Identification of Neoadjuvant Chemotherapy Response in Muscle-Invasive Bladder Cancer by Fourier-Transform Infrared Micro-Imaging
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation by Tissue Microarray (TMA)
2.2. Infrared Microimaging
2.3. Chemometric Processing of IR Data
2.3.1. Individual Kmeans Clustering for Associating IR Spectral Signatures with Tissue Structures
2.3.2. Partial Least Square-Discriminant Analysis (PLS-DA) for Automatic Selection of Image IR Pixels
2.3.3. Partial Least Square (PLS) Modeling for Scoring the Response to NAC Based on the IR Signatures of the Tissue Specimens
2.3.4. External Validation Set
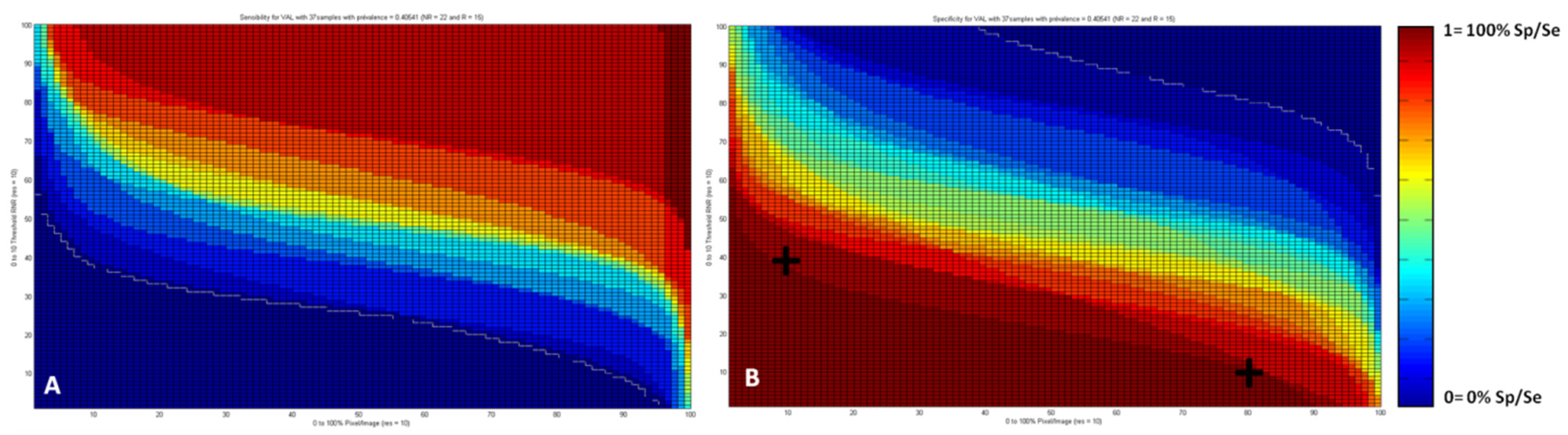
2.3.5. Sensitivity and Specificity of the IR Approach According to the Percentage of Pixels and the Responder/Non-Responder Score
3. Results
3.1. Patients Characteristics
3.2. IR Analysis of the Transurethral Resection of Bladder Tumor Samples and Constitution of Calibration and External Validation Sets
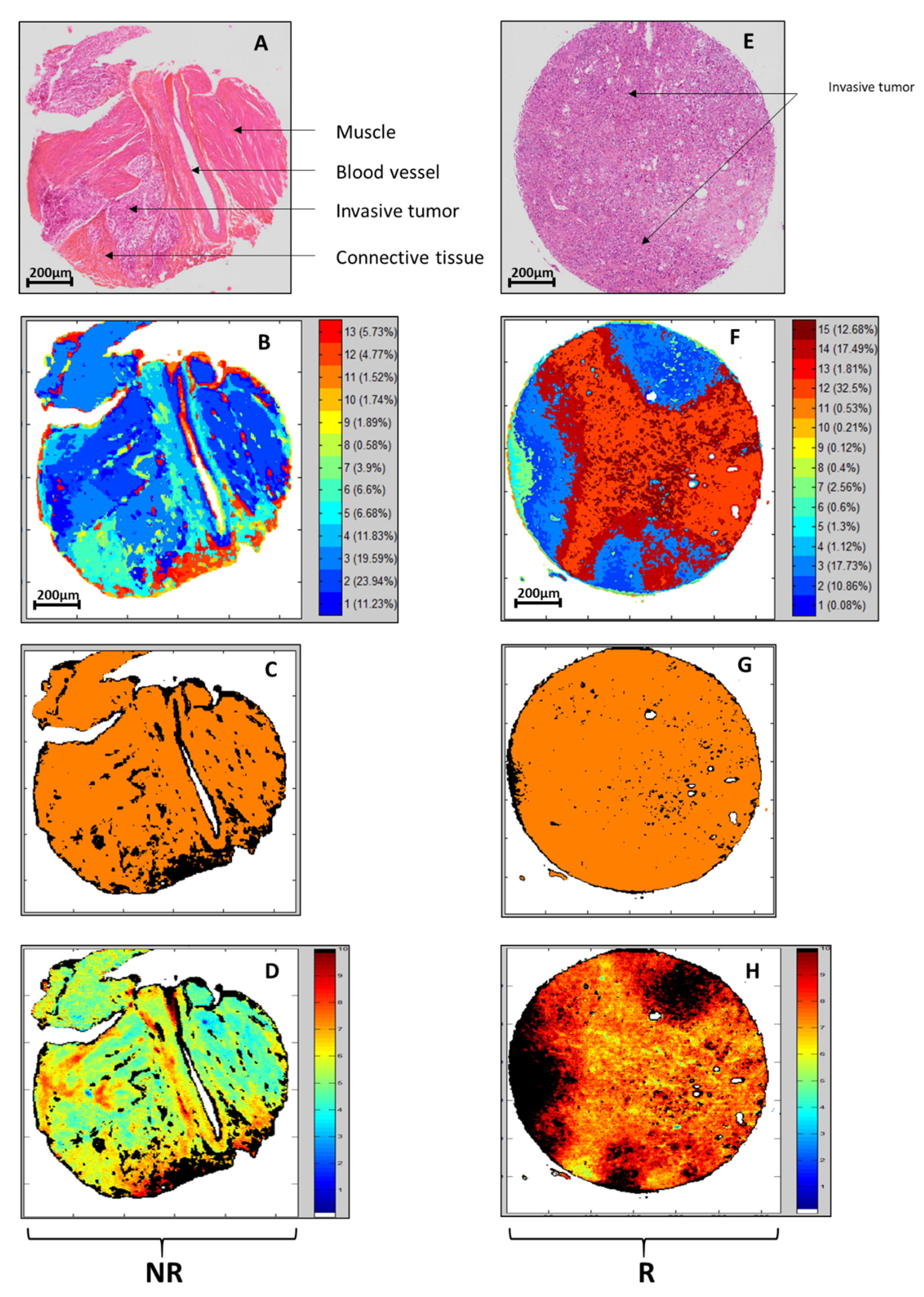
3.3. Recognition of Tissue Structures Using Individual KMeans Clustering and PLS-DA of Spectral Images
3.4. PLS Scoring of the R/NR Scale
3.5. Sensitivity and Specificity Maps
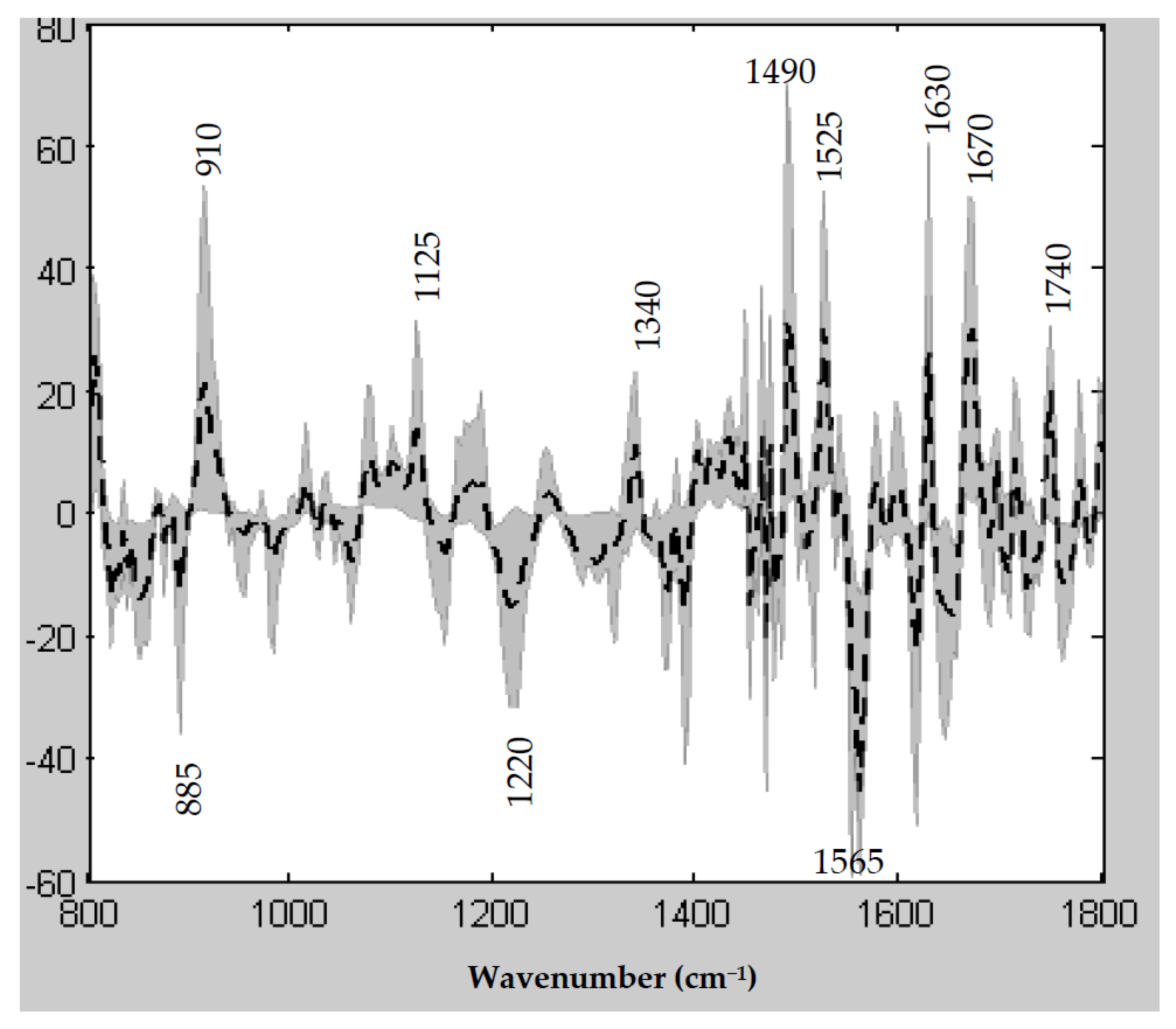
3.6. Spectral Features Underlying the PLS R/NR Scale
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rouprêt, M.; Neuzillet, Y.; Masson-Lecomte, A.; Colin, P.; Compérat, E.; Dubosq, F.; Houédé, N.; Larré, S.; Pignot, G.; Puech, P.; et al. CCAFU french national guidelines 2016–2018 on bladder cancer. Prog. Urol. 2016, 27, S67–S91. [Google Scholar] [CrossRef]
- Witjes, J.A.; Lebret, T.; Compérat, E.M.; Cowan, N.C.; De Santis, M.; Bruins, H.M.; Hernández, V.; Espinós, E.L.; Dunn, J.; Rouanne, M.; et al. Updated 2016 EAU Guidelines on muscle-invasive and metastatic bladder cancer. Eur. Urol. 2017, 71, 462–475. [Google Scholar] [CrossRef]
- Advanced Bladder Cancer Overview Collaboration. Neoadjuvant chemotherapy for invasive bladder cancer. Cochrane Database Syst. Rev. 2005, 2, CD005246. [Google Scholar]
- Sonpavde, G.; Goldman, B.H.; Speights, V.O.; Lerner, S.P.; Wood, D.P.; Vogelzang, N.J.; Trump, D.L.; Natale, R.B.; Grossman, H.B.; Crawford, E.D. Quality of pathologic response and surgery correlate with survival for patients with completely resected bladder cancer after neoadjuvant chemotherapy. Cancer 2009, 115, 4104–4109. [Google Scholar] [CrossRef] [Green Version]
- Petrelli, F.; Coinu, A.; Cabiddu, M.; Ghilardi, M.; Vavassori, I.; Barni, S. Correlation of pathologic complete response with survival after neoadjuvant chemotherapy in bladder cancer treated with cystectomy: A meta-analysis. Eur. Urol. 2014, 65, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Choueiri, T.K.; Jacobus, S.; Bellmunt, J.; Qu, A.; Appleman, L.J.; Tretter, C.; Bubley, G.J.; Stack, E.C.; Signoretti, S.; Walsh, M.; et al. Neoadjuvant dose-dense methotrexate; vinblastine; doxorubicin; and cisplatin with pegfilgrastim support in muscle-invasive urothelial cancer: Pathologic; radiologic; and biomarker correlates. J. Clin. Oncol. 2014, 32, 1889–1894. [Google Scholar] [CrossRef]
- Plimack, E.R.; Hoffman-Censits, J.H.; Viterbo, R.; Trabulsi, E.J.; Ross, E.A.; Greenberg, R.E.; Chen, D.Y.; Lallas, C.D.; Wong, Y.N.; Lin, J.; et al. Accelerated methotrexate; vinblastine; doxorubicin; and cisplatin is safe; effective; and efficient neoadjuvant treatment for muscle-invasive bladder cancer: Results of a multicenter phase II study with molecular correlates of response and toxicity. J. Clin. Oncol. 2014, 32, 1895–1901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef] [Green Version]
- Buttigliero, C.; Tucci, M.; Vignani, F.; Scagliotti, G.V.; Di Maio, M. Molecular biomarkers to predict response to neoadjuvant chemotherapy for bladder cancer. Cancer Treat Rev. 2017, 54, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.; DeFusco, P.A.; Ricci, A., Jr.; Cronin, E.B.; Hegde, P.U.; Kane, M.; Tavakoli, B.; Xu, Y.; Hart, J.; Tannenbaum, S.H. Breast cancer: Assessing response to neoadjuvant chemotherapy by using US-guided near-infrared tomograph. Radiology 2013, 266, 433–442. [Google Scholar] [CrossRef] [Green Version]
- Aine, M.; Eriksson, P.; Liedberg, F.; Sjödahl, G.; Höglund, M. Biological determinants of bladder cancer gene expression subtypes. Sci. Rep. 2015, 5, 10957. [Google Scholar] [CrossRef] [Green Version]
- Choi, W.; Porten, S.; Kim, S.; Willis, D.; Plimack, E.R.; Hoffman-Censits, J.; Roth, B.; Cheng, T.; Tran, M.; Lee, I.L.; et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014, 25, 152–165. [Google Scholar] [CrossRef] [Green Version]
- Kardos, J.; Chai, S.; Mose, L.E.; Selitsky, S.R.; Krishnan, B.; Saito, R.; Iglesia, M.D.; Milowsky, M.I.; Parker, J.S.; Kim, W.Y.; et al. Claudin-low bladder tumors are immune infiltrated and actively immune suppressed. JCI Insight 2016, 1, e85902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evers, D.; Hendriks, B.; Lucassen, G.; Ruers, T. Optical spectroscopy: Current advances and future applications in cancer diagnostics and therapy. Future Oncol. 2012, 8, 307–320. [Google Scholar] [CrossRef]
- Ly, E.; Piot, O.; Wolthuis, R.; Durlach, A.; Bernard, P.; Manfait, M. Combination of FTIR spectral imaging and chemometrics for tumour detection from paraffin-embedded biopsies. Analyst 2008, 133, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Mackanos, M.A.; Contag, C.H. FTIR microspectroscopy for improved prostate cancer diagnosis. Trends Biotechnol. 2009, 27, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Nallala, J.; Diebold, M.D.; Gobinet, C.; Bouché, O.; Sockalingum, G.D.; Piot, O.; Manfait, M. Infrared spectral histopathology for cancer diagnosis: A novel approach for automated pattern recognition of colon adenocarcinoma. Analyst 2014, 139, 4005–4015. [Google Scholar] [CrossRef]
- Verdonck, M.; Denayer, A.; Delvaux, B.; Garaud, S.; De Wind, R.; Desmedt, C.; Sotiriou, C.; Willard-Gallo, K.; Goormaghtigh, E. Characterization of human breast cancer tissues by infrared imaging. Analyst 2016, 141, 606–619. [Google Scholar] [CrossRef]
- Pounder, F.N.; Reddy, R.K.; Bhargava, R. Development of a practical spatial-spectral analysis protocol for breast histopathology using Fourier transform infrared spectroscopic imaging. Faraday Discuss. 2016, 187, 43–68. [Google Scholar] [CrossRef] [Green Version]
- Chrabaszcz, K.; Kaminska, K.; Augustyniak, K.; Kujdowicz, M.; Smeda, M.; Jaszta, A.; Stojak, M.; Marzec, K.M.; Malek, K. Tracking Extracellular Matrix Remodeling in Lungs Induced by Breast Cancer Metastasis. Fourier Transform Infrared Spectroscopic Studies. Molecules 2020, 25, 236. [Google Scholar] [CrossRef] [Green Version]
- Augustyniak, K.; Chrabaszcz, K.; Jasztal, A.; Smeda, M.; Quintas, G.; Kuligowski, J.; Marzec, K.M.; Malek, K. High and ultra-high definition of infrared spectral histopathology gives an insight into chemical environment of lung metastases in breast cancer. J. Biophotonics 2019, 12, e201800345. [Google Scholar] [CrossRef] [PubMed]
- Flower, K.R.; Khalifa, I.; Bassan, P.; Démoulin, D.; Jackson, E.; Lockyer, N.P.; McGown, A.T.; Miles, P.; Vaccari, L.; Gardner, P. Synchrotron FTIR analysis of drug treated ovarian A2780 cells: An ability to differentiate cell response to different drugs? Analyst 2011, 136, 498–507. [Google Scholar] [CrossRef]
- Derenne, A.; Gasper, R.; Goormaghtigh, E. The FTIR spectrum of prostate cancer cells allows the classification of anticancer drugs according to their mode of action. Analyst 2011, 136, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Gaydou, V.; Polette, M.; Gobinet, C.; Kileztky, C.; Angiboust, J.F.; Birembaut, P.; Vuiblet, V.; Piot, O. New insights into spectral histopathology: Infrared-based scoring of tumour aggressiveness of squamous cell lung carcinomas. Chem. Sci. 2019, 10, 4246–4258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolthuis, R.; Travo, A.; Nicolet, C.; Neuville, A.; Gaub, M.P.; Guenot, D.; Ly, E.; Manfait, M.; Jeannesson, P.; Piot, O. IR spectral imaging for histopathological characterization of xenografted human colon carcinomas. Anal. Chem. 2008, 80, 8461–8469. [Google Scholar] [CrossRef]
- Nguyen, T.N.Q.; Jeannesson, P.; Groh, A.; Piot, O.; Guenot, D.; Gobinet, C. Fully unsupervised inter-individual IR spectral histology of paraffinized tissue sections of normal colon. J. Biophotonics 2016, 9, 521–532. [Google Scholar] [CrossRef]
- Land, W.H.; Ford, W.; Park, J.W.; Mathur, R.; Hotchkiss, N.; Heine, J.; Eschrich, S.; Qiao, X.; Yeatman, T. Partial least squares (PLS) applied to medical bioinformatics. Procedia Comput.Sci. 2011, 6, 273–278. [Google Scholar] [CrossRef] [Green Version]
- Finlayson, D.; Rinaldi, C.; Baker, M.J. Is Infrared Spectroscopy Ready for the Clinic? Anal. Chem. 2019, 91, 12117–12128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paraskevaidi, M.; Morais, C.L.M.; Lima, K.M.G.; Snowden, J.S.; Saxon, J.A.; Richardson, A.M.T.; Jones, M.; Mann, D.M.A.; Allsop, D.; Martin-Hirsch, P.L.; et al. Differential diagnosis of Alzheimer’s disease using spectrochemical analysis of blood. Proc. Natl. Acad. Sci. USA 2017, 114, E7929–E7938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ami, D.; Mereghetti, P.; Foli, A.; Tasaki, M.; Milani, P.; Nuvolone, M.; Palladini, G.; Merlini, G.; Lavatelli, F.; Natalello, A. ATR-FTIR Spectroscopy Supported by Multivariate Analysis for the Characterization of Adipose Tissue Aspirates from Patients Affected by Systemic Amyloidosis. Anal. Chem. 2019, 91, 2894–2900. [Google Scholar] [CrossRef]
- Lechowicz, L.; Chrapek, M.; Gaweda, J.; Urbaniak, M.; Konieczna, I. Use of Fourier-transform infrared spectroscopy in the diagnosis of rheumatoid arthritis: A pilot study. Mol. Biol. Rep. 2016, 43, 1321–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.; Perez-Guaita, D.; Andrew, D.W.; Richards, J.S.; McNaughton, D.; Heraud, P.; Wood, B.R. Simultaneous ATR-FTIR Based Determination of Malaria Parasitemia, Glucose and Urea in Whole Blood Dried onto a Glass Slide. Anal. Chem. 2017, 89, 5238–5245. [Google Scholar] [CrossRef]
- Kumar, S.; Srinivasan, A.; Nikolajeff, F. Role of infrared spectroscopy and imaging in cancer diagnosis. Curr. Med. Chem. 2018, 25, 1055–1072. [Google Scholar] [CrossRef] [PubMed]
- De Bruyne, S.; Speeckaert, M.M.; Delanghe, J.R. Applications of mid-infrared spectroscopy in the clinical laboratory setting. Crit. Rev. Clin. Lab. Sci. 2018, 55, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Stanta, G.; Bonin, S. Overview on clinical relevance of intra-tumor heterogeneity. Front. Med. 2018, 5, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef]
- Mari, A.; D’Andrea, D.; Abufaraj, M.; Foerster, B.; Kimura, S.; Shariat, S.F. Genetic determinants for chemo- and radiotherapy resistance in bladder cancer. Transl. Androl. Urol. 2017, 6, 1081–1089. [Google Scholar] [CrossRef] [Green Version]
- Bellmunt, J.; Orsola, A.; Leow, J.J.; Wiegel, T.; De Santis, M.; Horwich, A. Bladder cancer: ESMO Practice Guidelines for diagnosis; treatment and follow-up. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2014, 25, iii40–iii48. [Google Scholar] [CrossRef]
- McConkey, D.J.; Choi, W.; Shen, Y.; Lee, I.L.; Porten, S.; Matin, S.F.; Kamat, A.M.; Corn, P.; Millikan, R.E.; Dinney, C.; et al. A prognostic gene expression signature in the molecular classification of chemotherapy-naïve urothelial cancer is predictive of clinical outcomes from neoadjuvant chemotherapy: A phase 2 trial of dose-dense methotrexate; vinblastine; doxorubicin; and cisplatin with bevacizumab in urothelial cancer. Eur. Urol. 2016, 69, 855–862. [Google Scholar] [PubMed] [Green Version]
- Shariat, S.F.; Palapattu, G.S.; Karakiewicz, P.I.; Rogers, C.G.; Vazina, A.; Bastian, P.J.; Schoenberg, M.P.; Lerner, S.P.; Sagalowsky, A.I.; Lotan, Y. Discrepancy between clinical and pathologic stage: Impact on prognosis after radical cystectomy. Eur. Urol. 2007, 51, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Turker, P.; Bostrom, P.J.; Wroclawski, M.L.; van Rhijn, B.; Kortekangas, H.; Kuk, C.; Mirtti, T.; Fleshner, N.E.; Jewett, M.A.; Finelli, A.; et al. Upstaging of urothelial cancer at the time of radical cystectomy: Factors associated with upstaging and its effect on outcome. BJU Int. 2012, 110, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Chappidi, M.R.; Kates, M.; Brant, A.; Baras, A.S.; Netto, G.J.; Pierorazio, P.M.; Hahn, N.M.; Bivalacqua, T.J. Assessing cancer progression and stable disease after neoadjuvant chemotherapy for organ-confined muscle-invasive bladder cancer. Urology 2017, 102, 148–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reardon, Z.D.; Patel, S.G.; Zaid, H.B.; Stimson, C.J.; Resnick, M.J.; Keegan, K.A.; Barocas, D.A.; Chang, S.S.; Cookson, M.S. Trends in the use of perioperative chemotherapy for localized and locally advanced muscle-invasive bladder cancer: A sign of changing tides. Eur. Urol. 2015, 67, 165–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borriello, L.; DeClerck, Y.A. Tumor microenvironment and therapeutic resistance process. Med. Sci. 2014, 30, 445–451. [Google Scholar]
- DeClerck, Y.A.; Pienta, K.J.; Woodhouse, E.C.; Singer, D.S.; Mohla, S. The Tumor Microenvironment at a Turning Point Knowledge Gained over the Last Decade; and Challenges and Opportunities ahead: A White Paper from the NCI TME Network. Cancer Res. 2017, 77, 1051–1059. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y. Tumor microenvironment and cancer therapy resistance. Cancer Lett. 2016, 380, 205–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Junttila, M.R.; de Sauvage, F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013, 501, 346–354. [Google Scholar] [CrossRef]

| Patients Characteristics at Diagnosis | Mean (Lower Quartile–Upper Quartile) |
|---|---|
| Age | 66 (48–78) |
| Sex | |
| Male | 33 (77%) |
| Female | 10 (23%) |
| OMS | |
| 0 | 17 (39%) |
| 1 | 16 (37%) |
| 2 | 3 (8%) |
| Missing data | 7 (16%) |
| Charlson score | 3 (2–6) |
| Smokers | 34 (85%) |
| Treatment and Tumor Characteristics | Number % |
|---|---|
| Tumor response | |
| Responders | 19 (44%) |
| Non responders | 24 (56%) |
| Chemotherapy | |
| MVAC-I | 10 (24%) |
| Gemcitabin cisplatin | 24 (57%) |
| Gemcitabin carboplatin | 8 (19%) |
| Mean number of chemotherapy cycles | 4 (3–6) |
| Toxicities (any grade) | 19 (44%) |
| Time between last chemotherapy and surgery (days) | 40 (7–69) |
| Relapse | |
| Number | 11 (34%) |
| Distance surgery-relapse (months) | 15 (2–37) |
| Metastatic | 10 (91%) |
| Missing data | 11 (26%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazza, C.; Gaydou, V.; Eymard, J.-C.; Birembaut, P.; Untereiner, V.; Côté, J.-F.; Brocheriou, I.; Coeffic, D.; Villena, P.; Larré, S.; et al. Identification of Neoadjuvant Chemotherapy Response in Muscle-Invasive Bladder Cancer by Fourier-Transform Infrared Micro-Imaging. Cancers 2022, 14, 21. https://doi.org/10.3390/cancers14010021
Mazza C, Gaydou V, Eymard J-C, Birembaut P, Untereiner V, Côté J-F, Brocheriou I, Coeffic D, Villena P, Larré S, et al. Identification of Neoadjuvant Chemotherapy Response in Muscle-Invasive Bladder Cancer by Fourier-Transform Infrared Micro-Imaging. Cancers. 2022; 14(1):21. https://doi.org/10.3390/cancers14010021
Chicago/Turabian StyleMazza, Camille, Vincent Gaydou, Jean-Christophe Eymard, Philippe Birembaut, Valérie Untereiner, Jean-François Côté, Isabelle Brocheriou, David Coeffic, Philippe Villena, Stéphane Larré, and et al. 2022. "Identification of Neoadjuvant Chemotherapy Response in Muscle-Invasive Bladder Cancer by Fourier-Transform Infrared Micro-Imaging" Cancers 14, no. 1: 21. https://doi.org/10.3390/cancers14010021
APA StyleMazza, C., Gaydou, V., Eymard, J.-C., Birembaut, P., Untereiner, V., Côté, J.-F., Brocheriou, I., Coeffic, D., Villena, P., Larré, S., Vuiblet, V., & Piot, O. (2022). Identification of Neoadjuvant Chemotherapy Response in Muscle-Invasive Bladder Cancer by Fourier-Transform Infrared Micro-Imaging. Cancers, 14(1), 21. https://doi.org/10.3390/cancers14010021






