The Claudin-Low Subtype of High-Grade Serous Ovarian Carcinoma Exhibits Stem Cell Features
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Cell Culture
2.3. Chemoresistance Assay
2.4. Soft Agar Assay
2.5. Quantitative PCR
2.6. Western Blot Analysis
2.7. Flow Cytometry and Cell Sorting
2.8. Metabolic Analysis
2.9. Data processing and Statistical Analysis
3. Results
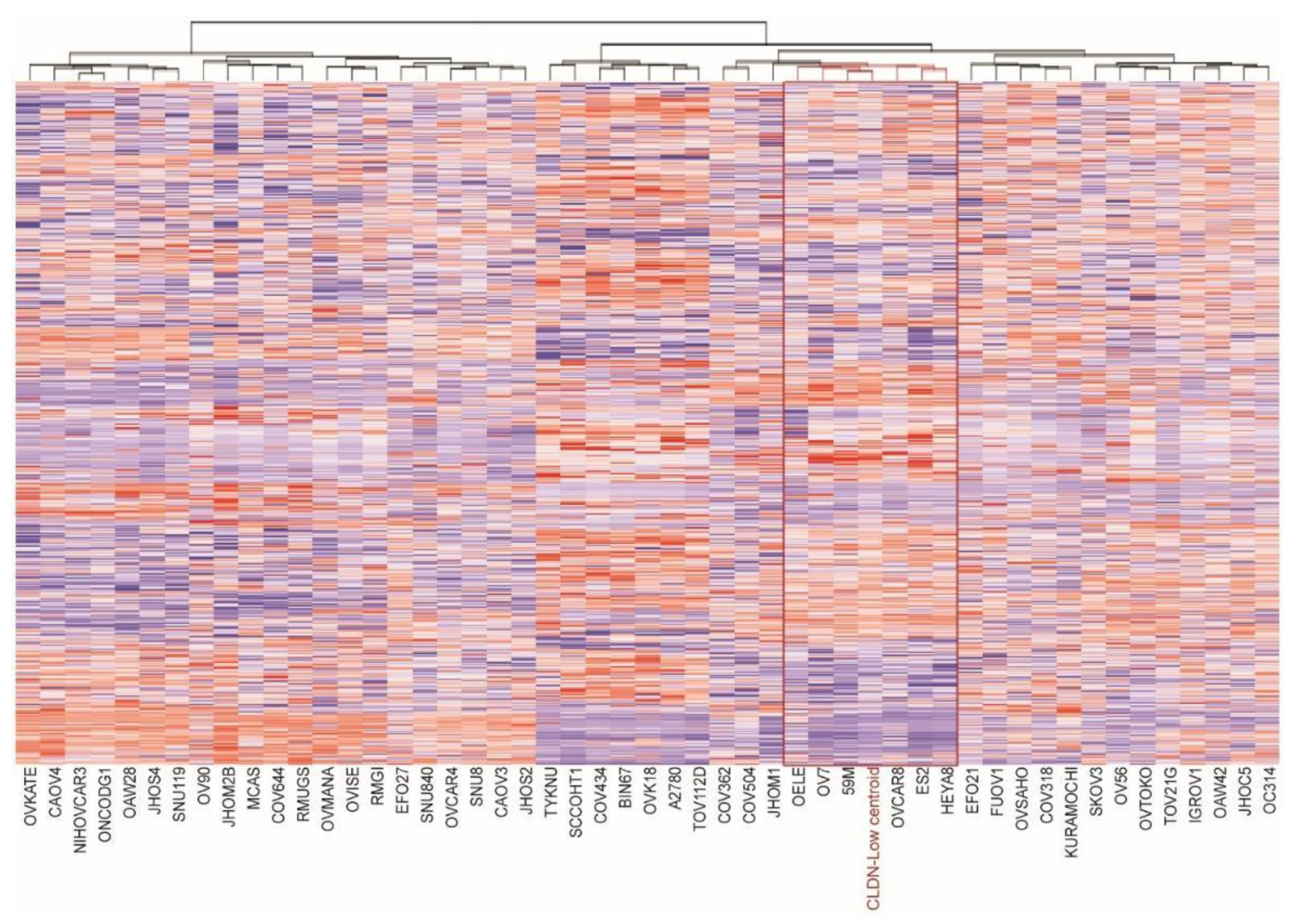
3.1. Identification of Claudin-Low Ovarian Cancer Cell Lines
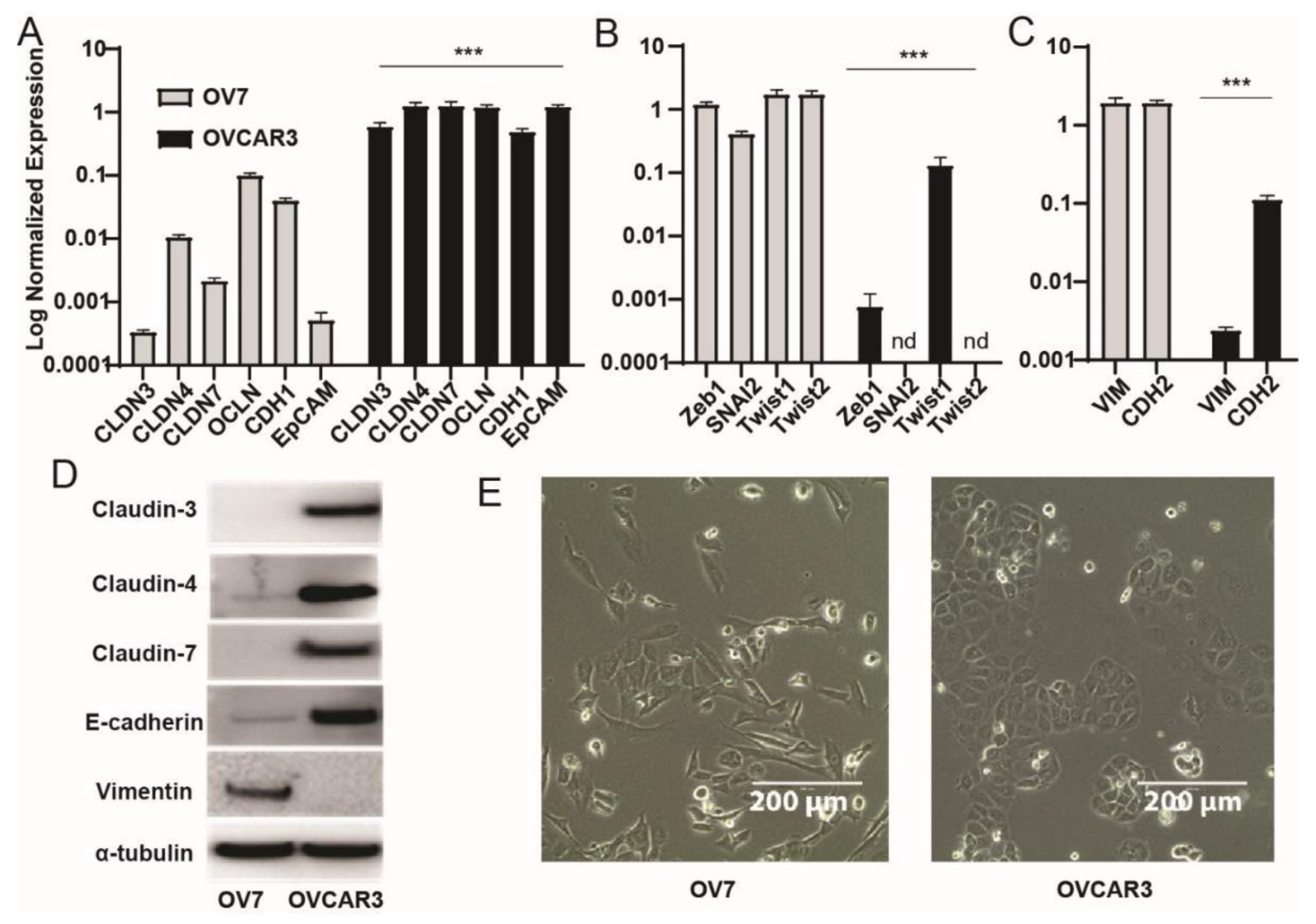
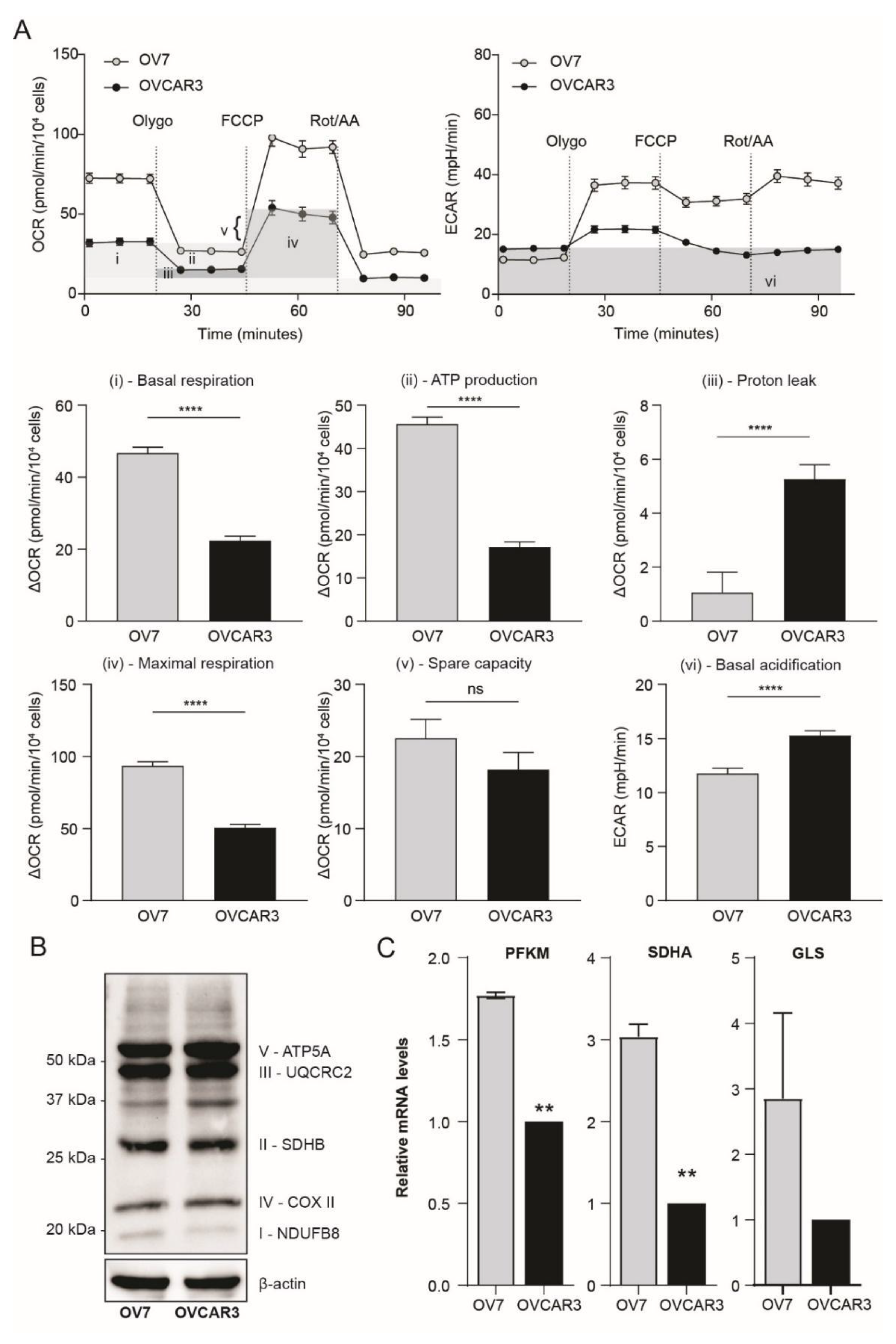
3.2. Ovarian Claudin-Low Cells Display Stem Cell Characteristics
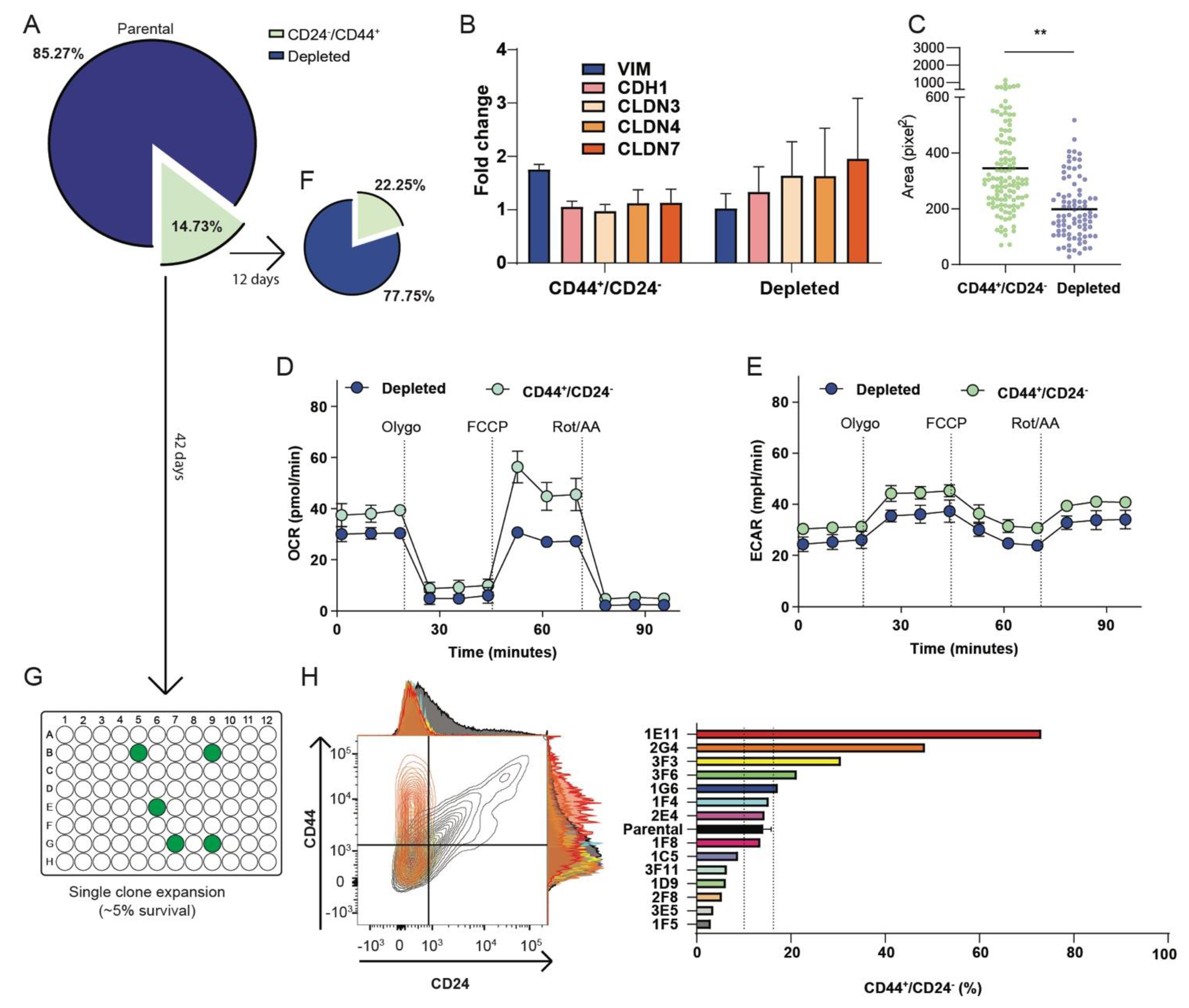
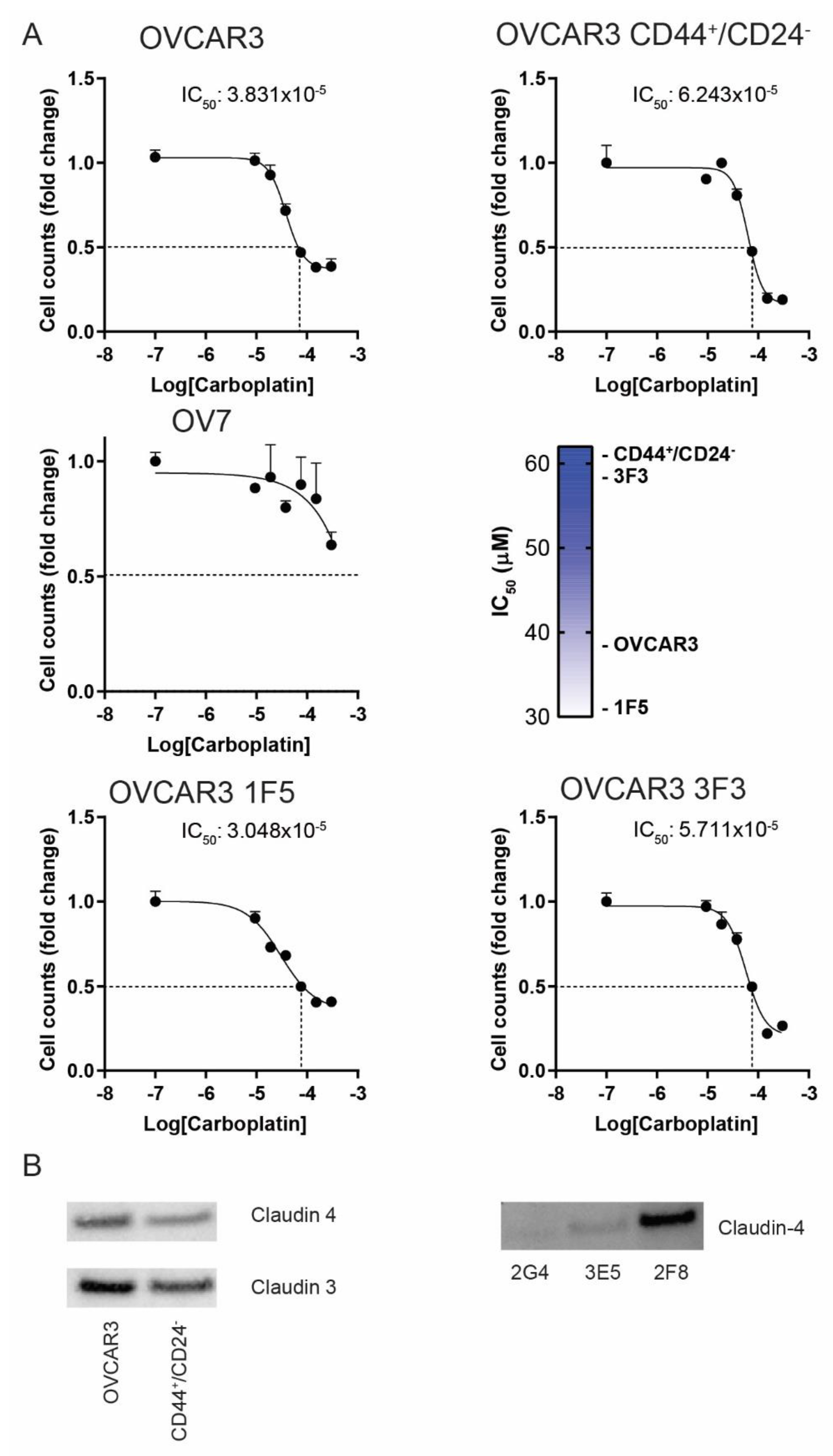
3.3. The CD44+/CD24− Subpopulation of OVCAR3 Cells Resembles CL OV7 Cells
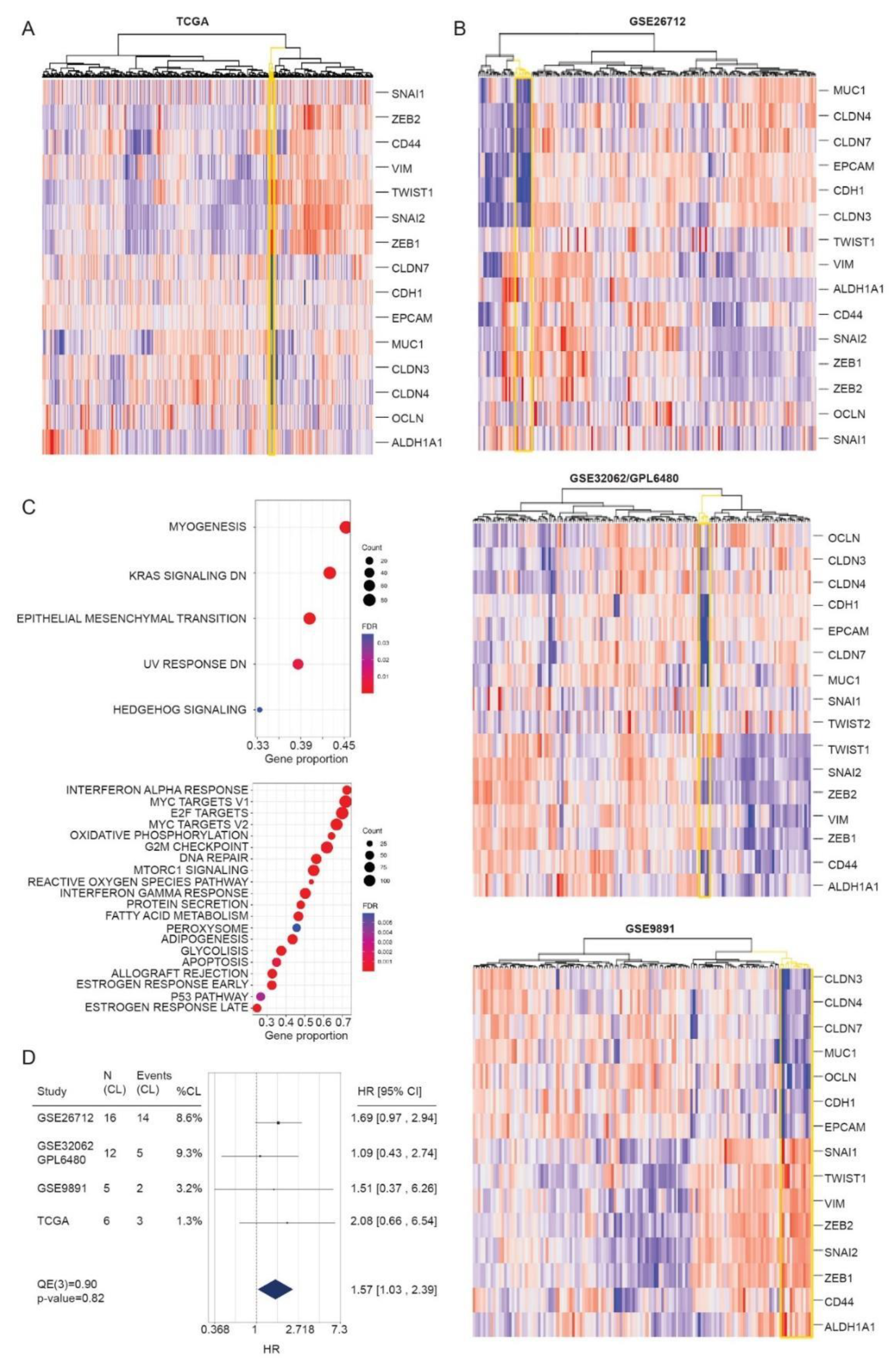
3.4. Identification of CL Molecular Profile in HGSOC Patient Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bowtell, D.D.; Boehm, S.; Ahmed, A.A.; Aspuria, P.-J.; Jr, R.C.B.; Beral, V.; Berek, J.S.; Birrer, M.J.; Blagden, S.; Bookman, M.A.; et al. Rethinking ovarian cancer II: Reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 2015, 15, 668–679. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef]
- Kurman, R.J.; Shih, I.-M. The Dualistic Model of Ovarian Carcinogenesis: Revisited, Revised, and Expanded. Am. J. Pathol. 2016, 186, 733–747. [Google Scholar] [CrossRef] [PubMed]
- Leong, H.S.; Galletta, L.; Etemadmoghadam, D.; George, J.; The Australian Ovarian Cancer Study; Köbel, M.; Ramus, S.J.; Bowtell, D. Efficient molecular subtype classification of high-grade serous ovarian cancer. J. Pathol. 2015, 236, 272–277. [Google Scholar] [CrossRef]
- Wang, C.; Armasu, S.M.; Kalli, K.R.; Maurer, M.J.; Heinzen, E.P.; Keeney, G.L.; Cliby, W.A.; Oberg, A.L.; Kaufmann, S.H.; Goode, E.L. Pooled Clustering of High-Grade Serous Ovarian Cancer Gene Expression Leads to Novel Consensus Subtypes Associated with Survival and Surgical Outcomes. Clin. Cancer Res. 2017, 23, 4077–4085. [Google Scholar] [CrossRef] [PubMed]
- Konstantinopoulos, P.A.; Spentzos, D.; Karlan, B.Y.; Taniguchi, T.; Fountzilas, E.; Francoeur, N.; Levine, D.A.; Cannistra, S.A. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J. Clin. Oncol. 2010, 28, 3555–3561. [Google Scholar] [CrossRef] [PubMed]
- Tucker, S.L.; Gharpure, K.; Herbrich, S.M.; Unruh, A.K.; Nick, A.M.; Crane, E.K.; Coleman, R.L.; Guenthoer, J.; Dalton, H.J.; Wu, S.Y.; et al. Molecular biomarkers of residual disease after surgical debulking of high-grade serous ovarian cancer. Clin. Cancer Res. 2014, 20, 3280–3288. [Google Scholar] [CrossRef]
- Haunschild, C.E.; Tewari, K.S. The current landscape of molecular profiling in the treatment of epithelial ovarian cancer. Gynecol. Oncol. 2021, 160, 333–345. [Google Scholar] [CrossRef]
- Miller, R.; Leary, A.; Scott, C.; Serra, V.; Lord, C.; Bowtell, D.; Chang, D.; Garsed, D.; Jonkers, J.; Ledermann, J.; et al. ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann. Oncol. 2020, 31, 1606–1622. [Google Scholar] [CrossRef]
- Chen, G.M.; Kannan, L.; Geistlinger, L.; Kofia, V.; Safikhani, Z.; Gendoo, D.M.; Parmigiani, G.; Birrer, M.J.; Haibe-Kains, B.; Waldron, L. Consensus on Molecular Subtypes of High-Grade Serous Ovarian Carcinoma. Clin. Cancer Res. 2018, 24, 5037–5047. [Google Scholar] [CrossRef]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Sørlie, T.; Tibshirani, R.; Parker, J.; Hastie, T.; Marron, J.S.; Nobel, A.; Deng, S.; Johnsen, H.; Pesich, R.; Geisler, S.; et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA 2003, 100, 8418–8423. [Google Scholar] [CrossRef] [PubMed]
- Calza, S.; Hall, P.; Auer, G.; Bjöhle, J.; Klaar, S.; Kronenwett, U.; Liu, E.T.; Miller, L.; Ploner, A.; Smeds, J.; et al. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res. 2006, 8, R34. [Google Scholar] [CrossRef]
- Nielsen, T.O.; Hsu, F.D.; Jensen, K.; Cheang, M.; Karaca, G.; Hu, Z.; Hernandez-Boussard, T.; Livasy, C.; Cowan, D.; Dressler, L.; et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin. Cancer Res. 2004, 10, 5367–5374. [Google Scholar] [CrossRef]
- Jönsson, J.-M.; Johansson, I.; Domínguez-Valentin, M.; Kimbung, S.; Jonsson, M.; Bonde, J.H.; Kannisto, P.; Måsbäck, A.; Malander, S.; Nilbert, M.; et al. Molecular subtyping of serous ovarian tumors reveals multiple connections to intrinsic breast cancer subtypes. PLoS ONE 2014, 9, e107643. [Google Scholar] [CrossRef]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef]
- Perou, C.M. Molecular stratification of triple-negative breast cancers. Oncologist 2010, 15 (Suppl. S5), 39–48. [Google Scholar] [CrossRef]
- Prat, A.; Parker, J.S.; Karginova, O.; Fan, C.; Livasy, C.; I Herschkowitz, J.; He, X.; Perou, C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010, 12, R68. [Google Scholar] [CrossRef]
- Sabatier, R.; Finetti, P.; Guille, A.; Adélaïde, J.; Chaffanet, M.; Viens, P.; Birnbaum, D.; Bertucci, F. Claudin-low breast cancers: Clinical, pathological, molecular and prognostic characterization. Mol. Cancer 2014, 13, 228. [Google Scholar] [CrossRef]
- Dias, K.; Dvorkin-Gheva, A.; Hallett, R.M.; Wu, Y.; Hassell, J.; Pond, G.R.; Levine, M.; Whelan, T.; Bane, A.L. Claudin-Low Breast Cancer; Clinical & Pathological Characteristics. PLoS ONE 2017, 12, e0168669. [Google Scholar] [CrossRef]
- Kardos, J.; Chai, S.; Mose, L.E.; Selitsky, S.R.; Krishnan, B.; Saito, R.; Iglesia, M.D.; Milowsky, M.I.; Parker, J.S.; Kim, W.Y.; et al. Claudin-low bladder tumors are immune infiltrated and actively immune suppressed. JCI Insight 2016, 1, e85902. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Ganzfried, B.F.; Riester, M.; Haibe-Kains, B.; Risch, T.; Tyekucheva, S.; Jazic, I.; Wang, X.V.; Ahmadifar, M.; Birrer, M.J.; Parmigiani, G.; et al. curatedOvarianData: Clinically annotated data for the ovarian cancer transcriptome. Database 2013, 2013, bat013. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Bonome, T.; Levine, D.A.; Shih, J.H.; Randonovich, M.; Pise-Masison, C.A.; Bogomolniy, F.; Ozbun, L.L.; Brady, J.N.; Barrett, J.C.; Boyd, J.; et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008, 68, 5478–5486. [Google Scholar] [CrossRef]
- Tothill, R.W.; Tinker, A.V.; George, J.; Brown, R.; Fox, S.B.; Lade, S.; Johnson, D.S.; Trivett, M.K.; Etemadmoghadam, D.; Locandro, B.; et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res. 2008, 14, 5198–5208. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Tsunoda, T.; Shigemizu, D.; Fujiwara, H.; Hatae, M.; Fujiwara, H.; Masuzaki, H.; Katabuchi, H.; Kawakami, Y.; Okamoto, A.; et al. High-risk ovarian cancer based on 126-gene expression signature is uniquely characterized by downregulation of antigen presentation pathway. Clin. Cancer Res. 2012, 18, 1374–1385. [Google Scholar] [CrossRef] [PubMed]
- Codenotti, S.; Faggi, F.; Ronca, R.; Chiodelli, P.; Grillo, E.; Guescini, M.; Megiorni, F.; Marampon, F.; Fanzani, A. Caveolin-1 enhances metastasis formation in a human model of embryonal rhabdomyosarcoma through Erk signaling cooperation. Cancer Lett. 2019, 449, 135–144. [Google Scholar] [CrossRef]
- Matarazzo, S.; Melocchi, L.; Rezzola, S.; Grillo, E.; Maccarinelli, F.; Giacomini, A.; Turati, M.; Taranto, S.; Zammataro, L.; Cerasuolo, M.; et al. Long Pentraxin-3 Follows and Modulates Bladder Cancer Progression. Cancers 2019, 11, 1277. [Google Scholar] [CrossRef]
- Romani, C.; Zizioli, V.; Silvestri, M.; Ardighieri, L.; Bugatti, M.; Corsini, M.; Todeschini, P.; Marchini, S.; D’Incalci, M.; Zanotti, L.; et al. Low Expression of Claudin-7 as Potential Predictor of Distant Metastases in High-Grade Serous Ovarian Carcinoma Patients. Front. Oncol. 2020, 10, 1287. [Google Scholar] [CrossRef]
- Ronca, R.; Ghedini, G.C.; Maccarinelli, F.; Sacco, A.; Locatelli, S.L.; Foglio, E.; Taranto, S.; Grillo, E.; Matarazzo, S.; Castelli, R.; et al. FGF Trapping Inhibits Multiple Myeloma Growth through c-Myc Degradation-Induced Mitochondrial Oxidative Stress. Cancer Res. 2020, 80, 2340–2354. [Google Scholar] [CrossRef] [PubMed]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Gendoo, D.M.A.; Ratanasirigulchai, N.; Schröder, M.S.; Paré, L.; Parker, J.S.; Prat, A.; Haibe-Kains, B. Genefu: An R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics 2016, 32, 1097–1099. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing. Vienna, Austria, 2020. Available online: https://www.R-project.org (accessed on 1 October 2020).
- Neve, R.M.; Chin, K.; Fridlyand, J.; Yeh, J.; Baehner, F.L.; Fevr, T.; Clark, L.; Bayani, N.; Coppe, J.-P.; Tong, F.; et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10, 515–527. [Google Scholar] [CrossRef]
- Prat, A.; Perou, C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2011, 5, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Meng, E.; Long, B.; Sullivan, P.; McClellan, S.; Finan, M.A.; Reed, E.; Shevde, L.; Rocconi, R.P. CD44+/CD24- ovarian cancer cells demonstrate cancer stem cell properties and correlate to survival. Clin. Exp. Metastasis 2012, 29, 939–948. [Google Scholar] [CrossRef]
- Liu, S.; Cong, Y.; Wang, D.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014, 2, 78–91. [Google Scholar] [CrossRef]
- Liu, M.; Liu, Y.; Deng, L.; Wang, D.; He, X.; Zhou, L.; Wicha, M.S.; Bai, F.; Liu, S. Transcriptional profiles of different states of cancer stem cells in triple-negative breast cancer. Mol. Cancer 2018, 17, 65. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-M.; Giltnane, J.M.; Balko, J.M.; Schwarz, L.J.; Guerrero-Zotano, A.L.; Hutchinson, K.E.; Nixon, M.J.; Estrada, M.V.; Sánchez, V.; Sanders, M.E.; et al. MYC and MCL1 Cooperatively Promote Chemotherapy-Resistant Breast Cancer Stem Cells via Regulation of Mitochondrial Oxidative Phosphorylation. Cell Metab. 2017, 26, 633–647.e7. [Google Scholar] [CrossRef]
- Nishijima, T.F.; Kardos, J.; Chai, S.; Smith, C.C.; Bortone, D.S.; Selitsky, S.R.; Parker, J.S.; Sanoff, H.K.; Lee, M.S.; Vincent, B.G. Molecular and Clinical Characterization of a Claudin-Low Subtype of Gastric Cancer. JCO Precis. Oncol. 2017, 1. [Google Scholar] [CrossRef]
- Pommier, R.M.; Sanlaville, A.; Tonon, L.; Kielbassa, J.; Thomas, E.; Ferrari, A.; Sertier, A.-S.; Hollande, F.; Martinez, P.; Tissier, A.; et al. Comprehensive characterization of claudin-low breast tumors reflects the impact of the cell-of-origin on cancer evolution. Nat. Commun. 2020, 11, 3431. [Google Scholar] [CrossRef] [PubMed]
- Morel, A.-P.; Hinkal, G.W.; Thomas, C.; Fauvet, F.; Courtois-Cox, S.; Wierinckx, A.; Devouassoux-Shisheboran, M.; Treilleux, I.; Tissier, A.; Gras, B.; et al. EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PLoS Genet. 2012, 8, e1002723. [Google Scholar] [CrossRef]
- Chung, V.Y.; Tan, T.Z.; Ye, J.; Huang, R.-L.; Lai, H.-C.; Kappei, D.; Wollmann, H.; Guccione, E.; Huang, R.Y.-J. The role of GRHL2 and epigenetic remodeling in epithelial-mesenchymal plasticity in ovarian cancer cells. Commun. Biol. 2019, 2, 272. [Google Scholar] [CrossRef]
- Corsini, M.; Ravaggi, A.; Odicino, F.; Santin, A.D.; Ravelli, C.; Presta, M.; Romani, C.; Mitola, S. Claudin3 is localized outside the tight junctions in human carcinomas. Oncotarget 2018, 9, 18446–18453. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-J.; Mannan, P.; Lu, M.; Udey, M.C. Epithelial cell adhesion molecule (EpCAM) regulates claudin dynamics and tight junctions. J. Biol. Chem. 2013, 288, 12253–12268. [Google Scholar] [CrossRef]
- Morel, A.-P.; Ginestier, C.; Pommier, R.M.; Cabaud, O.; Ruiz, E.; Wicinski, J.; Devouassoux-Shisheboran, M.; Combaret, V.; Finetti, P.; Chassot, C.; et al. A stemness-related ZEB1-MSRB3 axis governs cellular pliancy and breast cancer genome stability. Nat. Med. 2017, 23, 568–578. [Google Scholar] [CrossRef]
- Garziera, M.; Roncato, R.; Montico, M.; De Mattia, E.; Gagno, S.; Poletto, E.; Scalone, S.; Canzonieri, V.; Giorda, G.; Sorio, R.; et al. New Challenges in Tumor Mutation Heterogeneity in Advanced Ovarian Cancer by a Targeted Next-Generation Sequencing (NGS) Approach. Cells 2019, 8, 584. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, B.T.; Gonzalez-Angulo, A.-M.; Stemke-Hale, K.; Gilcrease, M.Z.; Krishnamurthy, S.; Lee, J.-S.; Fridlyand, J.; A Sahin, A.; Agarwal, R.; Joy, C.; et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009, 69, 4116–4124. [Google Scholar] [CrossRef]
- Fougner, C.; Bergholtz, H.; Norum, J.H.; Sørlie, T. Re-definition of claudin-low as a breast cancer phenotype. Nat. Commun. 2020, 11, 1787. [Google Scholar] [CrossRef] [PubMed]

| Target | TaqMan Gene Expression Assay ID |
|---|---|
| claudin-3 | Hs00265816-s1 |
| claudin-4 | Hs00976831-s1 |
| claudin-7 | Hs00600772-m1 |
| occludin | Hs00170162-m1 |
| E-cadherin | Hs01023895-m1 |
| Zeb1 | Hs00232783-m1 |
| Snai2 | Hs00950344-m1 |
| Twist1 | Hs00361186-m1 |
| Twist2 | Hs00382379-m1 |
| vimentin | Hs00958111-m1 |
| Gene Symbol | Gene Name | mRNA Expression Levels | References |
|---|---|---|---|
| CLDN3 | Claudin 3 | down | Prat, A.; et al. [19]; Sabatier, R.; et al. [20]; Perou, C.M. [18]. |
| CLDN4 | Claudin 4 | down | |
| CLDN7 | Claudin 7 | down | |
| CDH1 | E-cadherin | down | |
| OCLN | Occludin | down | |
| VIM | Vimentin | up | |
| SNAI1 | Snail-1 | up | |
| SNAI2 | Snail-2 | up | |
| TWIST1 | Twist1 | up | |
| TWIST2 | Twist2 | up | |
| ZEB1 | Zinc Finger E-Box Binding Homeobox 1 | up | |
| ZEB2 | Zinc Finger E-Box Binding Homeobox 2 | up | |
| CD44 | CD44 | up | Dias, K.; et al. [21]; Prat, A.; et al. [37]. |
| CD24 | CD24 | down | |
| EPCAM (alias TACSTD1) | Epithelial Cell Adhesion Molecule | down | |
| MUC1 | Mucin 1, Cell Surface Associated | down | |
| ALDH1A1 | Aldehyde Dehydrogenase 1 Family Member A1 | up |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romani, C.; Capoferri, D.; Grillo, E.; Silvestri, M.; Corsini, M.; Zanotti, L.; Todeschini, P.; Ravaggi, A.; Bignotti, E.; Odicino, F.; et al. The Claudin-Low Subtype of High-Grade Serous Ovarian Carcinoma Exhibits Stem Cell Features. Cancers 2021, 13, 906. https://doi.org/10.3390/cancers13040906
Romani C, Capoferri D, Grillo E, Silvestri M, Corsini M, Zanotti L, Todeschini P, Ravaggi A, Bignotti E, Odicino F, et al. The Claudin-Low Subtype of High-Grade Serous Ovarian Carcinoma Exhibits Stem Cell Features. Cancers. 2021; 13(4):906. https://doi.org/10.3390/cancers13040906
Chicago/Turabian StyleRomani, Chiara, Davide Capoferri, Elisabetta Grillo, Marco Silvestri, Michela Corsini, Laura Zanotti, Paola Todeschini, Antonella Ravaggi, Eliana Bignotti, Franco Odicino, and et al. 2021. "The Claudin-Low Subtype of High-Grade Serous Ovarian Carcinoma Exhibits Stem Cell Features" Cancers 13, no. 4: 906. https://doi.org/10.3390/cancers13040906
APA StyleRomani, C., Capoferri, D., Grillo, E., Silvestri, M., Corsini, M., Zanotti, L., Todeschini, P., Ravaggi, A., Bignotti, E., Odicino, F., Sartori, E., Calza, S., & Mitola, S. (2021). The Claudin-Low Subtype of High-Grade Serous Ovarian Carcinoma Exhibits Stem Cell Features. Cancers, 13(4), 906. https://doi.org/10.3390/cancers13040906








