The Impact of Biomarkers in Pancreatic Ductal Adenocarcinoma on Diagnosis, Surveillance and Therapy
Abstract
Simple Summary
Abstract
1. Introduction
2. The Role of Biomarkers in Diagnosis of Pancreatic Ductal Adenocarcinoma
2.1. CA19-9, CEA and Other Carbohydrate Antigens
2.2. Micro-RNA, Circulating Tumor DNA (ct-DNA) and Circulating Tumor Cells (CTCs)
2.3. Metabolomics
2.4. Further Biomarker Approaches
2.4.1. Multimarker Panels
2.4.2. Alternative Splicing Variants and Methylation
2.4.3. Exosomes
2.4.4. Radiomics
2.4.5. Outlook
3. Biomarkers as Predictors for Therapy Response and Targeted Therapy in PDAC
3.1. Prognostic Biomarkers for Cytostatic Chemotherapy
3.2. Predictive Biomarkers for Targeted Therapy in PDAC
4. The Role of Biomarkers in Surveillance after Curative Surgery
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ushio, J.; Kanno, A.; Ikeda, E.; Ando, K.; Nagai, H.; Miwata, T.; Kawasaki, Y.; Tada, Y.; Yokoyama, K.; Numao, N.; et al. Pancreatic Ductal Adenocarcinoma: Epidemiology and Risk Factors. Diagnostics 2021, 11, 562. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lok, V.; Ngai, C.H.; Zhang, L.; Yuan, J.; Lao, X.Q.; Ng, K.; Chong, C.; Zheng, Z.-J.; Wong, M.C. Worldwide Burden of, Risk Factors for, and Trends in Pancreatic Cancer. Gastroenterology 2021, 160, 744–754. [Google Scholar] [CrossRef]
- Maisonneuve, P.; Lowenfels, A.B. Risk factors for pancreatic cancer: A summary review of meta-analytical studies. Int. J. Epidemiol. 2014, 44, 186–198. [Google Scholar] [CrossRef]
- Gupta, S.; Wang, F.; Holly, E.A.; Bracci, P.M. Risk of pancreatic cancer by alcohol dose, duration, and pattern of consumption, including binge drinking: A population-based study. Cancer Causes Control. 2010, 21, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Arslan, A.A. Anthropometric Measures, Body Mass Index, and Pancreatic Cancer. Arch. Intern. Med. 2010, 170, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Wolpin, B.M.; Bao, Y.; Qian, Z.R.; Wu, C.; Kraft, P.; Ogino, S.; Stampfer, M.J.; Sato, K.; Ma, J.; Buring, J.E.; et al. Hyperglycemia, Insulin Resistance, Impaired Pancreatic β-Cell Function, and Risk of Pancreatic Cancer. J. Natl. Cancer Inst. 2013, 105, 1027–1035. [Google Scholar] [CrossRef]
- Chen, W.C.-Y.; Boursi, B.; Mamtani, R.; Yang, Y.-X. Total Serum Cholesterol and Pancreatic Cancer: A Nested Case–Control Study. Cancer Epidemiol. Biomark. Prev. 2018, 28, 363–369. [Google Scholar] [CrossRef]
- Groot, V.P.; Rezaee, N.; Wu, W.; Cameron, J.L.; Fishman, E.; Hruban, R.H.; Weiss, M.J.; Zheng, L.; Wolfgang, C.L.; He, J. Patterns, Timing, and Predictors of Recurrence Following Pancreatectomy for Pancreatic Ductal Adenocarcinoma. Ann. Surg. 2018, 267, 936–945. [Google Scholar] [CrossRef]
- Daamen, L.; Groot, V.P.; Intven, M.; Besselink, M.; Busch, O.; Koerkamp, B.G.; Mohammad, N.H.; Hermans, J.; Van Laarhoven, H.; Nuyttens, J.; et al. Postoperative surveillance of pancreatic cancer patients. Eur. J. Surg. Oncol. (EJSO) 2019, 45, 1770–1777. [Google Scholar] [CrossRef] [PubMed]
- Ducreux, M.; Cuhna, A.S.; Caramella, C.; Hollebecque, A.; Burtin, P.; Goéré, D.; Seufferlein, T.; Haustermans, K.; Van Laethem, J.L.; Conroy, T.; et al. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26 (Suppl. S5), v56–v68. [Google Scholar] [CrossRef]
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic cancer. Lancet 2011, 378, 607–620. [Google Scholar] [CrossRef]
- Matthaei, H.; Schulick, R.D.; Hruban, R.H.; Maitra, A. Cystic precursors to invasive pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Hong, S.-M. Precursor Lesions of Pancreatic Cancer. Oncol. Res. Treat. 2018, 41, 603–610. [Google Scholar] [CrossRef]
- Ren, B.; Liu, X.; Suriawinata, A.A. Pancreatic Ductal Adenocarcinoma and Its Precursor Lesions. Am. J. Pathol. 2019, 189, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Fernández-del Castillo, C.; Kamisawa, T.; Jang, J.Y.; Levy, P.; Ohtsuka, T.; Salvia, R.; Shimizu, Y.; Tada, M.; Wolfgang, C.L. Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology 2017, 17, 738–753. [Google Scholar] [CrossRef] [PubMed]
- Heckler, M.; Michalski, C.W.; Schaefle, S.; Kaiser, J.; Büchler, M.W.; Hackert, T. The Sendai and Fukuoka consensus criteria for the management of branch duct IPMN—A meta-analysis on their accuracy. Pancreatology 2017, 17, 255–262. [Google Scholar] [CrossRef]
- Keane, M.; Afghani, E. A Review of the Diagnosis and Management of Premalignant Pancreatic Cystic Lesions. J. Clin. Med. 2021, 10, 1284. [Google Scholar] [CrossRef]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouché, O.; Guimbaud, R.; Bécouarn, Y.; Adenis, A.; Raoul, J.-L.; Gourgou-Bourgade, S.; De La Fouchardière, C.; et al. FOLFIRINOX versus Gemcitabine for Metastatic Pancreatic Cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Livingston, P.O.; Ragupathi, G. Carbohydrate-Based Vaccines. In Encyclopedia of Cancer; Elsevier: Amsterdam, The Netherlands, 2002; pp. 337–344. [Google Scholar]
- Ballehaninna, U.K.; Chamberlain, R.S. Serum CA 19-9 as a Biomarker for Pancreatic Cancer—A Comprehensive Review. Indian J. Surg. Oncol. 2011, 2, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Goonetilleke, K.; Siriwardena, A. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur. J. Surg. Oncol. (EJSO) 2007, 33, 266–270. [Google Scholar] [CrossRef]
- Xing, H.; Wang, J.; Wang, Y.; Tong, M.; Hu, H.; Huang, C.; Li, D. Diagnostic Value of CA 19-9 and Carcinoembryonic Antigen for Pancreatic Cancer: A Meta-Analysis. Gastroenterol. Res. Pract. 2018, 2018, 8704751. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, J.; Li, H.; Wu, Y.; Zhang, H.; Chen, W. Tumor markers CA19-9, CA242 and CEA in the diagnosis of pancreatic cancer: A meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 11683–11691. [Google Scholar] [PubMed]
- Fahrmann, J.F.; Schmidt, C.M.; Mao, X.; Irajizad, E.; Loftus, M.; Zhang, J.; Patel, N.; Vykoukal, J.; Dennison, J.B.; Long, J.P.; et al. Lead-Time Trajectory of CA19-9 as an Anchor Marker for Pancreatic Cancer Early Detection. Gastroenterology 2021, 160, 1373–1383.e6. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Guo, M.; Jin, K.; Liu, Z.; Liu, C.; Cheng, H.; Lu, Y.; Long, J.; Liu, L.; Xu, J.; et al. Optimize CA19-9 in detecting pancreatic cancer by Lewis and Secretor genotyping. Pancreatology 2016, 16, 1057–1062. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Fan, Z.; Cheng, H.; Jin, K.; Guo, M.; Lu, Y.; Yang, C.; Fan, K.; Huang, Q.; Long, J.; et al. New observations on the utility of CA19-9 as a biomarker in Lewis negative patients with pancreatic cancer. Pancreatology 2018, 18, 971–976. [Google Scholar] [CrossRef]
- Loosen, S.H.; Neumann, U.P.; Trautwein, C.; Roderburg, C.; Luedde, T. Current and future biomarkers for pancreatic adenocarcinoma. Tumor Biol. 2017, 39, 1010428317692231. [Google Scholar] [CrossRef]
- Satake, K.; Takeuchi, T.; Homma, T.; Ozaki, H. CA19-9 as a Screening and Diagnostic Tool in Symptomatic Patients. Pancreas 1994, 9, 703–706. [Google Scholar] [CrossRef]
- Wang, Z.; Tian, Y.-P. Clinical value of serum tumor markers CA19-9, CA125 and CA72-4 in the diagnosis of pancreatic carcinoma. Mol. Clin. Oncol. 2013, 2, 265–268. [Google Scholar] [CrossRef]
- Lei, X.F.; Jia, S.Z.; Ye, J.; Qiao, Y.L.; Zhao, G.M.; Li, X.H.; Chang, H. Application values of detection of serum CA199, CA242 and CA50 in the diagnosis of pancreatic cancer. J. Boil. Regul. Homeost. Agents 2017, 31, 383–388. [Google Scholar]
- Jiang, J.-T.; Wu, C.-P.; Deng, H.-F.; Lu, M.-Y.; Wu, J.; Zhang, H.-Y.; Sun, W.-H.; Ji, M. Serum level of TSGF, CA242 and CA19-9 in pancreatic cancer. World J. Gastroenterol. 2004, 10, 1675–1677. [Google Scholar] [CrossRef]
- Dou, H.; Sun, G.; Zhang, L. CA242 as a biomarker for pancreatic cancer and other diseases. In Progress in Molecular Biology and Translational Science; Elsevier B.V.: Amsterdam, The Netherlands, 2019; Volume 162, pp. 229–239. ISBN 9780128177389. [Google Scholar]
- Nazli, O.; Bozdag, A.D.; Tansug, T.; Kir, R.; Kaymak, E. The diagnostic importance of CEA and CA 19-9 for the early diagnosis of pancreatic carcinoma. Hepatogastroenterology 2001, 47, 1750–1752. [Google Scholar]
- Van Manen, L.; Groen, J.V.; Putter, H.; Vahrmeijer, A.L.; Swijnenburg, R.-J.; Bonsing, B.A.; Mieog, J.S.D. Elevated CEA and CA19-9 serum levels independently predict advanced pancreatic cancer at diagnosis. Biomarkers 2020, 25, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef]
- Yonemori, K.; Kurahara, H.; Maemura, K.; Natsugoe, S. MicroRNA in pancreatic cancer. J. Hum. Genet. 2016, 62, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Gablo, N.A.; Prochazka, V.; Kala, Z.; Slaby, O.; Kiss, I. Cell-free microRNAs as Non-invasive Diagnostic and Prognostic Biomarkers in Pancreatic Cancer. Curr. Genom. 2020, 20, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Debernardi, S.; Massat, N.J.; Radon, T.P.; Sangaralingam, A.; Banissi, A.; Ennis, D.P.; Dowe, T.; Chelala, C.; Pereira, S.; Kocher, H.; et al. Noninvasive urinary miRNA biomarkers for early detection of pancreatic adenocarcinoma. Am. J. Cancer Res. 2015, 5, 3455–3466. [Google Scholar] [PubMed]
- Humeau, M.; Vignolle-Vidoni, A.; Sicard, F.; Martins, F.; Bournet, B.; Buscail, L.; Torrisani, J.; Cordelier, P. Salivary MicroRNA in Pancreatic Cancer Patients. PLoS ONE 2015, 10, e0130996. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-Y.; Sun, Y.-W.; Liu, D.-J.; Zhang, J.-F.; Li, J.; Hua, R. MicroRNAs in stool samples as potential screening biomarkers for pancreatic ductal adenocarcinoma cancer. Am. J. Cancer Res. 2014, 4, 663–673. [Google Scholar]
- Schultz, N.A.; Dehlendorff, C.; Jensen, B.V.; Bjerregaard, J.K.; Nielsen, K.R.; Bojesen, S.E.; Calatayud, D.; Nielsen, S.E.; Yilmaz, M.; Holländer, N.H.; et al. MicroRNA Biomarkers in Whole Blood for Detection of Pancreatic Cancer. JAMA 2014, 311, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Habbe, N.; Koorstra, J.-B.M.; Mendell, J.T.; Offerhaus, G.J.; Ryu, J.K.; Feldmann, G.; Mullendore, M.E.; Goggins, M.G.; Hong, S.-M.; Maitra, A. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol. Ther. 2009, 8, 340–346. [Google Scholar] [CrossRef]
- Ryu, J.K.; Hong, S.-M.; Karikari, C.A.; Hruban, R.H.; Goggins, M.G.; Maitra, A. Aberrant MicroRNA-155 Expression Is an Early Event in the Multistep Progression of Pancreatic Adenocarcinoma. Pancreatology 2010, 10, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Slater, E.P.; Strauch, K.; Rospleszcz, S.; Ramaswamy, A.; Esposito, I.; Klöppel, G.; Matthäi, E.; Heeger, K.; Fendrich, V.; Langer, P.; et al. MicroRNA-196a and -196b as Potential Biomarkers for the Early Detection of Familial Pancreatic Cancer. Transl. Oncol. 2014, 7, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Tayoun, A.N.A.; Abo, K.M.; Pipas, J.M.; Gordon, S.R.; Gardner, T.B.; Barth, R.J.; Suriawinata, A.A.; Tsongalis, G.J. MicroRNAs as diagnostic markers for pancreatic ductal adenocarcinoma and its precursor, pancreatic intraepithelial neoplasm. Cancer Genet. 2013, 206, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Takai, E.; Yachida, S. Circulating tumor DNA as a liquid biopsy target for detection of pancreatic cancer. World J. Gastroenterol. 2016, 22, 8480–8488. [Google Scholar] [CrossRef] [PubMed]
- Riva, F.; Dronov, O.I.; Khomenko, D.I.; Huguet, F.; Louvet, C.; Mariani, P.; Stern, M.-H.; Lantz, O.; Proudhon, C.; Pierga, J.-Y.; et al. Clinical applications of circulating tumor DNA and circulating tumor cells in pancreatic cancer. Mol. Oncol. 2016, 10, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.W.; Schwerdel, D.; Costa, I.G.; Hackert, T.; Strobel, O.; Lam, S.; Barth, T.F.; Schröppel, B.; Meining, A.; Büchler, M.W.; et al. Detection of Hot-Spot Mutations in Circulating Cell-Free DNA From Patients with Intraductal Papillary Mucinous Neoplasms of the Pancreas. Gastroenterology 2016, 151, 267–270. [Google Scholar] [CrossRef]
- Berger, A.W.; Schwerdel, D.; Reinacher-Schick, A.; Uhl, W.; Algül, H.; Friess, H.; Janssen, K.-P.; König, A.; Ghadimi, M.; Gallmeier, E.; et al. A Blood-Based Multi Marker Assay Supports the Differential Diagnosis of Early-Stage Pancreatic Cancer. Theranostics 2019, 9, 1280–1287. [Google Scholar] [CrossRef]
- Guler, G.D.; Ning, Y.; Ku, C.-J.; Phillips, T.; McCarthy, E.; Ellison, C.K.; Bergamaschi, A.; Collin, F.; Lloyd, P.; Scott, A.; et al. Detection of early stage pancreatic cancer using 5-hydroxymethylcytosine signatures in circulating cell free DNA. Nat. Commun. 2020, 11, 5270. [Google Scholar] [CrossRef]
- Li, S.; Wang, L.; Zhao, Q.; Wang, Z.; Lu, S.; Kang, Y.; Jin, G.; Tian, J. Genome-Wide Analysis of Cell-Free DNA Methylation Profiling for the Early Diagnosis of Pancreatic Cancer. Front. Genet. 2020, 11, 596078. [Google Scholar] [CrossRef] [PubMed]
- Vernucci, E.; Abrego, J.; Gunda, V.; Shukla, S.K.; Dasgupta, A.; Rai, V.; Chaika, N.; Buettner, K.; Illies, A.; Yu, F.; et al. Metabolic Alterations in Pancreatic Cancer Progression. Cancers 2019, 12, 2. [Google Scholar] [CrossRef]
- Sah, R.P.; Sharma, A.; Nagpal, S.; Patlolla, S.H.; Sharma, A.; Kandlakunta, H.; Anani, V.; Angom, R.S.; Kamboj, A.K.; Ahmed, N.; et al. Phases of Metabolic and Soft Tissue Changes in Months Preceding a Diagnosis of Pancreatic Ductal Adenocarcinoma. Gastroenterology 2019, 156, 1742–1752. [Google Scholar] [CrossRef] [PubMed]
- Sahni, S.; Pandya, A.R.; Hadden, W.J.; Nahm, C.B.; Maloney, S.; Cook, V.; Toft, J.A.; Wilkinson-White, L.; Gill, A.J.; Samra, J.S.; et al. A unique urinary metabolomic signature for the detection of pancreatic ductal adenocarcinoma. Int. J. Cancer 2020, 148, 1508–1518. [Google Scholar] [CrossRef] [PubMed]
- Mayerle, J.; Kalthoff, H.; Reszka, R.; Kamlage, B.; Peter, E.; Schniewind, B.; Maldonado, S.G.; Pilarsky, C.; Heidecke, C.-D.; Schatz, P.; et al. Metabolic biomarker signature to differentiate pancreatic ductal adenocarcinoma from chronic pancreatitis. Gut 2017, 67, 128–137. [Google Scholar] [CrossRef]
- Macias, R.I.R.; Muñoz-Bellvís, L.; Sánchez-Martín, A.; Arretxe, E.; Martínez-Arranz, I.; Lapitz, A.; Gutiérrez, M.L.; La La Casta, A.; Alonso, C.; González, L.M.; et al. A Novel Serum Metabolomic Profile for the Differential Diagnosis of Distal Cholangiocarcinoma and Pancreatic Ductal Adenocarcinoma. Cancers 2020, 12, 1433. [Google Scholar] [CrossRef]
- Yu, J.; Ploner, A.; Kordes, M.; Löhr, M.; Nilsson, M.; de Maturana, M.E.L.; Estudillo, L.; Renz, H.; Carrato, A.; Molero, X.; et al. Plasma protein biomarkers for early detection of pancreatic ductal adenocarcinoma. Int. J. Cancer 2021, 148, 2048–2058. [Google Scholar] [CrossRef]
- Kim, J.; Bamlet, W.R.; Oberg, A.L.; Chaffee, K.G.; Donahue, G.; Cao, X.-J.; Chari, S.; Garcia, B.A.; Petersen, G.M.; Zaret, K.S. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19-9 blood markers. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Nolen, B.M.; Brand, R.E.; Prosser, D.; Velikokhatnaya, L.; Allen, P.J.; Zeh, H.J.; Grizzle, W.E.; Lomakin, A.; Lokshin, A.E. Prediagnostic Serum Biomarkers as Early Detection Tools for Pancreatic Cancer in a Large Prospective Cohort Study. PLoS ONE 2014, 9, e94928. [Google Scholar] [CrossRef]
- Balasenthil, S.; Huang, Y.; Liu, S.; Marsh, T.; Chen, J.; Stass, S.A.; Kukuruga, D.; Brand, R.; Chen, N.; Frazier, M.L.; et al. A Plasma Biomarker Panel to Identify Surgically Resectable Early-Stage Pancreatic Cancer. J. Natl. Cancer Inst. 2017, 109, djw341. [Google Scholar] [CrossRef]
- Kruger, D.; Yako, Y.Y.; Devar, J.; Lahoud, N.; Smith, M. Inflammatory cytokines and combined biomarker panels in pancreatic ductal adenocarcinoma: Enhancing diagnostic accuracy. PLoS ONE 2019, 14, e0221169. [Google Scholar] [CrossRef] [PubMed]
- Omenn, G.S.; Yocum, A.K.; Menon, R. Alternative splice variants, a new class of protein cancer biomarker candidates: Findings in pancreatic cancer and breast cancer with systems biology implications. Dis. Markers 2010, 28, 241–251. [Google Scholar] [CrossRef]
- Yu, M.; Hong, W.; Ruan, S.; Guan, R.; Tu, L.; Huang, B.; Hou, B.; Jian, Z.; Ma, L.; Jin, H. Genome-Wide Profiling of Prognostic Alternative Splicing Pattern in Pancreatic Cancer. Front. Oncol. 2019, 9, 773. [Google Scholar] [CrossRef]
- Brancaccio, M.; Natale, F.; Falco, G.; Angrisano, T. Cell-Free DNA Methylation: The New Frontiers of Pancreatic Cancer Biomarkers’ Discovery. Genes 2019, 11, 14. [Google Scholar] [CrossRef]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct. Target. Ther. 2020, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kugeratski, F.G.; Hodge, K.; Lilla, S.; McAndrews, K.M.; Zhou, X.; Hwang, R.F.; Zanivan, S.; Kalluri, R. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat. Cell Biol. 2021, 23, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, T.; Taniuchi, K.; Tsuboi, M.; Sakaguchi, M.; Kohsaki, T.; Okabayashi, T.; Saibara, T. Circulating pancreatic cancer exosomal RNA s for detection of pancreatic cancer. Mol. Oncol. 2018, 13, 212–227. [Google Scholar] [CrossRef]
- Castillo, J.; Bernard, V.; Lucas, F.A.S.; Allenson, K.; Capello, M.; Kim, D.U.; Gascoyne, P.; Mulu, F.C.; Stephens, B.M.; Huang, J.; et al. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann. Oncol. 2018, 29, 223–229. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Bausch, D.; Thomas, S.; Mino-Kenudson, M.; Fernández-Del, C.C.; Bauer, T.W.; Williams, M.; Warshaw, A.L.; Thayer, S.P.; Kelly, K.A. Plectin-1 as a Novel Biomarker for Pancreatic Cancer. Clin. Cancer Res. 2010, 17, 302–309. [Google Scholar] [CrossRef]
- Lohrmann, C.; O’Reilly, E.M.; O’Donoghue, J.A.; Pandit-Taskar, N.; Carrasquillo, J.A.; Lyashchenko, S.K.; Ruan, S.; Teng, R.; Scholz, W.; Maffuid, P.W.; et al. Retooling a blood-based biomarker: Phase I assessment of the high-affinity CA19-9 antibody HuMAB-5B1 for immuno-PET imaging of pancreatic cancer. Clin. Cancer Res. 2019, 25, 7014–7023. [Google Scholar] [CrossRef]
- Hodolic, M.; Ambrosini, V.; Fanti, S. Potential use of radiolabelled neurotensin in PET imaging and therapy of patients with pancreatic cancer. Nucl. Med. Commun. 2020, 41, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Kwon, M.-S.; Kim, Y.; Lee, S.; Namkung, J.; Yun, T.; Yi, S.G.; Han, S.; Kang, M.; Kim, S.W.; Jang, J.-Y.; et al. Integrative analysis of multi-omics data for identifying multi-markers for diagnosing pancreatic cancer. BMC Genom. 2015, 16, S4. [Google Scholar] [CrossRef]
- Zhang, W.-H.; Wang, W.-Q.; Han, X.; Gao, H.-L.; Li, T.-J.; Xu, S.-S.; Li, S.; Xu, H.-X.; Li, H.; Ye, L.-Y.; et al. Advances on diagnostic biomarkers of pancreatic ductal adenocarcinoma: A systems biology perspective. Comput. Struct. Biotechnol. J. 2020, 18, 3606–3614. [Google Scholar] [CrossRef]
- Von Hoff, D.D.; Ervin, T.; Arena, F.P.; Chiorean, E.G.; Infante, J.; Moore, M.; Seay, T.; Tjulandin, S.A.; Ma, W.W.; Saleh, M.N.; et al. Increased Survival in Pancreatic Cancer with nab-Paclitaxel plus Gemcitabine. N. Engl. J. Med. 2013, 369, 1691–1703. [Google Scholar] [CrossRef]
- Wasan, H.S.; Springett, G.M.; Chodkiewicz, C.; Wong, R.; Maurel, J.; Barone, C.; Rosbrook, B.; Ricart, A.D.; Kim, S.; Spano, J.-P. CA 19-9 as a biomarker in advanced pancreatic cancer patients randomised to gemcitabine plus axitinib or gemcitabine alone. Br. J. Cancer 2009, 101, 1162–1167. [Google Scholar] [CrossRef][Green Version]
- Humphris, J.L.; Chang, D.K.; Johns, A.L.; Scarlett, C.J.; Pajic, M.; Jones, M.D.; Colvin, E.K.; Nagrial, A.; Chin, V.T.; Chantrill, L.A.; et al. The prognostic and predictive value of serum CA19.9 in pancreatic cancer. Ann. Oncol. 2012, 23, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Cohen, J.; Lipton, L.R.; Tie, J.; Javed, A.A.; Li, L.; Goldstein, D.; Cooray, P.; Nagrial, A.; Burge, M.E.; et al. Potential role of circulating tumor DNA (ctDNA) in the early diagnosis and post-operative management of localised pancreatic cancer. J. Clin. Oncol. 2017, 35, 4101. [Google Scholar] [CrossRef]
- Amrutkar, M.; Vethe, N.T.; Verbeke, C.S.; Aasrum, M.; Finstadsveen, A.V.; Sántha, P.; Gladhaug, I.P. Differential Gemcitabine Sensitivity in Primary Human Pancreatic Cancer Cells and Paired Stellate Cells Is Driven by Heterogenous Drug Uptake and Processing. Cancers 2020, 12, 3628. [Google Scholar] [CrossRef] [PubMed]
- Spratlin, J.; Sangha, R.; Glubrecht, D.; Dabbagh, L.; Young, J.D.; Dumontet, C.; Cass, C.; Lai, R.; Mackey, J.R. The Absence of Human Equilibrative Nucleoside Transporter 1 Is Associated with Reduced Survival in Patients with Gemcitabine-Treated Pancreas Adenocarcinoma. Clin. Cancer Res. 2004, 10, 6956–6961. [Google Scholar] [CrossRef]
- Farrell, J.J.; Elsaleh, H.; Garcia, M.; Lai, R.; Ammar, A.; Regine, W.F.; Abrams, R.; Benson, A.B.; Macdonald, J.; Cass, C.E.; et al. Human Equilibrative Nucleoside Transporter 1 Levels Predict Response to Gemcitabine in Patients with Pancreatic Cancer. Gastroenterology 2009, 136, 187–195. [Google Scholar] [CrossRef]
- Greenhalf, W.; Ghaneh, P.; Neoptolemos, J.; Palmer, D.H.; Cox, T.; Lamb, R.F.; Garner, E.; Campbell, F.; Mackey, J.R.; Costello, E.; et al. Pancreatic Cancer hENT1 Expression and Survival from Gemcitabine in Patients From the ESPAC-3 Trial. J. Natl. Cancer Inst. 2013, 106, djt347. [Google Scholar] [CrossRef]
- Giovannetti, E.; Del Tacca, M.; Mey, V.; Funel, N.; Nannizzi, S.; Ricci, S.; Orlandini, C.; Boggi, U.; Campani, D.; Del Chiaro, M.; et al. Transcription Analysis of Human Equilibrative Nucleoside Transporter-1 Predicts Survival in Pancreas Cancer Patients Treated with Gemcitabine. Cancer Res. 2006, 66, 3928–3935. [Google Scholar] [CrossRef]
- Mori, R.; Ishikawa, T.; Ichikawa, Y.; Taniguchi, K.; Matsuyama, R.; Ueda, M.; Fujii, Y.; Endo, I.; Togo, S.; Danenberg, P.V.; et al. Human equilibrative nucleoside transporter 1 is associated with the chemosensitivity of gemcitabine in human pancreatic adenocarcinoma and biliary tract carcinoma cells. Oncol. Rep. 2007, 17, 1201–1205. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yamada, R.; Mizuno, S.; Uchida, K.; Yoneda, M.; Kanayama, K.; Inoue, H.; Murata, Y.; Kuriyama, N.; Kishiwada, M.; Usui, M.; et al. Human Equilibrative Nucleoside Transporter 1 Expression in Endoscopic Ultrasonography-Guided Fine-Needle Aspiration Biopsy Samples Is a Strong Predictor of Clinical Response and Survival in the Patients with Pancreatic Ductal Adenocarcinoma Undergoing Gemcitabine-Based Chemoradiotherapy. Pancreas 2016, 45, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.H.; Gorgan, T.R.; Elashoff, D.A.; Hines, O.J.; Farrell, J.J.; Donahue, T.R. A Meta-Analysis of Gemcitabine Biomarkers in Patients with Pancreaticobiliary Cancers. Pancreas 2013, 42, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, V.; Ricci, F.; Rubio-Viquiera, B.; Kulesza, P.; Yeo, C.J.; Hidalgo, M.; Klein, A.; Laheru, D.; Iacobuzio-Donahue, C.A. Immunohistochemical and Genetic Evaluation of Deoxycytidine Kinase in Pancreatic Cancer: Relationship to Molecular Mechanisms of Gemcitabine Resistance and Survival. Clin. Cancer Res. 2006, 12, 2492–2497. [Google Scholar] [CrossRef] [PubMed]
- Nakahira, S.; Nakamori, S.; Tsujie, M.; Takahashi, Y.; Okami, J.; Yoshioka, S.; Yamasaki, M.; Marubashi, S.; Takemasa, I.; Miyamoto, A.; et al. Involvement of ribonucleotide reductase M1 subunit overexpression in gemcitabine resistance of human pancreatic cancer. Int. J. Cancer 2006, 120, 1355–1363. [Google Scholar] [CrossRef]
- Minami, K.; Shinsato, Y.; Yamamoto, M.; Takahashi, H.; Zhang, S.; Nishizawa, Y.; Tabata, S.; Ikeda, R.; Kawahara, K.; Tsujikawa, K.; et al. Ribonucleotide reductase is an effective target to overcome gemcitabine resistance in gemcitabine-resistant pancreatic cancer cells with dual resistant factors. J. Pharmacol. Sci. 2015, 127, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Weizman, N.; Krelin, Y.; Shabtayorbach, A.; Amit, M.; Binenbaum, Y.; Wong, R.J.; Gil, Z. Macrophages mediate gemcitabine resistance of pancreatic adenocarcinoma by upregulating cytidine deaminase. Oncogene 2013, 33, 3812–3819. [Google Scholar] [CrossRef] [PubMed]
- Vaz, J.; Ansari, D.; Sasor, A.; Andersson, R. SPARC: A potential prognostic and therapeutic target in pancreatic cancer. Pancreas 2015, 44, 1024–1035. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, M.; Plaza, C.; Musteanu, M.; Illei, P.B.; Brachmann, C.B.; Heise, C.; Pierce, D.; Lopez-Casas, P.P.; Menéndez, M.D.C.; Tabernero, J.; et al. SPARC Expression Did Not Predict Efficacy of nab-Paclitaxel plus Gemcitabine or Gemcitabine Alone for Metastatic Pancreatic Cancer in an Exploratory Analysis of the Phase III MPACT Trial. Clin. Cancer Res. 2015, 21, 4811–4818. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Samuel, S.; Lopez-Casas, P.P.; Grizzle, W.; Hidalgo, M.; Kovar, J.; Oelschlager, D.; Zinn, K.; Warram, J.; Buchsbaum, D. SPARC-Independent Delivery of Nab-Paclitaxel without Depleting Tumor Stroma in Patient-Derived Pancreatic Cancer Xenografts. Mol. Cancer Ther. 2016, 15, 680–688. [Google Scholar] [CrossRef] [PubMed]
- Ormanns, S.; Haas, M.; Baechmann, S.; Altendorf-Hofmann, A.; Remold, A.; Quietzsch, D.; Clemens, M.R.; Bentz, M.; Geissler, M.; Lambertz, H.; et al. Impact of SPARC expression on outcome in patients with advanced pancreatic cancer not receiving nab-paclitaxel: A pooled analysis from prospective clinical and translational trials. Br. J. Cancer 2016, 115, 1520–1529. [Google Scholar] [CrossRef]
- Sinn, M.; Sinn, B.V.; Striefler, J.K.; Lindner, J.L.; Stieler, J.M.; Lohneis, P.; Bischoff, S.; Bläker, H.; Pelzer, U.; Bahra, M.; et al. SPARC expression in resected pancreatic cancer patients treated with gemcitabine: Results from the CONKO-001 study. Ann. Oncol. 2014, 25, 1025–1032. [Google Scholar] [CrossRef]
- Nambaru, P.K.; Hübner, T.; Köck, K.; Mews, S.; Grube, M.; Payen, L.; Guitton, J.; Sendler, M.; Jedlitschky, G.; Rimmbach, C.; et al. Drug Efflux Transporter Multidrug Resistance-Associated Protein 5 Affects Sensitivity of Pancreatic Cancer Cell Lines to the Nucleoside Anticancer Drug 5-Fluorouracil. Drug Metab. Dispos. 2010, 39, 132–139. [Google Scholar] [CrossRef]
- Wang, W.-B.; Yang, Y.; Zhao, Y.-P.; Zhang, T.-P.; Liao, Q.; Shu, H. Recent studies of 5-fluorouracil resistance in pancreatic cancer. World J. Gastroenterol. 2014, 20, 15682–15690. [Google Scholar] [CrossRef]
- Elander, N.O.; The European Study Group for Pancreatic Cancer; Aughton, K.; Ghaneh, P.; Neoptolemos, J.P.; Palmer, D.H.; Cox, T.F.; Campbell, F.; Costello, E.; Halloran, C.; et al. Expression of dihydropyrimidine dehydrogenase (DPD) and hENT1 predicts survival in pancreatic cancer. Br. J. Cancer 2018, 118, 947–954. [Google Scholar] [CrossRef]
- Tasaki, Y.; Suzuki, M.; Katsushima, K.; Shinjo, K.; Iijima, K.; Murofushi, Y.; Naiki-Ito, A.; Hayashi, K.; Qiu, C.; Takahashi, A.; et al. Cancer-Specific Targeting of Taurine-Upregulated Gene 1 Enhances the Effects of Chemotherapy in Pancreatic Cancer. Cancer Res. 2021, 81, 1654–1666. [Google Scholar] [CrossRef]
- Johnson, M.R.; Diasio, R.B. Importance of dihydropyrimidine dehydrogenase (DPD) deficiency in patients exhibiting toxicity following treatment with 5-fluorouracil. Adv. Enzym. Regul. 2001, 41, 151–157. [Google Scholar] [CrossRef]
- Wörmann, B.; Bokemeyer, C.; Burmeister, T.; Köhne, C.-H.; Schwab, M.; Arnold, D.; Blohmer, J.-U.; Borner, M.; Brucker, S.; Cascorbi, I.; et al. Dihydropyrimidine Dehydrogenase Testing prior to Treatment with 5-Fluorouracil, Capecitabine, and Tegafur: A Consensus Paper. Oncol. Res. Treat. 2020, 43, 628–636. [Google Scholar] [CrossRef]
- Capello, M.; Lee, M.; Wang, H.; Babel, I.; Katz, M.H.; Fleming, J.B.; Maitra, A.; Wang, H.; Tian, W.; Taguchi, A.; et al. Carboxylesterase 2 as a Determinant of Response to Irinotecan and Neoadjuvant FOLFIRINOX Therapy in Pancreatic Ductal Adenocarcinoma. J. Natl. Cancer Inst. 2015, 107, djv132. [Google Scholar] [CrossRef]
- Merz, V.; Zecchetto, C.; Santoro, R.; Simionato, F.; Sabbadini, F.; Mangiameli, D.; Piro, G.; Cavaliere, A.; Deiana, M.; Valenti, M.T.; et al. Plasma IL8 Is a Biomarker for TAK1 Activation and Predicts Resistance to Nanoliposomal Irinotecan in Patients with Gemcitabine-Refractory Pancreatic Cancer. Clin. Cancer Res. 2020, 26, 4661–4669. [Google Scholar] [CrossRef]
- Alcindor, T.; Beauger, N. Oxaliplatin: A Review in the Era of Molecularly Targeted Therapy. Curr. Oncol. 2011, 18, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, J.D.; Wimberger, P.; Bankfalvi, A.; Keller, T.; Schöler, S.; Aktas, B.; Buderath, P.; Hauch, S.; Otterbach, F.; Kimmig, R.; et al. ERCC1-Positive Circulating Tumor Cells in the Blood of Ovarian Cancer Patients as a Predictive Biomarker for Platinum Resistance. Clin. Chem. 2014, 60, 1282–1289. [Google Scholar] [CrossRef]
- Reed, E. ERCC1 and Clinical Resistance to Platinum-Based Therapy. Clin. Cancer Res. 2005, 11, 6100–6102. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Perkhofer, L.; Gout, J.; Roger, E.; de Almeida, F.K.; Simões, C.B.; Wiesmüller, L.; Seufferlein, T.; Kleger, A. DNA damage repair as a target in pancreatic cancer: State-of-the-art and future perspectives. Gut 2020, 70, 606–617. [Google Scholar] [CrossRef]
- Fang, Y.; Zhou, W.; Rong, Y.; Kuang, T.; Xu, X.; Wu, W.; Wang, D.; Lou, W. Exosomal miRNA-106b from cancer-associated fibroblast promotes gemcitabine resistance in pancreatic cancer. Exp. Cell Res. 2019, 383, 111543. [Google Scholar] [CrossRef]
- Wattenberg, M.; Asch, D.; Yu, S.; O’Dwyer, P.J.; Domchek, S.M.; Nathanson, K.L.; Rosen, M.A.; Beatty, G.L.; Siegelman, E.S.; Reiss, K.A. Platinum response characteristics of patients with pancreatic ductal adenocarcinoma and a germline BRCA1, BRCA2 or PALB2 mutation. Br. J. Cancer 2019, 122, 333–339. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.-M.; Gingras, M.-C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.C.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.S.; Fakih, M.G.; Strickler, J.H.; Desai, J.; Durm, G.A.; Shapiro, G.I.; Falchook, G.S.; Price, T.J.; Sacher, A.; Denlinger, C.S.; et al. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020, 383, 1207–1217. [Google Scholar] [CrossRef]
- Johnson, M.; Ou, S.; Barve, M.; Rybkin, I.; Papadopoulos, K.; Leal, T.; Velastegui, K.; Christensen, J.; Kheoh, T.; Chao, R.; et al. KRYSTAL-1: Activity and Safety of Adagrasib (MRTX849) in Patients with Colorectal Cancer (CRC) and Other Solid Tumors Harboring a KRAS G12C Mutation. Eur. J. Cancer 2020, 138, S2. [Google Scholar] [CrossRef]
- Zeitouni, D.; Pylayeva-Gupta, Y.; Der, C.J.; Bryant, K.L. KRAS Mutant Pancreatic Cancer: No Lone Path to an Effective Treatment. Cancers 2016, 8, 45. [Google Scholar] [CrossRef]
- Qian, Y.; Gong, Y.; Fan, Z.; Luo, G.; Huang, Q.; Deng, S.; Cheng, H.; Jin, K.; Ni, Q.; Yu, X.; et al. Molecular alterations and targeted therapy in pancreatic ductal adenocarcinoma. J. Hematol. Oncol. 2020, 13, 130. [Google Scholar] [CrossRef]
- Singh, R.; O’Reilly, E.M. New Treatment Strategies for Metastatic Pancreatic Ductal Adenocarcinoma. Drugs 2020, 80, 647–669. [Google Scholar] [CrossRef]
- Nevala-Plagemann, C.; Hidalgo, M.; Garrido-Laguna, I. From state-of-the-art treatments to novel therapies for advanced-stage pancreatic cancer. Nat. Rev. Clin. Oncol. 2019, 17, 108–123. [Google Scholar] [CrossRef]
- Golan, T.; Hammel, P.; Reni, M.; Van Cutsem, E.; Macarulla, T.; Hall, M.J.; Park, J.-O.; Hochhauser, D.; Arnold, D.; Oh, D.-Y.; et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N. Engl. J. Med. 2019, 381, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Humphris, J.L.; Patch, A.-M.; Nones, K.; Bailey, P.J.; Johns, A.L.; McKay, S.; Chang, D.K.; Miller, D.K.; Pajic, M.; Kassahn, K.S.; et al. Hypermutation in Pancreatic Cancer. Gastroenterology 2017, 152, 68–74.e2. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.J.; Goldstein, D.; Hamm, J.; Figer, A.; Hecht, J.R.; Gallinger, S.; Au, H.J.; Murawa, P.; Walde, D.; Wolff, R.A.; et al. Erlotinib Plus Gemcitabine Compared with Gemcitabine Alone in Patients With Advanced Pancreatic Cancer: A Phase III Trial of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 2007, 25, 1960–1966. [Google Scholar] [CrossRef] [PubMed]
- Okamura, R.; Boichard, A.; Kato, S.; Sicklick, J.K.; Bazhenova, L.; Kurzrock, R. Analysis of NTRK Alterations in Pan-Cancer Adult and Pediatric Malignancies: Implications for NTRK-Targeted Therapeutics. JCO Precis. Oncol. 2018, 2, 1–20. [Google Scholar] [CrossRef]
- Pishvaian, M.J.; Rolfo, C.D.; Liu, S.V.; Multani, P.S.; Maneval, E.C.; Garrido-Laguna, I. Clinical benefit of entrectinib for patients with metastatic pancreatic cancer who harbor NTRK and ROS1 fusions. J. Clin. Oncol. 2018, 36, 521. [Google Scholar] [CrossRef]
- Drilon, A.; Laetsch, T.W.; Kummar, S.; Dubois, S.G.; Lassen, U.N.; Demetri, G.D.; Nathenson, M.; Doebele, R.C.; Farago, A.F.; Pappo, A.S.; et al. Efficacy of Larotrectinib inTRKFusion–Positive Cancers in Adults and Children. N. Engl. J. Med. 2018, 378, 731–739. [Google Scholar] [CrossRef]
- Singhi, A.D.; Ali, S.M.; Lacy, J.; Hendifar, A.; Nguyen, K.; Koo, J.; Chung, J.H.; Greenbowe, J.; Ross, J.S.; Nikiforova, M.N.; et al. Identification of Targetable ALK Rearrangements in Pancreatic Ductal Adenocarcinoma. J. Natl. Compr. Cancer Netw. 2017, 15, 555–562. [Google Scholar] [CrossRef]
- Fountzilas, C.; Adjei, A.; Opyrchal, M.; Evans, R.; Ghasemi, M.; Attwood, K.; Groman, A.; Bshara, W.; Goey, A.; Wilton, J.; et al. A phase I study of the anaplastic lymphoma kinase inhibitor ceritinib in combination with gemcitabine-based chemotherapy in patients with advanced solid tumors. Int. J. Cancer 2021, 149, 2063–2074. [Google Scholar] [CrossRef]
- Heining, C.; Horak, P.; Uhrig, S.; Codo, P.L.; Klink, B.; Hutter, B.; Fröhlich, M.; Bonekamp, D.; Richter, D.; Steiger, K.; et al. NRG1 Fusions in KRAS Wild-Type Pancreatic Cancer. Cancer Discov. 2018, 8, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Chou, A.; Froio, D.; Nagrial, A.M.; Parkin, A.; Murphy, K.J.; Chin, V.T.; Wohl, D.; Steinmann, A.; Stark, R.; Drury, A.; et al. Tailored first-line and second-line CDK4-targeting treatment combinations in mouse models of pancreatic cancer. Gut 2017, 67, 2142–2155. [Google Scholar] [CrossRef]
- Sasankan, S.; Rebuck, L.; Darrah, G.; Turquie, M.H.; Rabinowitz, I. Metastatic Pancreatic Cancer with BRAF and P53 Mutations: Case Report of Therapeutic Response to Doublet Targeted Therapy. Case Rep. Oncol. 2020, 13, 1239–1243. [Google Scholar] [CrossRef]
- Daamen, L.A.; Groot, V.P.; Heerkens, H.D.; Intven, M.P.; Van Santvoort, H.C.; Molenaar, I.Q. Systematic review on the role of serum tumor markers in the detection of recurrent pancreatic cancer. HPB 2018, 20, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Rieser, C.J.; Zenati, M.; Hamad, A.; Al Abbas, A.; Bahary, N.; Zureikat, A.; Zeh, H.J.; Hogg, M.E. CA19-9 on Postoperative Surveillance in Pancreatic Ductal Adenocarcinoma: Predicting Recurrence and Changing Prognosis over Time. Ann. Surg. Oncol. 2018, 25, 3483–3491. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Z.; Kan, H.; Sun, Z.; Xing, J.; Cheng, Y.; Bai, C. CA19-9 elevation as an indication to start salvage treatment in surveillance after pancreatic cancer resection. Pancreatology 2019, 19, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Groot, V.P.; Gemenetzis, G.; Blair, A.; Rivero-Soto, R.J.; Yu, J.; Javed, A.A.; Burkhart, R.A.; Rinkes, I.H.M.B.; Molenaar, I.Q.; Cameron, J.L.; et al. Defining and Predicting Early Recurrence in 957 Patients with Resected Pancreatic Ductal Adenocarcinoma. Ann. Surg. 2019, 269, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Sausen, M.; Phallen, J.; Adleff, V.; Jones, S.; Leary, R.J.; Barrett, M.T.; Anagnostou, V.; Parpart-Li, S.; Murphy, D.; Kay Li, Q.; et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat. Commun. 2015, 6, 7686. [Google Scholar] [CrossRef] [PubMed]
- Creemers, A.; Krausz, S.; Strijker, M.; Van Der Wel, M.; Soer, E.; Reinten, R.; Besselink, M.; Wilmink, J.; van de Vijver, M.; Van Noesel, C.; et al. Clinical value of ctDNA in upper-GI cancers: A systematic review and meta-analysis. Biochim. Biophys. Acta (BBA) Bioenergy 2017, 1868, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.; Lindeman, N.; Lockwood, C.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients with Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, H.; Chen, N.; Hao, J.; Jin, H.; Ma, X. Diagnostic value of various liquid biopsy methods for pancreatic cancer: A systematic review and meta-analysis. Medicine 2020, 99, e18581. [Google Scholar] [CrossRef]
- Märten, A.; Büchler, M.W.; Werft, W.; Wente, M.N.; Kirschfink, M.; Schmidt, J. Soluble iC3b as an Early Marker for Pancreatic Adenocarcinoma Is Superior to CA19.9 and Radiology. J. Immunother. 2010, 33, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhong, X.; Zhou, L.; Lu, J.; Jiang, B.; Liu, C.; Guo, J. Prognostic Biomarkers for Pancreatic Ductal Adenocarcinoma: An Umbrella Review. Front. Oncol. 2020, 10, 1466. [Google Scholar] [CrossRef] [PubMed]
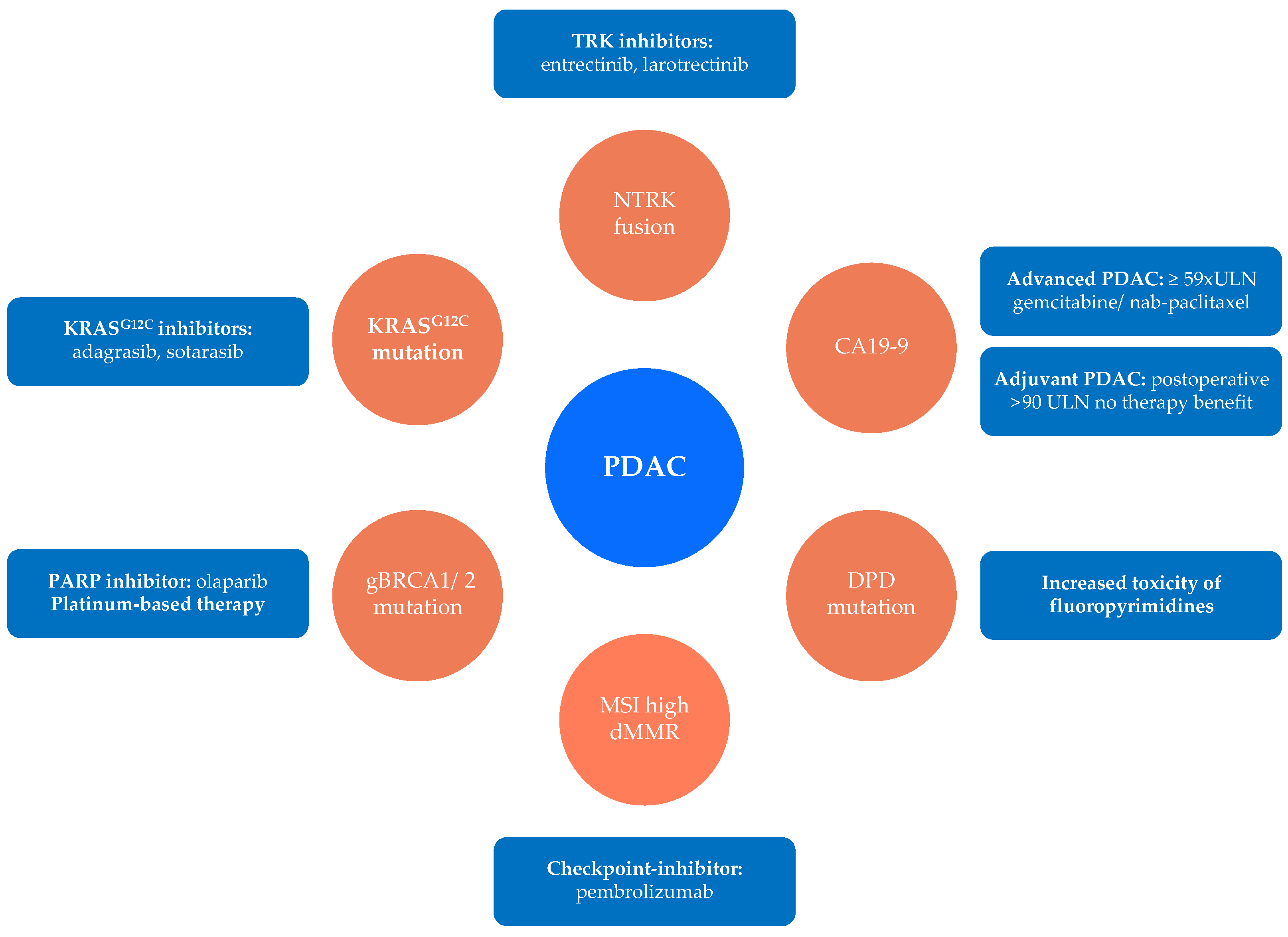
| Drug | Biomarker | Source | Prognosis | Evidence | Ref. |
|---|---|---|---|---|---|
| Gemcitabine | hENT1-high | Tumor | ↑ | IB | [84] |
| Exosomal miRNA-106b | Blood | ↓ | Preclinical | [110] | |
| Deoxycytidine kinase | Tumor | ↓ | Preclinical | [89] | |
| SPARC | Tumor | ↓ | Preclinical | [96] | |
| Nab-Paclitaxel | CA19-9 | Blood | ↓ | IB | [77] |
| 5-FU | Multidrug resistance associated protein 5 | Tumor | ↓ | Preclinical | [98] |
| Dihydropyrimidine dehydrogenase | Tumor | ↓ | IB | [100] | |
| Irinotecan | Carboxylesterase 2 | Tumor | ↑ | III | [104] |
| Nal-Irinotecan | IL8 | Blood | ↓ | Preclinical | [105] |
| Oxaliplatin | ERCC1 | Tumor | ↓ | III | [107] |
| BRCA1/2, PALB2 | Serum | ↑ | III | [111] |
| Drug | Target | Biomarker | Frequency | Evidence | Ref. |
|---|---|---|---|---|---|
| Pembrolizumab | PD-1 | MSI-high, dMMR, TMB-high | <1% | IIa | [120] |
| Olaparib | PARP | BRCA1/2, PALB2, ATM/ATR, CHEK2, RAD51 mutation | 14–24% | Ib | [119] |
| Larotrectinib Entrectinib | TRK | NTRK-fusion | <1% | IIa | [125] |
| Ceritinib Crizotinib Alecitinib | ALK | ALK rearrangement | <1% | IV | [126] |
| Afatinib Erlotinib | EGFR | NRG1 fusion | <1% | IV/III | [128] |
| Trametinib | BRAF | BRAF deletion | 1% | IV | [130] |
| Sotorasib | KRAS G12C | KRAS G12C mutation | 1% | IV | [113] |
| Palbociclib Ribociclib Abemaciclib | CDK4/6 | Loss of CDKN2a | 47% | IV | [129] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sturm, N.; Ettrich, T.J.; Perkhofer, L. The Impact of Biomarkers in Pancreatic Ductal Adenocarcinoma on Diagnosis, Surveillance and Therapy. Cancers 2022, 14, 217. https://doi.org/10.3390/cancers14010217
Sturm N, Ettrich TJ, Perkhofer L. The Impact of Biomarkers in Pancreatic Ductal Adenocarcinoma on Diagnosis, Surveillance and Therapy. Cancers. 2022; 14(1):217. https://doi.org/10.3390/cancers14010217
Chicago/Turabian StyleSturm, Niklas, Thomas J. Ettrich, and Lukas Perkhofer. 2022. "The Impact of Biomarkers in Pancreatic Ductal Adenocarcinoma on Diagnosis, Surveillance and Therapy" Cancers 14, no. 1: 217. https://doi.org/10.3390/cancers14010217
APA StyleSturm, N., Ettrich, T. J., & Perkhofer, L. (2022). The Impact of Biomarkers in Pancreatic Ductal Adenocarcinoma on Diagnosis, Surveillance and Therapy. Cancers, 14(1), 217. https://doi.org/10.3390/cancers14010217






