Low BACH2 Expression Predicts Adverse Outcome in Chronic Lymphocytic Leukaemia
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
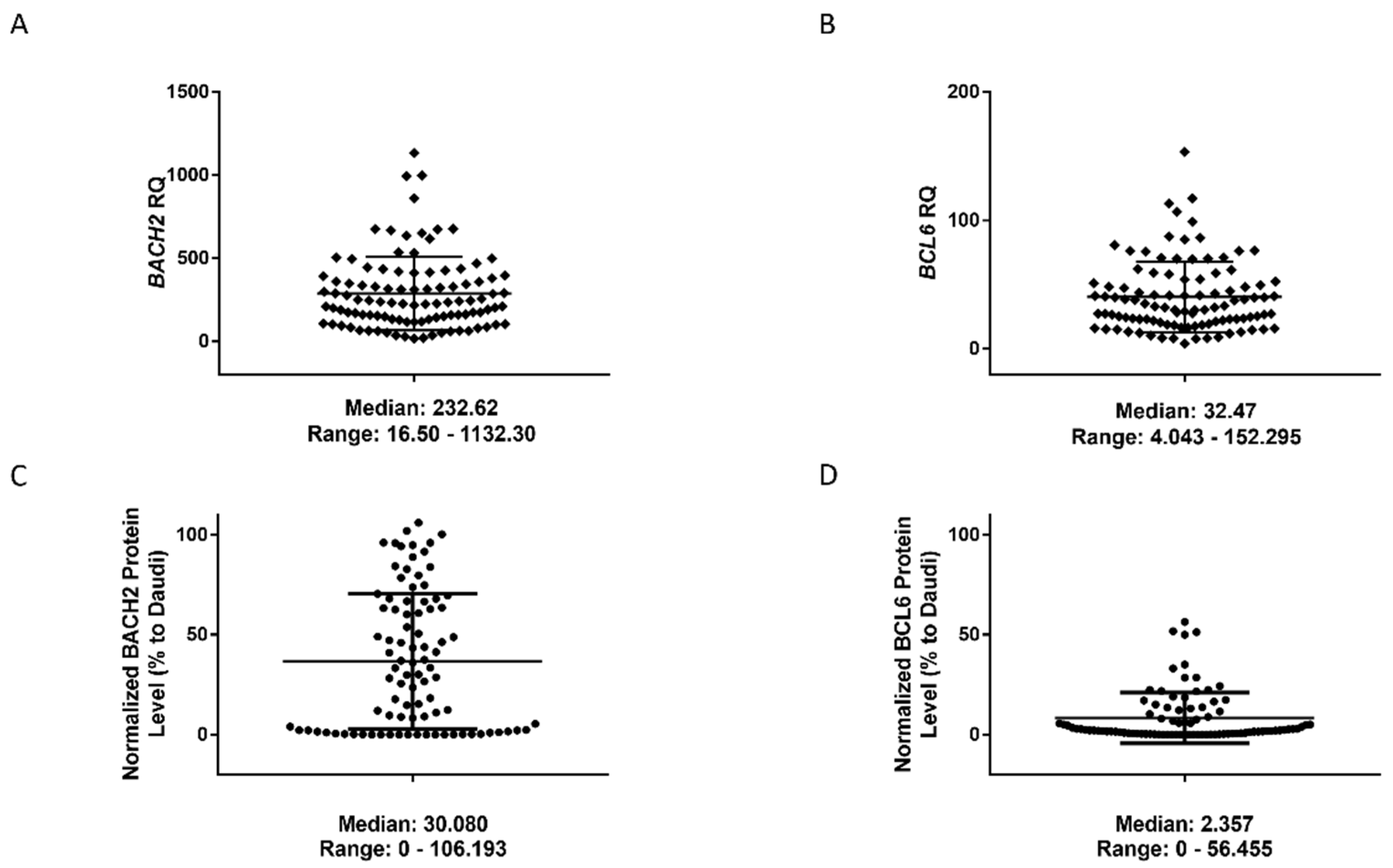
3. Results
3.1. BACH2 and BCL6 Low RNA Expression Predicts Shorter Overall Survival in CLL
3.2. BACH2 and BCL6 Low RNA Expression Predicts Shorter Overall Survival in IGHV-M, 13q-, and 11q-Deleted Patient Subgroups of CLL
3.3. BACH2 Is a Promising Independent and Integrative Predictor of Outcome for CLL
3.4. Mapping of BACH2 and BCL6 Mediated Signalling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zenz, T.; Mertens, D.; Kuppers, R.; Dohner, H.; Stilgenbauer, S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat. Rev. Cancer 2010, 10, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Kipps, T.J. Immunoglobulin genes in chronic lymphocytic leukemia. Blood Cells 1993, 19, 615–625; discussion 612–631. [Google Scholar] [PubMed]
- Fais, F.; Ghiotto, F.; Hashimoto, S.; Sellars, B.; Valetto, A.; Allen, S.L.; Schulman, P.; Vinciguerra, V.P.; Rai, K.; Rassenti, L.Z.; et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J. Clin. Investig. 1998, 102, 1515–1525. [Google Scholar] [CrossRef] [PubMed]
- Seifert, M.; Sellmann, L.; Bloehdorn, J.; Wein, F.; Stilgenbauer, S.; Durig, J.; Kuppers, R. Cellular origin and pathophysiology of chronic lymphocytic leukemia. J. Exp. Med. 2012, 209, 2183–2198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuppers, R.; Sousa, A.B.; Baur, A.S.; Strickler, J.G.; Rajewsky, K.; Hansmann, M.L. Common germinal-center B-cell origin of the malignant cells in two composite lymphomas, involving classical Hodgkin’s disease and either follicular lymphoma or B-CLL. Mol. Med. 2001, 7, 285–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kay, N.E.; O’Brien, S.M.; Pettitt, A.R.; Stilgenbauer, S. The role of prognostic factors in assessing ‘high-risk’ subgroups of patients with chronic lymphocytic leukemia. Leukemia 2007, 21, 1885–1891. [Google Scholar] [CrossRef] [Green Version]
- Roychoudhuri, R.; Hirahara, K.; Mousavi, K.; Clever, D.; Klebanoff, C.A.; Bonelli, M.; Sciumè, G.; Zare, H.; Vahedi, G.; Dema, B.; et al. BACH2 represses effector programs to stabilize Treg-mediated immune homeostasis. Nature 2013, 498, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Muto, A.; Ochiai, K.; Kimura, Y.; Itoh-Nakadai, A.; Calame, K.L.; Ikebe, D.; Tashiro, S.; Igarashi, K. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 2010, 29, 4048–4061. [Google Scholar] [CrossRef] [Green Version]
- Itoh-Nakadai, A.; Hikota, R.; Muto, A.; Kometani, K.; Watanabe-Matsui, M.; Sato, Y.; Kobayashi, M.; Nakamura, A.; Miura, Y.; Yano, Y.; et al. The transcription repressors Bach2 and Bach1 promote B cell development by repressing the myeloid program. Nat. Immunol. 2014, 15, 1171–1180. [Google Scholar] [CrossRef]
- Huang, C.; Geng, H.; Boss, I.; Wang, L.; Melnick, A. Cooperative transcriptional repression by BCL6 and BACH2 in germinal center B-cell differentiation. Blood 2014, 123, 1012–1020. [Google Scholar] [CrossRef]
- Hatzi, K.; Melnick, A. Breaking bad in the germinal center: How deregulation of BCL6 contributes to lymphomagenesis. Trends Mol. Med. 2014, 20, 343–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swaminathan, S.; Huang, C.; Geng, H.; Chen, Z.; Harvey, R.; Kang, H.; Ng, C.; Titz, B.; Hurtz, C.; Sadiyah, M.F.; et al. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat. Med. 2013, 19, 1014–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Grubor, V.; Love, C.L.; Banerjee, A.; Richards, K.L.; Mieczkowski, P.A.; Dunphy, C.; Choi, W.; Au, W.Y.; Srivastava, G.; et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1398–1403. [Google Scholar] [CrossRef] [Green Version]
- Ichikawa, S.; Fukuhara, N.; Katsushima, H.; Takahashi, T.; Yamamoto, J.; Yokoyama, H.; Sasaki, O.; Fukuhara, O.; Nomura, J.; Ishizawa, K.; et al. Association between BACH2 expression and clinical prognosis in diffuse large B-cell lymphoma. Cancer Sci. 2014, 105, 437–444. [Google Scholar] [CrossRef]
- Hunter, J.E.; Butterworth, J.A.; Zhao, B.; Sellier, H.; Campbell, K.J.; Thomas, H.D.; Bacon, C.M.; Cockell, S.J.; Gewurz, B.E.; Perkins, N.D. The NF-kappaB subunit c-Rel regulates Bach2 tumour suppressor expression in B-cell lymphoma. Oncogene 2016, 35, 3476–3484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, V.L.D.; Garaud, S.; De Silva, P.; Thibaud, V.; Stamatopoulos, B.; Berehad, M.; Gu-Trantien, C.; Krayem, M.; Duvillier, H.; Lodewyckx, J.-N.; et al. Age-related changes in the BACH2 and PRDM1 genes in lymphocytes from healthy donors and chronic lymphocytic leukemia patients. BMC Cancer 2019, 19, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jantus Lewintre, E.; Reinoso Martin, C.; Garcia Ballesteros, C.; Pendas, J.; Benet Campos, C.; Mayans Ferrer, J.R.; Garcia-Conde, J. BCL6: Somatic mutations and expression in early-stage chronic lymphocytic leukemia. Leuk. Lymphoma 2009, 50, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.M.; Peters, A.; Baron, B.; Zhu, X.; Storb, U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science 1998, 280, 1750–1752. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Neri, A.; Baldini, L.; Dalla-Favera, R.; Migliazza, A. BCL-6 mutations are associated with immunoglobulin variable heavy chain mutations in B-cell chronic lymphocytic leukemia. Cancer Res. 2000, 60, 5644–5648. [Google Scholar]
- Smonskey, M.T.; Block, A.W.; Deeb, G.; Chanan-Khan, A.A.; Bernstein, Z.P.; Miller, K.C.; Wallace, P.K.; Starostik, P. Monoallelic and biallelic deletions of 13q14.3 in chronic lymphocytic leukemia: FISH vs. miRNA RT-qPCR detection. Am. J. Clin. Pathol. 2012, 137, 641–646. [Google Scholar] [CrossRef]
- Lanham, S.; Hamblin, T.; Oscier, D.; Ibbotson, R.; Stevenson, F.; Packham, G. Differential signaling via surface IgM is associated with VH gene mutational status and CD38 expression in chronic lymphocytic leukemia. Blood 2003, 101, 1087–1093. [Google Scholar] [CrossRef]
- Phan, R.T.; Dalla-Favera, R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 2004, 432, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Matthias, P.D. The Transcriptional Regulation of Germinal Center Formation. Front. Immunol. 2018, 9, 2026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.K.; Song, J.Y.; Koh, D.I.; Kim, J.Y.; Hatano, M.; Jeon, B.N.; Kim, M.Y.; Cho, S.Y.; Kim, K.S.; Hur, M.W. Reciprocal negative regulation between the tumor suppressor protein p53 and B cell CLL/lymphoma 6 (BCL6) via control of caspase-1 expression. J. Biol. Chem. 2019, 294, 299–313. [Google Scholar] [CrossRef] [Green Version]
- Rousseau, A.; Bertolotti, A. Regulation of proteasome assembly and activity in health and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 697–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masdehors, P.; Merle-Beral, H.; Magdelenat, H.; Delic, J. Ubiquitin-proteasome system and increased sensitivity of B-CLL lymphocytes to apoptotic death activation. Leuk. Lymphoma 2000, 38, 499–504. [Google Scholar] [CrossRef]
- Drake, J.R. The immunobiology of ubiquitin-dependent B cell receptor functions. Mol. Immunol. 2018, 101, 146–154. [Google Scholar] [CrossRef]
- Sahasrabuddhe, A.A.; Elenitoba-Johnson, K.S. Role of the ubiquitin proteasome system in hematologic malignancies. Immunol. Rev. 2015, 263, 224–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masdehors, P.; Omura, S.; Merle-Beral, H.; Mentz, F.; Cosset, J.M.; Dumont, J.; Magdelenat, H.; Delic, J. Increased sensitivity of CLL-derived lymphocytes to apoptotic death activation by the proteasome-specific inhibitor lactacystin. Br. J. Haematol. 1999, 105, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Pahler, J.C.; Ruiz, S.; Niemer, I.; Calvert, L.R.; Andreeff, M.; Keating, M.; Faderl, S.; McConkey, D.J. Effects of the proteasome inhibitor, bortezomib, on apoptosis in isolated lymphocytes obtained from patients with chronic lymphocytic leukemia. Clin. Cancer Res. 2003, 9, 4570–4577. [Google Scholar]
- Duan, S.; Cermak, L.; Pagan, J.K.; Rossi, M.; Martinengo, C.; di Celle, P.F.; Chapuy, B.; Shipp, M.; Chiarle, R.; Pagano, M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature 2012, 481, 90–93. [Google Scholar] [CrossRef] [Green Version]
- Deegan, T.D.; Diffley, J.F. MCM: One ring to rule them all. Curr. Opin. Struct. Biol. 2016, 37, 145–151. [Google Scholar] [CrossRef]
- Wei, R.; Liu, S.; Zhang, S.; Min, L.; Zhu, S. Cellular and Extracellular Components in Tumor Microenvironment and Their Application in Early Diagnosis of Cancers. Anal. Cell. Pathol. 2020, 2020, 6283796. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Y.; Li, N.; Jiang, H.; Huang, X.; Gao, N.; Tye, B.K. Unique Roles of the Non-identical MCM Subunits in DNA Replication Licensing. Mol. Cell 2017, 67, 168–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obermann, E.C.; Went, P.; Zimpfer, A.; Tzankov, A.; Wild, P.J.; Stoehr, R.; Pileri, S.A.; Dirnhofer, S. Expression of minichromosome maintenance protein 2 as a marker for proliferation and prognosis in diffuse large B-cell lymphoma: A tissue microarray and clinico-pathological analysis. BMC Cancer 2005, 5, 162. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, X.; Li, Y.; Zhang, Y.; Wang, X. Suppression of minichromosome maintenance 7 expression sensitizes chronic lymphocytic leukemia cells to fludarabine. Leuk. Lymphoma 2019, 60, 1266–1274. [Google Scholar] [CrossRef] [PubMed]
- Uittenboogaard, L.M.; Payan-Gomez, C.; Pothof, J.; van Ijcken, W.; Mastroberardino, P.G.; van der Pluijm, I.; Hoeijmakers, J.H.; Tresini, M. BACH2: A marker of DNA damage and ageing. DNA Repair. 2013, 12, 982–992. [Google Scholar] [CrossRef] [Green Version]
- Schauber, C.; Chen, L.; Tongaonkar, P.; Vega, I.; Lambertson, D.; Potts, W.; Madura, K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 1998, 391, 715–718. [Google Scholar] [CrossRef]
- Wade, S.L.; Auble, D.T. The Rad23 ubiquitin receptor, the proteasome and functional specificity in transcriptional control. Transcription 2010, 1, 22–26. [Google Scholar] [CrossRef] [Green Version]
- Grzanka, M.; Piekielko-Witkowska, A. The Role of TCOF1 Gene in Health and Disease: Beyond Treacher Collins Syndrome. Int. J. Mol. Sci. 2021, 22, 2482. [Google Scholar] [CrossRef] [PubMed]
- Brumbaugh, J.; Di Stefano, B.; Wang, X.; Borkent, M.; Forouzmand, E.; Clowers, K.J.; Ji, F.; Schwarz, B.A.; Kalocsay, M.; Elledge, S.J.; et al. Nudt21 Controls Cell Fate by Connecting Alternative Polyadenylation to Chromatin Signaling. Cell 2018, 172, 629–631. [Google Scholar] [CrossRef]
- Debaize, L.; Troadec, M.B. The master regulator FUBP1: Its emerging role in normal cell function and malignant development. Cell. Mol. Life Sci. 2019, 76, 259–281. [Google Scholar] [CrossRef]
- McIver, S.C.; Kang, Y.A.; DeVilbiss, A.W.; O’Driscoll, C.A.; Ouellette, J.N.; Pope, N.J.; Camprecios, G.; Chang, C.J.; Yang, D.; Bouhassira, E.E.; et al. The exosome complex establishes a barricade to erythroid maturation. Blood 2014, 124, 2285–2297. [Google Scholar] [CrossRef] [Green Version]
- An, X. Exosome complex and erythropoiesis. Blood 2014, 124, 2169–2171. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.H.; Gasper, D.J.; Lee, S.H.; Plisch, E.H.; Svaren, J.; Suresh, M. Bach2 regulates homeostasis of Foxp3+ regulatory T cells and protects against fatal lung disease in mice. J. Immunol. 2014, 192, 985–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiorazzi, N.; Ferrarini, M. Cellular origin(s) of chronic lymphocytic leukemia: Cautionary notes and additional considerations and possibilities. Blood 2011, 117, 1781–1791. [Google Scholar] [CrossRef]
- Crassini, K.; Stevenson, W.S.; Mulligan, S.P.; Best, O.G. Molecular pathogenesis of chronic lymphocytic leukaemia. Br. J. Haematol. 2019, 186, 668–684. [Google Scholar] [CrossRef] [PubMed]
- Norgaard, C.H.; Jakobsen, L.H.; Gentles, A.J.; Dybkaer, K.; El-Galaly, T.C.; Bodker, J.S.; Schmitz, A.; Johansen, P.; Herold, T.; Spiekermann, K.; et al. Subtype assignment of CLL based on B-cell subset associated gene signatures from normal bone marrow—A proof of concept study. PLoS ONE 2018, 13, e0193249. [Google Scholar] [CrossRef]
- Nutt, S.L.; Taubenheim, N.; Hasbold, J.; Corcoran, L.M.; Hodgkin, P.D. The genetic network controlling plasma cell differentiation. Semin. Immunol. 2011, 23, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, S.; Duy, C.; Muschen, M. BACH2-BCL6 balance regulates selection at the pre-B cell receptor checkpoint. Trends Immunol. 2014, 35, 131–137. [Google Scholar] [CrossRef] [Green Version]
- Shinnakasu, R.; Kurosaki, T. Regulation of memory B and plasma cell differentiation. Curr. Opin. Immunol. 2017, 45, 126–131. [Google Scholar] [CrossRef]
- De Silva, N.S.; Klein, U. Dynamics of B cells in germinal centres. Nat. Rev. Immunol. 2015, 15, 137–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beauchemin, C.; Johnston, J.B.; Lapierre, M.È.; Aissa, F.; Lachaine, J. Relationship between progression-free survival and overall survival in chronic lymphocytic leukemia: A literature-based analysis. Curr. Oncol. 2015, 22, e148–e156. [Google Scholar] [CrossRef] [Green Version]
- Bosch, F.; Dalla-Favera, R. Chronic lymphocytic leukaemia: From genetics to treatment. Nat. Rev. Clin. Oncol. 2019, 16, 684–701. [Google Scholar] [CrossRef] [PubMed]
- Dohner, H.; Stilgenbauer, S.; Benner, A.; Leupolt, E.; Krober, A.; Bullinger, L.; Dohner, K.; Bentz, M.; Lichter, P. Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 2000, 343, 1910–1916. [Google Scholar] [CrossRef] [Green Version]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalachikov, S.; Migliazza, A.; Cayanis, E.; Fracchiolla, N.S.; Bonaldo, M.F.; Lawton, L.; Jelenc, P.; Ye, X.; Qu, X.; Chien, M.; et al. Cloning and gene mapping of the chromosome 13q14 region deleted in chronic lymphocytic leukemia. Genomics 1997, 42, 369–377. [Google Scholar] [CrossRef]
- Palamarchuk, A.; Efanov, A.; Nazaryan, N.; Santanam, U.; Alder, H.; Rassenti, L.; Kipps, T.; Croce, C.M.; Pekarsky, Y. 13q14 deletions in CLL involve cooperating tumor suppressors. Blood 2010, 115, 3916–3922. [Google Scholar] [CrossRef] [Green Version]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [Green Version]
- Klein, U.; Lia, M.; Crespo, M.; Siegel, R.; Shen, Q.; Mo, T.; Ambesi-Impiombato, A.; Califano, A.; Migliazza, A.; Bhagat, G.; et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 2010, 17, 28–40. [Google Scholar] [CrossRef] [Green Version]
- Tamahara, T.; Ochiai, K.; Muto, A.; Kato, Y.; Sax, N.; Matsumoto, M.; Koseki, T.; Igarashi, K. The mTOR-Bach2 Cascade Controls Cell Cycle and Class Switch Recombination during B Cell Differentiation. Mol. Cell. Biol. 2017, 37, e00418-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarsotti, E.; Marugan, I.; Benet, I.; Terol, M.J.; Sanchez-Izquierdo, D.; Tormo, M.; Rubio-Moscardo, F.; Martinez-Climent, J.A.; Garcia-Conde, J. Bcl-6 mutation status provides clinically valuable information in early-stage B-cell chronic lymphocytic leukemia. Leukemia 2004, 18, 743–746. [Google Scholar] [CrossRef]
- Capello, D.; Fais, F.; Vivenza, D.; Migliaretti, G.; Chiorazzi, N.; Gaidano, G.; Ferrarini, M. Identification of three subgroups of B cell chronic lymphocytic leukemia based upon mutations of BCL-6 and IgV genes. Leukemia 2000, 14, 811–815. [Google Scholar] [CrossRef] [Green Version]
- Capello, D.; Vitolo, U.; Pasqualucci, L.; Quattrone, S.; Migliaretti, G.; Fassone, L.; Ariatti, C.; Vivenza, D.; Gloghini, A.; Pastore, C.; et al. Distribution and pattern of BCL-6 mutations throughout the spectrum of B-cell neoplasia. Blood 2000, 95, 651–659. [Google Scholar] [PubMed]
- Darwiche, W.; Gubler, B.; Marolleau, J.P.; Ghamlouch, H. Chronic Lymphocytic Leukemia B-Cell Normal Cellular Counterpart: Clues From a Functional Perspective. Front. Immunol. 2018, 9, 683. [Google Scholar] [CrossRef]
- Tchourine, K.; Poultney, C.S.; Wang, L.; Silva, G.M.; Manohar, S.; Mueller, C.L.; Bonneau, R.; Vogel, C. One third of dynamic protein expression profiles can be predicted by a simple rate equation. Mol. Biosyst. 2014, 10, 2850–2862. [Google Scholar] [CrossRef] [Green Version]
- Schwanhausser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [Green Version]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef] [PubMed]
- De Sousa Abreu, R.; Penalva, L.O.; Marcotte, E.M.; Vogel, C. Global signatures of protein and mRNA expression levels. Mol. Biosyst. 2009, 5, 1512–1526. [Google Scholar] [CrossRef] [Green Version]
- Han, L.; Lam, E.W.F.; Sun, Y. Extracellular vesicles in the tumor microenvironment: Old stories, but new tales. Mol. Cancer 2019, 18, 59. [Google Scholar] [CrossRef] [Green Version]
- Duechler, M.; Shehata, M.; Schwarzmeier, J.D.; Hoelbl, A.; Hilgarth, M.; Hubmann, R. Induction of apoptosis by proteasome inhibitors in B-CLL cells is associated with downregulation of CD23 and inactivation of Notch2. Leukemia 2005, 19, 260–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, W.; Wang, L.; Pan, M.; Zheng, J. The regulatory role of exosomes in leukemia and their clinical significance. J. Int. Med. Res. 2020, 48, 0300060520950135. [Google Scholar] [CrossRef] [PubMed]
- Gargiulo, E.; Morande, P.E.; Largeot, A.; Moussay, E.; Paggetti, J. Diagnostic and Therapeutic Potential of Extracellular Vesicles in B-Cell Malignancies. Front. Oncol. 2020, 10, 580874. [Google Scholar] [CrossRef] [PubMed]






| Characteristics, n = 102 Unless Stated | N | % |
|---|---|---|
| Age ≥ 65 | 31 | 62 |
| Male | 71 | 69.6 |
| TP53 gene status n = 100 | ||
| M | 12 | 12 |
| UM | 88 | 88 |
| ZAP70 status n = 19 | ||
| >20% | 9 | 47.4 |
| <20% | 10 | 52.6 |
| CD38 status n = 36 | ||
| >20% | 13 | 36.1 |
| <20% | 23 | 63.9 |
| Treatment status n = 102 | ||
| Treated | 35 | 34.3 |
| Untreated | 67 | 65.7 |
| Binet Stage n = 90 | N | % | Treated | % | Untreated | % |
|---|---|---|---|---|---|---|
| A | 49 | 54.4 | 10 | 20.4 | 39 | 79.6 |
| B | 16 | 17.8 | 7 | 43.8 | 9 | 56.3 |
| C | 25 | 27.8 | 16 | 64.0 | 9 | 36.0 |
| IGHV status n = 63 | Treated | Untreated | ||||
| M | 38 | 60.3 | 17 | 44.7 | 21 | 55.3 |
| UM | 25 | 39.7 | 7 | 28.0 | 18 | 72.0 |
| Cytogenetics n = 101 | Treated | Untreated | ||||
| 13q | 62 | 61.4 | 21 | 33.9 | 41 | 66.1 |
| 11q | 20 | 19.8 | 11 | 55.0 | 9 | 45.0 |
| 12+ | 6 | 5.9 | 4 | 66.7 | 2 | 33.3 |
| 17p | 8 | 7.9 | 1 | 12.5 | 7 | 87.5 |
| Normal karyotype | 24 | 23.8 | 18 | 75.0 | 6 | 25.0 |
| Cytogenetics n = 101 | BACH2 High | BCL6 High | ||||
| 13q | 62 | 61.4 | 35 | 56.5 | 42 | 67.7 |
| 11q | 20 | 19.8 | 12 | 60.0 | 14 | 70.0 |
| 12+ | 6 | 5.9 | 2 | 33.3 | 4 | 66.7 |
| 17p | 8 | 7.9 | 3 | 37.5 | 4 | 50.0 |
| Normal karyotype | 24 | 23.8 | 12 | 50.0 | 15 | 62.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciardullo, C.; Szoltysek, K.; Zhou, P.; Pietrowska, M.; Marczak, L.; Willmore, E.; Enshaei, A.; Walaszczyk, A.; Ho, J.Y.; Rand, V.; et al. Low BACH2 Expression Predicts Adverse Outcome in Chronic Lymphocytic Leukaemia. Cancers 2022, 14, 23. https://doi.org/10.3390/cancers14010023
Ciardullo C, Szoltysek K, Zhou P, Pietrowska M, Marczak L, Willmore E, Enshaei A, Walaszczyk A, Ho JY, Rand V, et al. Low BACH2 Expression Predicts Adverse Outcome in Chronic Lymphocytic Leukaemia. Cancers. 2022; 14(1):23. https://doi.org/10.3390/cancers14010023
Chicago/Turabian StyleCiardullo, Carmela, Katarzyna Szoltysek, Peixun Zhou, Monika Pietrowska, Lukasz Marczak, Elaine Willmore, Amir Enshaei, Anna Walaszczyk, Jia Yee Ho, Vikki Rand, and et al. 2022. "Low BACH2 Expression Predicts Adverse Outcome in Chronic Lymphocytic Leukaemia" Cancers 14, no. 1: 23. https://doi.org/10.3390/cancers14010023
APA StyleCiardullo, C., Szoltysek, K., Zhou, P., Pietrowska, M., Marczak, L., Willmore, E., Enshaei, A., Walaszczyk, A., Ho, J. Y., Rand, V., Marshall, S., Hall, A. G., Harrison, C. J., Soundararajan, M., & Eswaran, J. (2022). Low BACH2 Expression Predicts Adverse Outcome in Chronic Lymphocytic Leukaemia. Cancers, 14(1), 23. https://doi.org/10.3390/cancers14010023







