutz, M.F.; Rabbitts, T.H. The Rhombotin Family of Cysteine-Rich LIM-Domain Oncogenes: Distinct Members Are Involved in T-Cell Translocations to Human Chromosomes 11p15 and 11p13. Proc. Natl. Acad. Sci. USA 1991, 88, 4367–4371. [Google Scholar] [CrossRef] [PubMed]
- Royer-Pokora, B.; Loos, U.; Ludwig, W.D. TTG-2, a New Gene Encoding a Cysteine-Rich Protein with the LIM Motif, Is Overexpressed in Acute T-Cell Leukaemia with the t(11;14)(P13;Q11). Oncogene 1991, 6, 1887–1893. [Google Scholar]
- Dube, I.; Kamel-Reid, S.; Yuan, C.; Lu, M.; Wu, X.; Corpus, G.; Raimondi, S.; Crist, W.; Carroll, A.; Minowada, J. A Novel Human Homeobox Gene Lies at the Chromosome 10 Breakpoint in Lymphoid Neoplasias with Chromosomal Translocation t(10;14). Blood 1991, 78, 2996–3003. [Google Scholar] [CrossRef] [PubMed]
- Hatano, M.; Roberts, C.W.; Minden, M.; Crist, W.M.; Korsmeyer, S.J. Deregulation of a Homeobox Gene, HOX11, by the t(10;14) in T Cell Leukemia. Science 1991, 253, 79–82. [Google Scholar] [CrossRef]
- Bernard, O.A.; Busson-LeConiat, M.; Ballerini, P.; Mauchauffé, M.; Della Valle, V.; Monni, R.; Nguyen Khac, F.; Mercher, T.; Penard-Lacronique, V.; Pasturaud, P.; et al. A New Recurrent and Specific Cryptic Translocation, t(5;14)(Q35;Q32), Is Associated with Expression of the Hox11L2 Gene in T Acute Lymphoblastic Leukemia. Leukemia 2001, 15, 1495–1504. [Google Scholar] [CrossRef]
- Ferrando, A.A.; Neuberg, D.S.; Staunton, J.; Loh, M.L.; Huard, C.; Raimondi, S.C.; Behm, F.G.; Pui, C.H.; Downing, J.R.; Gilliland, D.G.; et al. Gene Expression Signatures Define Novel Oncogenic Pathways in T Cell Acute Lymphoblastic Leukemia. Cancer Cell 2002, 1, 75–87. [Google Scholar] [CrossRef]
- Soulier, J.; Clappier, E.; Cayuela, J.-M.; Regnault, A.; García-Peydró, M.; Dombret, H.; Baruchel, A.; Toribio, M.-L.; Sigaux, F. HOXA Genes Are Included in Genetic and Biologic Networks Defining Human Acute T-Cell Leukemia (T-ALL). Blood 2005, 106, 274–286. [Google Scholar] [CrossRef]
- Homminga, I.; Pieters, R.; Langerak, A.W.; de Rooi, J.J.; Stubbs, A.; Verstegen, M.; Vuerhard, M.; Buijs-Gladdines, J.; Kooi, C.; Klous, P.; et al. Integrated Transcript and Genome Analyses Reveal NKX2-1 and MEF2C as Potential Oncogenes in T Cell Acute Lymphoblastic Leukemia. Cancer Cell 2011, 19, 484–497. [Google Scholar] [CrossRef]
- Gutierrez, A.; Dahlberg, S.E.; Neuberg, D.S.; Zhang, J.; Grebliunaite, R.; Sanda, T.; Protopopov, A.; Tosello, V.; Kutok, J.; Larson, R.S.; et al. Absence of Biallelic TCRγ Deletion Predicts Early Treatment Failure in Pediatric T-Cell Acute Lymphoblastic Leukemia. JCO 2010, 28, 3816–3823. [Google Scholar] [CrossRef]
- Coustan-Smith, E.; Mullighan, C.G.; Onciu, M.; Behm, F.G.; Raimondi, S.C.; Pei, D.; Cheng, C.; Su, X.; Rubnitz, J.E.; Basso, G.; et al. Early T-Cell Precursor Leukaemia: A Subtype of Very High-Risk Acute Lymphoblastic Leukaemia. Lancet Oncol. 2009, 10, 147–156. [Google Scholar] [CrossRef]
- Van Vlierberghe, P.; Ambesi-Impiombato, A.; De Keersmaecker, K.; Hadler, M.; Paietta, E.; Tallman, M.S.; Rowe, J.M.; Forne, C.; Rue, M.; Ferrando, A.A. Prognostic Relevance of Integrated Genetic Profiling in Adult T-Cell Acute Lymphoblastic Leukemia. Blood 2013, 122, 74–82. [Google Scholar] [CrossRef]
- Vicente, C.; Schwab, C.; Broux, M.; Geerdens, E.; Degryse, S.; Demeyer, S.; Lahortiga, I.; Elliott, A.; Chilton, L.; La Starza, R.; et al. Targeted Sequencing Identifies Associations between IL7R-JAK Mutations and Epigenetic Modulators in T-Cell Acute Lymphoblastic Leukemia. Haematologica 2015, 100, 1301–1310. [Google Scholar] [CrossRef]
- Genescà, E.; Lazarenkov, A.; Morgades, M.; Berbis, G.; Ruíz-Xivillé, N.; Gómez-Marzo, P.; Ribera, J.; Juncà, J.; González-Pérez, A.; Mercadal, S.; et al. Frequency and Clinical Impact of CDKN2A/ARF/CDKN2B Gene Deletions as Assessed by in-Depth Genetic Analyses in Adult T Cell Acute Lymphoblastic Leukemia. J. Hematol. Oncol. 2018, 11, 96. [Google Scholar] [CrossRef]
- Van Vlierberghe, P.; Ambesi-Impiombato, A.; Perez-Garcia, A.; Haydu, J.E.; Rigo, I.; Hadler, M.; Tosello, V.; Della Gatta, G.; Paietta, E.; Racevskis, J.; et al. ETV6 Mutations in Early Immature Human T Cell Leukemias. J. Exp. Med. 2011, 208, 2571–2579. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ding, L.; Holmfeldt, L.; Wu, G.; Heatley, S.L.; Payne-Turner, D.; Easton, J.; Chen, X.; Wang, J.; Rusch, M.; et al. The Genetic Basis of Early T-Cell Precursor Acute Lymphoblastic Leukaemia. Nature 2012, 481, 157–163. [Google Scholar] [CrossRef]
- Neumann, M.; Heesch, S.; Schlee, C.; Schwartz, S.; Gökbuget, N.; Hoelzer, D.; Konstandin, N.P.; Ksienzyk, B.; Vosberg, S.; Graf, A.; et al. Whole-Exome Sequencing in Adult ETP-ALL Reveals a High Rate of DNMT3A Mutations. Blood 2013, 121, 4749–4752. [Google Scholar] [CrossRef] [PubMed]
- Bond, J.; Graux, C.; Lhermitte, L.; Lara, D.; Cluzeau, T.; Leguay, T.; Cieslak, A.; Trinquand, A.; Pastoret, C.; Belhocine, M.; et al. Early Response-Based Therapy Stratification Improves Survival in Adult Early Thymic Precursor Acute Lymphoblastic Leukemia: A Group for Research on Adult Acute Lymphoblastic Leukemia Study. J. Clin. Oncol. 2017, 35, 2683–2691. [Google Scholar] [CrossRef]
- Liu, Y.; Easton, J.; Shao, Y.; Maciaszek, J.; Wang, Z.; Wilkinson, M.R.; McCastlain, K.; Edmonson, M.; Pounds, S.B.; Shi, L.; et al. The Genomic Landscape of Pediatric and Young Adult T-Lineage Acute Lymphoblastic Leukemia. Nat. Genet. 2017, 49, 1211–1218. [Google Scholar] [CrossRef]
- Gachet, S.; El-Chaar, T.; Avran, D.; Genesca, E.; Catez, F.; Quentin, S.; Delord, M.; Thérizols, G.; Briot, D.; Meunier, G.; et al. Deletion 6q Drives T-Cell Leukemia Progression by Ribosome Modulation. Cancer Discov. 2018, 8, 1614–1631. [Google Scholar] [CrossRef] [PubMed]
- Herranz, D.; Ambesi-Impiombato, A.; Palomero, T.; Schnell, S.A.; Belver, L.; Wendorff, A.A.; Xu, L.; Castillo-Martin, M.; Llobet-Navás, D.; Cordon-Cardo, C.; et al. A NOTCH1-Driven MYC Enhancer Promotes T Cell Development, Transformation and Acute Lymphoblastic Leukemia. Nat. Med. 2014, 20, 1130–1137. [Google Scholar] [CrossRef]
- Mansour, M.R.; Abraham, B.J.; Anders, L.; Berezovskaya, A.; Gutierrez, A.; Durbin, A.D.; Etchin, J.; Lawton, L.; Sallan, S.E.; Silverman, L.B.; et al. Oncogene Regulation. An Oncogenic Super-Enhancer Formed through Somatic Mutation of a Noncoding Intergenic Element. Science 2014, 346, 1373–1377. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Qian, M.; Zhang, H.; Guo, Y.; Yang, J.; Zhao, X.; He, H.; Lu, J.; Pan, J.; Chang, M.; et al. Whole-Genome Noncoding Sequence Analysis in T-Cell Acute Lymphoblastic Leukemia Identifies Oncogene Enhancer Mutations. Blood 2017, 129, 3264–3268. [Google Scholar] [CrossRef]
- Li, Z.; Abraham, B.J.; Berezovskaya, A.; Farah, N.; Liu, Y.; Leon, T.; Fielding, A.; Tan, S.H.; Sanda, T.; Weintraub, A.S.; et al. APOBEC Signature Mutation Generates an Oncogenic Enhancer That Drives LMO1 Expression in T-ALL. Leukemia 2017, 31, 2057–2064. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Magnussen, M.; León, T.E.; Farah, N.; Li, Z.; Abraham, B.J.; Alapi, K.Z.; Mitchell, R.J.; Naughton, T.; Fielding, A.K.; et al. Activation of the LMO2 Oncogene through a Somatically Acquired Neomorphic Promoter in T-Cell Acute Lymphoblastic Leukemia. Blood 2017, 129, 3221–3226. [Google Scholar] [CrossRef] [PubMed]
- Tottone, L.; Lancho, O.; Loh, J.-W.; Singh, A.; Kimura, S.; Roels, J.; Kuchmiy, A.; Strubbe, S.; Lawlor, M.A.; da Silva-Diz, V.; et al. A Tumor Suppressor Enhancer of PTEN in T-Cell Development and Leukemia. Blood Cancer Discov. 2021, 2, 92–109. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; IARC Publications: Lyon, France, 2018; ISBN 978-92-832-4494-3. [Google Scholar]
- Morita, K.; Jain, N.; Kantarjian, H.; Takahashi, K.; Fang, H.; Konopleva, M.; El Hussein, S.; Wang, F.; Short, N.J.; Maiti, A.; et al. Outcome of T-Cell Acute Lymphoblastic Leukemia/Lymphoma: Focus on near-ETP Phenotype and Differential Impact of Nelarabine. Am. J. Hematol. 2021, 96, 589–598. [Google Scholar] [CrossRef]
- Bond, J.; Touzart, A.; Leprêtre, S.; Graux, C.; Bargetzi, M.; Lhermitte, L.; Hypolite, G.; Leguay, T.; Hicheri, Y.; Guillerm, G.; et al. DNMT3A Mutation Is Associated with Increased Age and Adverse Outcome in Adult T-Cell Acute Lymphoblastic Leukemia. Haematologica 2019, 104, 1617–1625. [Google Scholar] [CrossRef]
- González-Gil, C.; Morgades, M.; Fuster-Tormo, F.; García-Chica, J.; Montesinos, P.; Torrent, A.; Diaz-Beyá, M.; Coll, R.; Ribera, J.; Zhao, R.; et al. Genomic Data Improves Prognostic Stratification in Adult T-Cell Acute Lymphoblastic Leukemia Patients Enrolled in Measurable Residual Disease-Oriented Trials. Blood 2021, 138, 3486. [Google Scholar] [CrossRef]
- Bond, J.; Marchand, T.; Touzart, A.; Cieslak, A.; Trinquand, A.; Sutton, L.; Radford-Weiss, I.; Lhermitte, L.; Spicuglia, S.; Dombret, H.; et al. An Early Thymic Precursor Phenotype Predicts Outcome Exclusively in HOXA-Overexpressing Adult T-Cell Acute Lymphoblastic Leukemia: A Group for Research in Adult Acute Lymphoblastic Leukemia Study. Haematologica 2016, 101, 732–740. [Google Scholar] [CrossRef]
- Ben Abdelali, R.; Asnafi, V.; Petit, A.; Micol, J.-B.; Callens, C.; Villarese, P.; Delabesse, E.; Reman, O.; Lepretre, S.; Cahn, J.-Y.; et al. The Prognosis of CALM-AF10-Positive Adult T-Cell Acute Lymphoblastic Leukemias Depends on the Stage of Maturation Arrest. Haematologica 2013, 98, 1711–1717. [Google Scholar] [CrossRef] [PubMed]
- Montefiori, L.E.; Bendig, S.; Gu, Z.; Chen, X.; Pölönen, P.; Ma, X.; Murison, A.; Zeng, A.; Garcia-Prat, L.; Dickerson, K.; et al. Enhancer Hijacking Drives Oncogenic BCL11B Expression in Lineage-Ambiguous Stem Cell Leukemia. Cancer Discov. 2021, 11, 2846–2867. [Google Scholar] [CrossRef] [PubMed]
- Di Giacomo, D.; La Starza, R.; Gorello, P.; Pellanera, F.; Kalender Atak, Z.; De Keersmaecker, K.; Pierini, V.; Harrison, C.J.; Arniani, S.; Moretti, M.; et al. 14q32 Rearrangements Deregulating BCL11B Mark a Distinct Subgroup of T-Lymphoid and Myeloid Immature Acute Leukemia. Blood 2021, 138, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Alexander, T.B.; Gu, Z.; Iacobucci, I.; Dickerson, K.; Choi, J.K.; Xu, B.; Payne-Turner, D.; Yoshihara, H.; Loh, M.L.; Horan, J.; et al. The Genetic Basis and Cell of Origin of Mixed Phenotype Acute Leukaemia. Nature 2018, 562, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Wang, F.; Morita, K.; Yan, Y.; Hu, P.; Zhao, P.; Zhar, A.A.; Wu, C.J.; Gumbs, C.; Little, L.; et al. Integrative Genomic Analysis of Adult Mixed Phenotype Acute Leukemia Delineates Lineage Associated Molecular Subtypes. Nat. Commun. 2018, 9, 2670. [Google Scholar] [CrossRef]
- Huether, R.; Dong, L.; Chen, X.; Wu, G.; Parker, M.; Wei, L.; Ma, J.; Edmonson, M.N.; Hedlund, E.K.; Rusch, M.C.; et al. The Landscape of Somatic Mutations in Epigenetic Regulators across 1,000 Paediatric Cancer Genomes. Nat. Commun. 2014, 5, 3630. [Google Scholar] [CrossRef]
- Christman, J.K. 5-Azacytidine and 5-Aza-2’-Deoxycytidine as Inhibitors of DNA Methylation: Mechanistic Studies and Their Implications for Cancer Therapy. Oncogene 2002, 21, 5483–5495. [Google Scholar] [CrossRef]
- Batova, A.; Diccianni, M.B.; Yu, J.C.; Nobori, T.; Link, M.P.; Pullen, J.; Yu, A.L. Frequent and Selective Methylation of P15 and Deletion of Both P15 and P16 in T-Cell Acute Lymphoblastic Leukemia. Cancer Res. 1997, 57, 832–836. [Google Scholar] [PubMed]
- Tsellou, E.; Troungos, C.; Moschovi, M.; Athanasiadou-Piperopoulou, F.; Polychronopoulou, S.; Kosmidis, H.; Kalmanti, M.; Hatzakis, A.; Dessypris, N.; Kalofoutis, A.; et al. Hypermethylation of CpG Islands in the Promoter Region of the P15INK4B Gene in Childhood Acute Leukaemia. Eur. J. Cancer 2005, 41, 584–589. [Google Scholar] [CrossRef]
- Takeuchi, S.; Matsushita, M.; Zimmermann, M.; Ikezoe, T.; Komatsu, N.; Seriu, T.; Schrappe, M.; Bartram, C.R.; Koeffler, H.P. Clinical Significance of Aberrant DNA Methylation in Childhood Acute Lymphoblastic Leukemia. Leuk. Res. 2011, 35, 1345–1349. [Google Scholar] [CrossRef][Green Version]
- Chim, C.S.; Tam, C.Y.; Liang, R.; Kwong, Y.L. Methylation of P15 and P16 Genes in Adult Acute Leukemia: Lack of Prognostic Significance. Cancer 2001, 91, 2222–2229. [Google Scholar] [CrossRef]
- Garcia-Manero, G.; Bueso-Ramos, C.; Daniel, J.; Williamson, J.; Kantarjian, H.M.; Issa, J.-P.J. DNA Methylation Patterns at Relapse in Adult Acute Lymphocytic Leukemia. Clin. Cancer Res. 2002, 8, 1897–1903. [Google Scholar]
- Bueso-Ramos, C.; Xu, Y.; McDonnell, T.J.; Brisbay, S.; Pierce, S.; Kantarjian, H.; Rosner, G.; Garcia-Manero, G. Protein Expression of a Triad of Frequently Methylated Genes, P73, P57Kip2, and P15, Has Prognostic Value in Adult Acute Lymphocytic Leukemia Independently of Its Methylation Status. J. Clin. Oncol. 2005, 23, 3932–3939. [Google Scholar] [CrossRef]
- Grossmann, V.; Haferlach, C.; Weissmann, S.; Roller, A.; Schindela, S.; Poetzinger, F.; Stadler, K.; Bellos, F.; Kern, W.; Haferlach, T.; et al. The Molecular Profile of Adult T-Cell Acute Lymphoblastic Leukemia: Mutations in RUNX1 and DNMT3A Are Associated with Poor Prognosis in T-ALL. Genes Chromosom. Cancer 2013, 52, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.; Park, J.; Kwon, A.; Choi, H.; Kim, J.; Lee, G.D.; Han, E.; Jekarl, D.W.; Chae, H.; Han, K.; et al. CDKN2B Downregulation and Other Genetic Characteristics in T-Acute Lymphoblastic Leukemia. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Borssén, M.; Palmqvist, L.; Karrman, K.; Abrahamsson, J.; Behrendtz, M.; Heldrup, J.; Forestier, E.; Roos, G.; Degerman, S. Promoter DNA Methylation Pattern Identifies Prognostic Subgroups in Childhood T-Cell Acute Lymphoblastic Leukemia. PLoS ONE 2013, 8, e65373. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Seki, M.; Kawai, T.; Goto, H.; Yoshida, K.; Isobe, T.; Sekiguchi, M.; Watanabe, K.; Kubota, Y.; Nannya, Y.; et al. DNA Methylation-Based Classification Reveals Difference between Pediatric T-Cell Acute Lymphoblastic Leukemia and Normal Thymocytes. Leukemia 2020, 34, 1163–1168. [Google Scholar] [CrossRef]
- Touzart, A.; Boissel, N.; Belhocine, M.; Smith, C.; Graux, C.; Latiri, M.; Lhermitte, L.; Mathieu, E.-L.; Huguet, F.; Lamant, L.; et al. Low Level CpG Island Promoter Methylation Predicts a Poor Outcome in Adult T-Cell Acute Lymphoblastic Leukemia. Haematologica 2020, 105, 1575–1581. [Google Scholar] [CrossRef]
- Roels, J.; Thénoz, M.; Szarzyńska, B.; Landfors, M.; De Coninck, S.; Demoen, L.; Provez, L.; Kuchmiy, A.; Strubbe, S.; Reunes, L.; et al. Aging of Preleukemic Thymocytes Drives CpG Island Hypermethylation in T-Cell Acute Lymphoblastic Leukemia. Blood Cancer Discov. 2020, 1, 274–289. [Google Scholar] [CrossRef] [PubMed]
- Haider, Z.; Larsson, P.; Landfors, M.; Köhn, L.; Schmiegelow, K.; Flægstad, T.; Kanerva, J.; Heyman, M.; Hultdin, M.; Degerman, S. An Integrated Transcriptome Analysis in T-cell Acute Lymphoblastic Leukemia Links DNA Methylation Subgroups to Dysregulated TAL1 and ANTP Homeobox Gene Expression. Cancer Med. 2018, 8, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Maćkowska, N.; Drobna-Śledzińska, M.; Witt, M.; Dawidowska, M. DNA Methylation in T-Cell Acute Lymphoblastic Leukemia: In Search for Clinical and Biological Meaning. Int. J. Mol. Sci. 2021, 22, 1388. [Google Scholar] [CrossRef]
- Newman, S.; Nakitandwe, J.; Kesserwan, C.A.; Azzato, E.M.; Wheeler, D.A.; Rusch, M.; Shurtleff, S.; Hedges, D.J.; Hamilton, K.V.; Foy, S.G.; et al. Genomes for Kids: The Scope of Pathogenic Mutations in Pediatric Cancer Revealed by Comprehensive DNA and RNA Sequencing. Cancer Discov. 2021, 11, 3008–3027. [Google Scholar] [CrossRef]
- Bodmer, W.; Tomlinson, I. Rare Genetic Variants and the Risk of Cancer. Curr. Opin. Genet. Dev. 2010, 20, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Rahman, N. Realizing the Promise of Cancer Predisposition Genes. Nature 2014, 505, 302–308. [Google Scholar] [CrossRef]
- Huang, K.-L.; Mashl, R.J.; Wu, Y.; Ritter, D.I.; Wang, J.; Oh, C.; Paczkowska, M.; Reynolds, S.; Wyczalkowski, M.A.; Oak, N.; et al. Pathogenic Germline Variants in 10,389 Adult Cancers. Cell 2018, 173, 355–370.e14. [Google Scholar] [CrossRef]
- Qian, M.; Zhao, X.; Devidas, M.; Yang, W.; Gocho, Y.; Smith, C.; Gastier-Foster, J.M.; Li, Y.; Xu, H.; Zhang, S.; et al. Genome-Wide Association Study of Susceptibility Loci for T-Cell Acute Lymphoblastic Leukemia in Children. J. Natl. Cancer Inst. 2019, 111, 1350–1357. [Google Scholar] [CrossRef]
- Liberzon, E.; Avigad, S.; Stark, B.; Zilberstein, J.; Freedman, L.; Gorfine, M.; Gavriel, H.; Cohen, I.J.; Goshen, Y.; Yaniv, I.; et al. Germ-Line ATM Gene Alterations Are Associated with Susceptibility to Sporadic T-Cell Acute Lymphoblastic Leukemia in Children. Genes Chromosom. Cancer 2004, 39, 161–166. [Google Scholar] [CrossRef]
- Yoshida, N.; Sakaguchi, H.; Muramatsu, H.; Okuno, Y.; Song, C.; Dovat, S.; Shimada, A.; Ozeki, M.; Ohnishi, H.; Teramoto, T.; et al. Germline IKAROS Mutation Associated with Primary Immunodeficiency That Progressed to T-Cell Acute Lymphoblastic Leukemia. Leukemia 2017, 31, 1221–1223. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.L.; Arts, P.; Carmichael, C.L.; Babic, M.; Dobbins, J.; Chong, C.-E.; Schreiber, A.W.; Feng, J.; Phillips, K.; Wang, P.P.S.; et al. RUNX1-Mutated Families Show Phenotype Heterogeneity and a Somatic Mutation Profile Unique to Germline Predisposed AML. Blood Adv. 2020, 4, 1131–1144. [Google Scholar] [CrossRef]
- Li, Y.; Yang, W.; Devidas, M.; Winter, S.S.; Kesserwan, C.; Yang, W.; Dunsmore, K.P.; Smith, C.; Qian, M.; Zhao, X.; et al. Germline RUNX1 Variation and Predisposition to Childhood Acute Lymphoblastic Leukemia. J. Clin. Invest. 2021, 131, e147898. [Google Scholar] [CrossRef]
- Tulstrup, M.; Grosjean, M.; Nielsen, S.N.; Grell, K.; Wolthers, B.O.; Wegener, P.S.; Jonsson, O.G.; Lund, B.; Harila-Saari, A.; Abrahamsson, J.; et al. NT5C2 Germline Variants Alter Thiopurine Metabolism and Are Associated with Acquired NT5C2 Relapse Mutations in Childhood Acute Lymphoblastic Leukaemia. Leukemia 2018, 32, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Yang, W.; Moriyama, T.; Liu, C.; Smith, C.; Yang, W.; Qian, M.; Li, Z.; Tulstrup, M.; Schmiegelow, K.; et al. Effects of NT5C2 Germline Variants on 6-Mecaptopurine Metabolism in Children With Acute Lymphoblastic Leukemia. Clin. Pharmacol. Ther. 2021, 109, 1538–1545. [Google Scholar] [CrossRef] [PubMed]
- Godley, L.A. Inherited Predisposition to Acute Myeloid Leukemia. Semin. Hematol. 2014, 51, 306–321. [Google Scholar] [CrossRef]
- Antony-Debré, I.; Duployez, N.; Bucci, M.; Geffroy, S.; Micol, J.-B.; Renneville, A.; Boissel, N.; Dhédin, N.; Réa, D.; Nelken, B.; et al. Somatic Mutations Associated with Leukemic Progression of Familial Platelet Disorder with Predisposition to Acute Myeloid Leukemia. Leukemia 2016, 30, 999–1002. [Google Scholar] [CrossRef]
- Spychała, J.; Madrid-Marina, V.; Fox, I.H. High Km Soluble 5’-Nucleotidase from Human Placenta. Properties and Allosteric Regulation by IMP and ATP. J. Biol. Chem. 1988, 263, 18759–18765. [Google Scholar] [CrossRef]
- Gazziola, C.; Ferraro, P.; Moras, M.; Reichard, P.; Bianchi, V. Cytosolic High K(m) 5’-Nucleotidase and 5’(3’)-Deoxyribonucleotidase in Substrate Cycles Involved in Nucleotide Metabolism. J. Biol. Chem. 2001, 276, 6185–6190. [Google Scholar] [CrossRef]
- Moriyama, T.; Liu, S.; Li, J.; Meyer, J.; Zhao, X.; Yang, W.; Shao, Y.; Heath, R.; Hnízda, A.; Carroll, W.L.; et al. Mechanisms of NT5C2-Mediated Thiopurine Resistance in Acute Lymphoblastic Leukemia. Mol. Cancer Ther. 2019, 18, 1887–1895. [Google Scholar] [CrossRef]
- Cooper, J.N.; Young, N.S. Clonality in Context: Hematopoietic Clones in Their Marrow Environment. Blood 2017, 130, 2363–2372. [Google Scholar] [CrossRef] [PubMed]
- Welch, J.S.; Ley, T.J.; Link, D.C.; Miller, C.A.; Larson, D.E.; Koboldt, D.C.; Wartman, L.D.; Lamprecht, T.L.; Liu, F.; Xia, J.; et al. The Origin and Evolution of Mutations in Acute Myeloid Leukemia. Cell 2012, 150, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Zink, F.; Stacey, S.N.; Norddahl, G.L.; Frigge, M.L.; Magnusson, O.T.; Jonsdottir, I.; Thorgeirsson, T.E.; Sigurdsson, A.; Gudjonsson, S.A.; Gudmundsson, J.; et al. Clonal Hematopoiesis, with and without Candidate Driver Mutations, Is Common in the Elderly. Blood 2017, 130, 742–752. [Google Scholar] [CrossRef] [PubMed]
- Silver, A.J.; Jaiswal, S. Clonal Hematopoiesis: Pre-Cancer PLUS. Adv. Cancer Res. 2019, 141, 85–128. [Google Scholar] [CrossRef]
- Steensma, D.P.; Bejar, R.; Jaiswal, S.; Lindsley, R.C.; Sekeres, M.A.; Hasserjian, R.P.; Ebert, B.L. Clonal Hematopoiesis of Indeterminate Potential and Its Distinction from Myelodysplastic Syndromes. Blood 2015, 126, 9–16. [Google Scholar] [CrossRef]
- Busque, L.; Buscarlet, M.; Mollica, L.; Levine, R.L. Age-Related Clonal Hematopoiesis: Stem Cells Tempting the Devil. Stem Cells 2018, 36, 1287–1294. [Google Scholar] [CrossRef]
- Abelson, S.; Collord, G.; Ng, S.W.K.; Weissbrod, O.; Mendelson Cohen, N.; Niemeyer, E.; Barda, N.; Zuzarte, P.C.; Heisler, L.; Sundaravadanam, Y.; et al. Prediction of Acute Myeloid Leukaemia Risk in Healthy Individuals. Nature 2018, 559, 400–404. [Google Scholar] [CrossRef]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-Related Clonal Hematopoiesis Associated with Adverse Outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef]
- Desai, P.; Mencia-Trinchant, N.; Savenkov, O.; Simon, M.S.; Cheang, G.; Lee, S.; Samuel, M.; Ritchie, E.K.; Guzman, M.L.; Ballman, K.V.; et al. Somatic Mutations Precede Acute Myeloid Leukemia Years before Diagnosis. Nat. Med. 2018, 24, 1015–1023. [Google Scholar] [CrossRef]
- Oshima, K.; Khiabanian, H.; da Silva-Almeida, A.C.; Tzoneva, G.; Abate, F.; Ambesi-Impiombato, A.; Sanchez-Martin, M.; Carpenter, Z.; Penson, A.; Perez-Garcia, A.; et al. Mutational Landscape, Clonal Evolution Patterns, and Role of RAS Mutations in Relapsed Acute Lymphoblastic Leukemia. Proc. Natl. Acad. Sci. USA 2016, 113, 11306–11311. [Google Scholar] [CrossRef]
- Tzoneva, G.; Perez-Garcia, A.; Carpenter, Z.; Khiabanian, H.; Tosello, V.; Allegretta, M.; Paietta, E.; Racevskis, J.; Rowe, J.M.; Tallman, M.S.; et al. Activating Mutations in the NT5C2 Nucleotidase Gene Drive Chemotherapy Resistance in Relapsed ALL. Nat. Med. 2013, 19, 368–371. [Google Scholar] [CrossRef]
- Oshima, K.; Zhao, J.; Pérez-Durán, P.; Brown, J.A.; Patiño-Galindo, J.A.; Chu, T.; Quinn, A.; Gunning, T.; Belver, L.; Ambesi-Impiombato, A.; et al. Mutational and Functional Genetics Mapping of Chemotherapy Resistance Mechanisms in Relapsed Acute Lymphoblastic Leukemia. Nat. Cancer 2020, 1, 1113–1127. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.J.R.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.-L.; et al. Signatures of Mutational Processes in Human Cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Brady, S.W.; Ma, X.; Shen, S.; Zhang, Y.; Li, Y.; Szlachta, K.; Dong, L.; Liu, Y.; Yang, F.; et al. Therapy-Induced Mutations Drive the Genomic Landscape of Relapsed Acute Lymphoblastic Leukemia. Blood 2020, 135, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Richter-Pechańska, P.; Kunz, J.B.; Hof, J.; Zimmermann, M.; Rausch, T.; Bandapalli, O.R.; Orlova, E.; Scapinello, G.; Sagi, J.C.; Stanulla, M.; et al. Identification of a Genetically Defined Ultra-High-Risk Group in Relapsed Pediatric T-Lymphoblastic Leukemia. Blood Cancer J. 2017, 7, e523. [Google Scholar] [CrossRef]
- Sentís, I.; Gonzalez, S.; Genescà, E.; García-Hernández, V.; Muiños, F.; Gonzalez, C.; López-Arribillaga, E.; Gonzalez, J.; Fernandez-Ibarrondo, L.; Mularoni, L.; et al. The Evolution of Relapse of Adult T Cell Acute Lymphoblastic Leukemia. Genome Biol. 2020, 21, 1–24. [Google Scholar] [CrossRef]
- Waanders, E.; Gu, Z.; Dobson, S.M.; Antić, Ž.; Crawford, J.C.; Ma, X.; Edmonson, M.N.; Payne-Turner, D.; van de Vorst, M.; Jongmans, M.C.J.; et al. Mutational Landscape and Patterns of Clonal Evolution in Relapsed Pediatric Acute Lymphoblastic Leukemia. Blood Cancer Discov. 2020, 1, 96–111. [Google Scholar] [CrossRef]
- Gaynon, P.S.; Orgel, E.; Ji, L. Preexisting or Therapy-Induced Mutations in Relapsed Acute Lymphoblastic Leukemia? Blood 2020, 136, 2233–2235. [Google Scholar] [CrossRef]
- Pellegrino, M.; Sciambi, A.; Yates, J.L.; Mast, J.D.; Silver, C.; Eastburn, D.J. RNA-Seq Following PCR-Based Sorting Reveals Rare Cell Transcriptional Signatures. BMC Genom. 2016, 17, 361. [Google Scholar] [CrossRef]
- Pellegrino, M.; Sciambi, A.; Treusch, S.; Durruthy-Durruthy, R.; Gokhale, K.; Jacob, J.; Chen, T.X.; Geis, J.A.; Oldham, W.; Matthews, J.; et al. High-Throughput Single-Cell DNA Sequencing of Acute Myeloid Leukemia Tumors with Droplet Microfluidics. Genome Res. 2018, 28, 1345–1352. [Google Scholar] [CrossRef]
- Albertí-Servera, L.; Demeyer, S.; Govaerts, I.; Swings, T.; De Bie, J.; Gielen, O.; Brociner, M.; Michaux, L.; Maertens, J.; Uyttebroeck, A.; et al. Single-Cell DNA Amplicon Sequencing Reveals Clonal Heterogeneity and Evolution in T-Cell Acute Lymphoblastic Leukemia. Blood 2021, 137, 801–811. [Google Scholar] [CrossRef] [PubMed]
| Gene | Affected Region | Variant | Alteration | Functional Impact | Frequency | Reference |
|---|---|---|---|---|---|---|
| MYC | 1427 kb downstream of MYC | Focal duplications | Creation of binding site for NOTCH1 | MYC expression | 8/160 (5%) Adult and pediatric | [34] |
| TAL1 | 8 kb upstream of the transcription start site of TAL1 | Heterozygous indel (2–18 bp) | Creation of binding motifs for the MYB TF | TAL1 overexpression | 8/146 (5.5%) pediatric | [35,36] |
| LMO1 | 4 kb upstream of the transcriptional start site of LMO1 | SNV: C → T | Creation of binding motifs for the MYB TF | LMO1 overexpression | 4/187 (2.14%) pediatric | [36,37] |
| LMO2 | Non-coding region of the exon 2 of LMO2 | Heterozygous indel | Creation of binding motifs for the MYB TF | Activating LMO2 function | 6/160 (3.75%) pediatric 9/163 (5.52%) adult | [38] |
| PTEN | 550 kb downstream of transcription start site of PTEN | Focal deletions | Deletion of PTEN enhancer region | Reduced levels of PTEN | 5/398 (1.25%) | [39] |
| Gene | Type of Alteration | SNP ID | Alteration | Association with T-ALL: Odds Ratio (P) | Functional Impact | Reference |
|---|---|---|---|---|---|---|
| Predisposition alterations contributing to the development of T-ALL | ||||||
| USP7 | allele | rs74010351 | wt allele → A Risk allele → G | Discovery cohort → 1.44 (4.51 × 10−8) | Downregulation of USP7 transcription. Risk allele associated with Higher levels of African ancestry and older age at diagnosis | [70] |
| Validation cohort → 1.51 (0.04) | ||||||
| ATM | variant | Truncation mutations: R35X 30del215 228delCT | 12.9 (0.004) | Aberrant ATM protein production → prone to T-ALL | [71] | |
| Missense mutations: V410A F582L F143C | 4.9 (0.03) | |||||
| IKZF1 | variant | N159S | - | Impaired recognition of the target DNA sequences by IKAROS | [72] | |
| RUNX1 | variant | 36171607G > A 36231773 C > T | Risk to develop T-ALL | [73] | ||
| - | p.K117* p.A142fs p.S213fs p.R233fs p.Y287* G365R | - | Loss of transcription factor activity | [74] | ||
| Predisposition alterations affecting drug response and treatment | ||||||
| NT5C2 | allele | rs72846714 | wt allele → A Risk allele → G | - | Higher level of expression of NT5C2 and lower level of TGN in erythrocytes. | [75,76] |
| rs58700372 | wt allele → T Risk allele → C | Activation of NT5C2 transcription and reduction of 6-MP metabolism | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Genescà, E.; González-Gil, C. Latest Contributions of Genomics to T-Cell Acute Lymphoblastic Leukemia (T-ALL). Cancers 2022, 14, 2474. https://doi.org/10.3390/cancers14102474
Genescà E, González-Gil C. Latest Contributions of Genomics to T-Cell Acute Lymphoblastic Leukemia (T-ALL). Cancers. 2022; 14(10):2474. https://doi.org/10.3390/cancers14102474
Chicago/Turabian StyleGenescà, Eulàlia, and Celia González-Gil. 2022. "Latest Contributions of Genomics to T-Cell Acute Lymphoblastic Leukemia (T-ALL)" Cancers 14, no. 10: 2474. https://doi.org/10.3390/cancers14102474
APA StyleGenescà, E., & González-Gil, C. (2022). Latest Contributions of Genomics to T-Cell Acute Lymphoblastic Leukemia (T-ALL). Cancers, 14(10), 2474. https://doi.org/10.3390/cancers14102474






