Expression of Genomic Instability-Related Molecules: Cyclin F, RRM2 and SPDL1 and Their Prognostic Significance in Pancreatic Adenocarcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Material
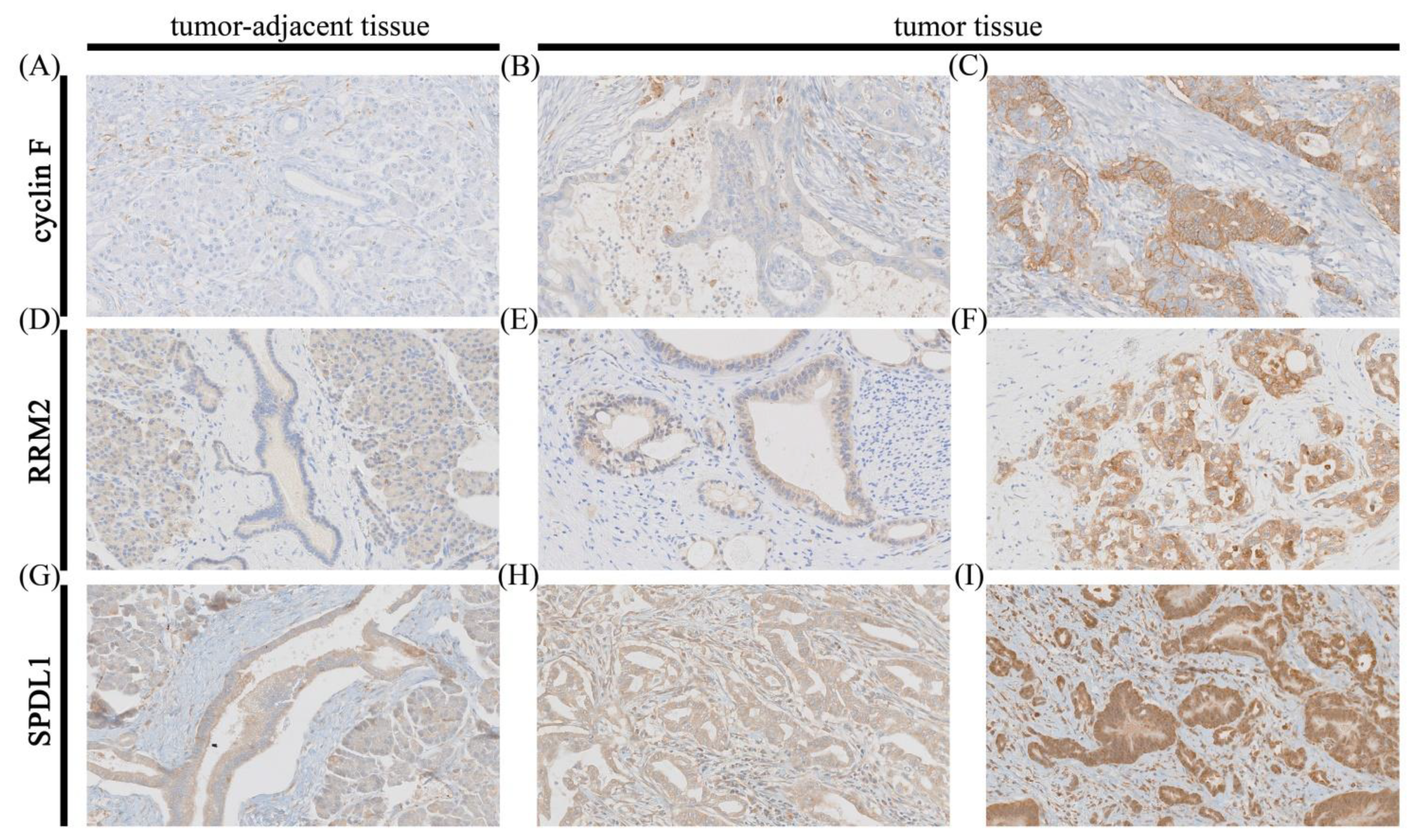
2.2. Immunohistochemistry on Tissue Macroarrays
2.3. Immunostaining Evaluation
2.4. Database Analysis
2.5. Statistical Analysis
3. Results
3.1. Analysis of Immunohistochemical and In Silico Gene Expression Data—Association with Clinicopathological Features and Overall Survival
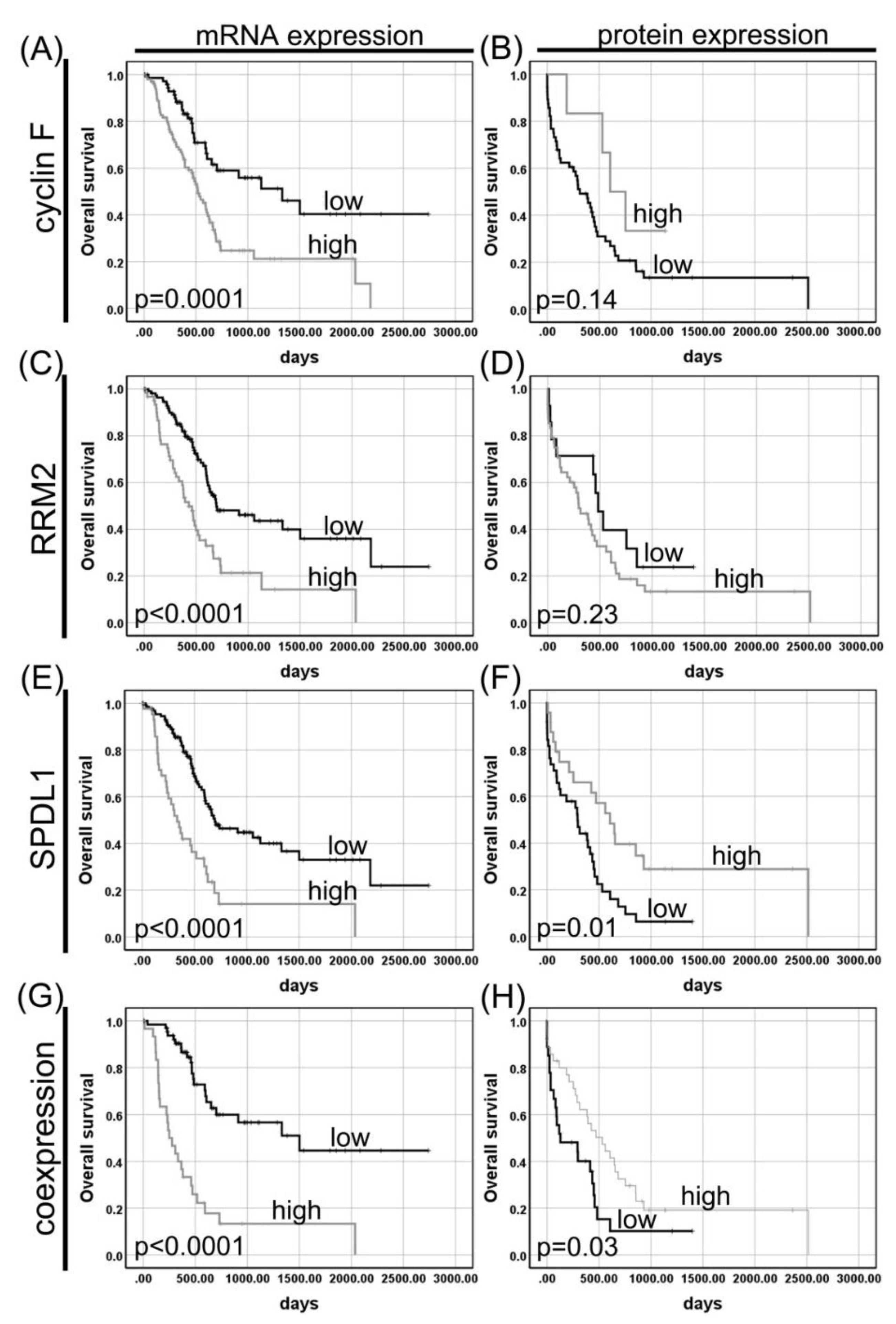
3.1.1. Cyclin F
3.1.2. RRM2
3.1.3. SPDL1 (Spindly)
3.2. Coexpression of Cyclin F, RRM2, and SPDL1 in Pancreatic Adenocarcinoma and Its Impact on Patient Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yao, Y.; Dai, W. Genomic Instability and Cancer. J. Carcinog. Mutagen. 2014, 5, 1000165. [Google Scholar] [CrossRef]
- Sahin, I.H.; Lowery, M.A.; Stadler, Z.K.; Salo-Mullen, E.; Iacobuzio-Donahue, C.A.; Kelsen, D.P.; O’Reilly, E.M. Genomic Instability in Pancreatic Adenocarcinoma: A New Step towards Precision Medicine and Novel Therapeutic Approaches. Expert Rev. Gastroenterol. Hepatol. 2016, 10, 893–905. [Google Scholar] [CrossRef]
- Grossberg, A.J.; Chu, L.C.; Deig, C.R.; Fishman, E.K.; Hwang, W.L.; Maitra, A.; Marks, D.L.; Mehta, A.; Nabavizadeh, N.; Simeone, D.M.; et al. Multidisciplinary Standards of Care and Recent Progress in Pancreatic Ductal Adenocarcinoma. CA Cancer J. Clin. 2020, 70, 375–403. [Google Scholar] [CrossRef] [PubMed]
- Notta, F.; Chan-Seng-Yue, M.; Lemire, M.; Li, Y.; Wilson, G.W.; Connor, A.A.; Denroche, R.E.; Liang, S.-B.; Brown, A.M.K.; Kim, J.C.; et al. A Renewed Model of Pancreatic Cancer Evolution Based on Genomic Rearrangement Patterns. Nature 2016, 538, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Juiz, N.A.; Iovanna, J.; Dusetti, N. Pancreatic Cancer Heterogeneity Can Be Explained Beyond the Genome. Front. Oncol. 2019, 9, 246. [Google Scholar] [CrossRef]
- Ogłuszka, M.; Orzechowska, M.; Jędroszka, D.; Witas, P.; Bednarek, A.K. Evaluate Cutpoints: Adaptable Continuous Data Distribution System for Determining Survival in Kaplan-Meier Estimator. Comput. Methods Programs Biomed. 2019, 177, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Soca-Chafre, G.; Montiel-Dávalos, A.; Rosa-Velázquez, I.A.D.L.; Caro-Sánchez, C.H.S.; Peña-Nieves, A.; Arrieta, O. Multiple Molecular Targets Associated with Genomic Instability in Lung Cancer. Int. J. Genom. 2019, 2019, 9584504. [Google Scholar] [CrossRef]
- D’Angiolella, V.; Donato, V.; Vijayakumar, S.; Saraf, A.; Florens, L.; Washburn, M.P.; Dynlacht, B.; Pagano, M. SCF(Cyclin F) Controls Centrosome Homeostasis and Mitotic Fidelity through CP110 Degradation. Nature 2010, 466, 138–142. [Google Scholar] [CrossRef] [PubMed]
- D’Angiolella, V.; Donato, V.; Forrester, F.M.; Jeong, Y.-T.; Pellacani, C.; Kudo, Y.; Saraf, A.; Florens, L.; Washburn, M.P.; Pagano, M. Cyclin F-Mediated Degradation of Ribonucleotide Reductase M2 Controls Genome Integrity and DNA Repair. Cell 2012, 149, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- D’Angiolella, V.; Esencay, M.; Pagano, M. A Cyclin without Cyclin-Dependent Kinases: Cyclin F Controls Genome Stability through Ubiquitin-Mediated Proteolysis. Trends Cell Biol. 2013, 23, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Emanuele, M.J.; Elia, A.E.H.; Xu, Q.; Thoma, C.R.; Izhar, L.; Leng, Y.; Guo, A.; Chen, Y.-N.; Rush, J.; Hsu, P.W.-C.; et al. Global Identification of Modular Cullin-RING Ligase Substrates. Cell 2011, 147, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Walter, D.; Hoffmann, S.; Komseli, E.-S.; Rappsilber, J.; Gorgoulis, V.; Sørensen, C.S. SCF(Cyclin F)-Dependent Degradation of CDC6 Suppresses DNA Re-Replication. Nat. Commun. 2016, 7, 10530. [Google Scholar] [CrossRef] [PubMed]
- Gagat, M.; Krajewski, A.; Grzanka, D.; Grzanka, A. Potential Role of Cyclin F MRNA Expression in the Survival of Skin Melanoma Patients: Comprehensive Analysis of the Pathways Altered Due to Cyclin F Upregulation. Oncol. Rep. 2018, 40, 123–144. [Google Scholar] [CrossRef]
- Thakur, A.; Bollig, A.; Wu, J.; Liao, D.J. Gene Expression Profiles in Primary Pancreatic Tumors and Metastatic Lesions of Ela-c-Myc Transgenic Mice. Mol. Cancer 2008, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Pan, B.; Qu, W.; Cao, Y.; Li, J.; Zhao, H. Systematic Analysis of the Expression and Prognosis Relevance of FBXO Family Reveals the Significance of FBXO1 in Human Breast Cancer; In Review; 2020. Available online: https://www.researchsquare.com/article/rs-86575/v1 (accessed on 8 October 2020).
- Wang, X.; Zhang, T.; Zhang, S.; Shan, J. Prognostic Values of F-Box Members in Breast Cancer: An Online Database Analysis and Literature Review. Biosci. Rep. 2019, 39, BSR20180949. [Google Scholar] [CrossRef]
- Li, N.; Jiang, S.; Shi, J.; Fu, R.; Wu, H.; Lu, M. Construction of a Potential MicroRNA, Transcription Factor and MRNA Regulatory Network in Hepatocellular Carcinoma. Transl. Cancer Res. TCR 2020, 9, 5528–5543. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Z.; Shen, X.; Cui, X.; Guo, Y. Identification of Novel Biomarkers and Small Molecule Drugs in Human Colorectal Cancer by Microarray and Bioinformatics Analysis. Mol. Genet. Genom. Med. 2019, 7, e00713. [Google Scholar] [CrossRef]
- Fu, J.; Qiu, H.; Cai, M.; Pan, Y.; Cao, Y.; Liu, L.; Yun, J.; Zhang, C.Z. Low Cyclin F Expression in Hepatocellular Carcinoma Associates with Poor Differentiation and Unfavorable Prognosis. Cancer Sci. 2013, 104, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, R.; Bonacci, T.; Wang, X.; Truong, A.; Arceci, A.; Zhang, Y.; Mills, C.A.; Kernan, J.L.; Liu, P.; Emanuele, M.J. The E3 Ubiquitin Ligase SCF(Cyclin F) Transmits AKT Signaling to the Cell-Cycle Machinery. Cell Rep. 2017, 20, 3212–3222. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Chen, Y.; Du, Y.; Ren, Z.; Li, X.; Hu, B. Methylation of Promoter of RBL1 Enhances the Radioresistance of Three Dimensional Cultured Carcinoma Cells. Oncotarget 2017, 8, 4422–4435. [Google Scholar] [CrossRef] [PubMed]
- Comisso, E.; Scarola, M.; Rosso, M.; Piazza, S.; Marzinotto, S.; Ciani, Y.; Orsaria, M.; Mariuzzi, L.; Schneider, C.; Schoeftner, S.; et al. OCT4 Controls Mitotic Stability and Inactivates the RB Tumor Suppressor Pathway to Enhance Ovarian Cancer Aggressiveness. Oncogene 2017, 36, 4253–4266. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Zhang, J.; Liu, S.; Lou, X.; Liao, D.J. The Other Side of the Coin: The Tumor-Suppressive Aspect of Oncogenes and the Oncogenic Aspect of Tumor-Suppressive Genes, Such as Those along the CCND–CDK4/6–RB Axis. Cell Cycle 2014, 13, 1677–1693. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Beattie, A.; Baradaran, B.; Dray, E.; Duijf, P.H.G. Contradictory MRNA and Protein Misexpression of EEF1A1 in Ductal Breast Carcinoma Due to Cell Cycle Regulation and Cellular Stress. Sci. Rep. 2018, 8, 13904. [Google Scholar] [CrossRef]
- Ginestier, C.; Charafe-Jauffret, E.; Bertucci, F.; Eisinger, F.; Geneix, J.; Bechlian, D.; Conte, N.; Adélaïde, J.; Toiron, Y.; Nguyen, C.; et al. Distinct and Complementary Information Provided by Use of Tissue and DNA Microarrays in the Study of Breast Tumor Markers. Am. J. Pathol. 2002, 161, 1223–1233. [Google Scholar] [CrossRef]
- Fisher, S.B.; Patel, S.H.; Bagci, P.; Kooby, D.A.; El-Rayes, B.F.; Staley, C.A.; Adsay, N.V.; Maithel, S.K. An Analysis of Human Equilibrative Nucleoside Transporter-1, Ribonucleoside Reductase Subunit M1, Ribonucleoside Reductase Subunit M2, and Excision Repair Cross-Complementing Gene-1 Expression in Patients with Resected Pancreas Adenocarcinoma: Implications for Adjuvant Treatment. Cancer 2013, 119, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Lin, J.; Thomas, D.G.; Jiang, W.; Liu, X. Ribonucleotide Reductase M2 Does Not Predict Survival in Patients with Resectable Pancreatic Adenocarcinoma. Int. J. Clin. Exp. Pathol. 2012, 5, 347–355. [Google Scholar]
- Itoi, T.; Sofuni, A.; Fukushima, N.; Itokawa, F.; Tsuchiya, T.; Kurihara, T.; Moriyasu, F.; Tsuchida, A.; Kasuya, K. Ribonucleotide Reductase Subunit M2 MRNA Expression in Pretreatment Biopsies Obtained from Unresectable Pancreatic Carcinomas. J. Gastroenterol. 2007, 42, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Giovannetti, E.; Del Tacca, M.; Mey, V.; Funel, N.; Nannizzi, S.; Ricci, S.; Orlandini, C.; Boggi, U.; Campani, D.; Del Chiaro, M.; et al. Transcription Analysis of Human Equilibrative Nucleoside Transporter-1 Predicts Survival in Pancreas Cancer Patients Treated with Gemcitabine. Cancer Res. 2006, 66, 3928–3935. [Google Scholar] [CrossRef] [PubMed]
- Fujita, H.; Ohuchida, K.; Mizumoto, K.; Itaba, S.; Ito, T.; Nakata, K.; Yu, J.; Kayashima, T.; Souzaki, R.; Tajiri, T.; et al. Gene Expression Levels as Predictive Markers of Outcome in Pancreatic Cancer after Gemcitabine-Based Adjuvant Chemotherapy. Neoplasia 2010, 12, 807–817. [Google Scholar] [CrossRef] [PubMed]
- Sierzega, M.; Pach, R.; Kulig, P.; Legutko, J.; Kulig, J. Prognostic Implications of Expression Profiling for Gemcitabine-Related Genes (HENT1, DCK, RRM1, RRM2) in Patients With Resectable Pancreatic Adenocarcinoma Receiving Adjuvant Chemotherapy. Pancreas 2017, 46, 684–689. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Jiao, Y.; Ji, J.; Jiang, W.; Ni, W.; Wu, Y.; Ni, R.; Lu, C.; Qu, L.; Ni, H.; et al. Identification of Prognostic Risk Factors for Pancreatic Cancer Using Bioinformatics Analysis. PeerJ 2020, 8, e9301. [Google Scholar] [CrossRef]
- Griffis, E.R.; Stuurman, N.; Vale, R.D. Spindly, a Novel Protein Essential for Silencing the Spindle Assembly Checkpoint, Recruits Dynein to the Kinetochore. J. Cell Biol. 2007, 177, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, R.; Essex, A.; Hu, J.-S.; Maddox, P.S.; Motegi, F.; Sugimoto, A.; O’Rourke, S.M.; Bowerman, B.; McLeod, I.; Yates, J.R.; et al. A New Mechanism Controlling Kinetochore-Microtubule Interactions Revealed by Comparison of Two Dynein-Targeting Components: SPDL-1 and the Rod/Zwilch/Zw10 Complex. Genes Dev. 2008, 22, 2385–2399. [Google Scholar] [CrossRef]
- Barisic, M.; Sohm, B.; Mikolcevic, P.; Wandke, C.; Rauch, V.; Ringer, T.; Hess, M.; Bonn, G.; Geley, S. Spindly/CCDC99 Is Required for Efficient Chromosome Congression and Mitotic Checkpoint Regulation. Mol. Biol. Cell 2010, 21, 1968–1981. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.M.A.; Delgado, M.L.; Ribeiro, N.; Florindo, C.; Tavares, Á.A.; Ribeiro, D.; Lopes, C.; do Amaral, B.; Bousbaa, H.; Monteiro, L.S. Spindly and Bub3 Expression in Oral Cancer: Prognostic and Therapeutic Implications. Oral Dis. 2019, 25, 1291–1301. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Wang, N. Upregulation of ASPM, BUB1B and SPDL1 in Tumor Tissues Predicts Poor Survival in Patients with Pancreatic Ductal Adenocarcinoma. Oncol. Lett. 2020, 19, 3307–3315. [Google Scholar] [CrossRef] [PubMed]
- Kodama, T.; Marian, T.A.; Lee, H.; Kodama, M.; Li, J.; Parmacek, M.S.; Jenkins, N.A.; Copeland, N.G.; Wei, Z. MRTFB Suppresses Colorectal Cancer Development through Regulating SPDL1 and MCAM. Proc. Natl. Acad. Sci. USA 2019, 116, 23625–23635. [Google Scholar] [CrossRef] [PubMed]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between Differentially Expressed MRNA and MRNA-Protein Correlations in a Xenograft Model System. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef] [PubMed]
- Wennemers, M.; Bussink, J.; Grebenchtchikov, N.; Sweep, F.C.G.J.; Span, P.N. TRIB3 Protein Denotes a Good Prognosis in Breast Cancer Patients and Is Associated with Hypoxia Sensitivity. Radiother. Oncol. 2011, 101, 198–202. [Google Scholar] [CrossRef]
- Wennemers, M.; Bussink, J.; van den Beucken, T.; Sweep, F.C.G.J.; Span, P.N. Regulation of TRIB3 MRNA and Protein in Breast Cancer. PLoS ONE 2012, 7, e49439. [Google Scholar] [CrossRef]

| Variables | n (%) | Cyclin F Expression | p | RRM2 Expression | p | SPDL1 Expression | p | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Low n = 62 | High n = 6 | Low n = 17 | High n = 51 | Low n = 40 | High n = 28 | |||||
| Age (years) | ||||||||||
| ≤60 | 29 (42.65) | 28 (96.55) | 1 (3.45) | 0.23 | 7 (24.14) | 22 (75.86) | >0.99 | 19 (65.52) | 10 (34.48) | 0.31 |
| >60 | 39 (57.35) | 34 (87.18) | 5 (12.82) | 10 (25.64) | 29 (74.36) | 18 (50.00) | 18 (50.00) | |||
| Gender | ||||||||||
| Male | 34 (50.00) | 30 (88.24) | 4 (11.76) | 0.67 | 7 (20.59) | 27 (79.41) | 0.58 | 20 (55.82) | 14 (44.18) | >0.99 |
| Female | 34 (50.00) | 32 (94.12) | 2 (5.88) | 10 (29.41) | 24 (70.59) | 20 (55.82) | 14 (44.18) | |||
| Grading | ||||||||||
| G1 | 5 (7.35) | 4 (80.00) | 1 (20.00) | 0.80 | 1 (20.00) | 4 (80.00) | 0.67 | 2 (40.00) | 3 (60.00) | 0.45 |
| G2 | 55 (80.88) | 51 (92.73) | 4 (7.27) | 13 (23.64) | 42 (76.36) | 32 (58.18) | 23 (41.82) | |||
| G3 | 8 (11.77) | 7 (87.50) | 1 (12.50) | 3 (37.50) | 5 (62.50) | 6 (75.00) | 2 (25.00) | |||
| pT status | ||||||||||
| T1-T2 | 53 (84.13) | 50 (94.34) | 3 (5.66) | 0.51 | 13 (24.53) | 40 (75.47) | 0.71 | 34 (64.15) | 19 (35.85) | 0.18 |
| T3-T4 | 10 (15.87) | 9 (90.00) | 1 (10.00) | 3 (30.00) | 7 (70.00) | 4 (40.00) | 6 (60.00) | |||
| pN status | ||||||||||
| N0 | 30 (45.46) | 28 (93.33) | 2 (6.67) | 0.68 | 8 (26.67) | 22 (73.33) | >0.99 | 15 (50.00) | 15 (50.00) | 0.21 |
| N1-N2 | 36 (54.54) | 32 (88.89) | 4 (11.11) | 9 (25.00) | 27 (75.00) | 24 (66.67) | 12 (33.33) | |||
| TNM stage | ||||||||||
| I | 24 (38.71) | 23 (95.83) | 1 (4.17) | 0.56 | 5 (20.83) | 19 (79.17) | 0.56 | 12 (50.00) | 12 (50.00) | 0.46 |
| II | 24 (38.71) | 21 (87.50) | 3 (12.50) | 8 (33.33) | 16 (66.67) | 16 (66.67) | 8 (33.33) | |||
| III-IV | 14 (22.58) | 13 (92.86) | 1 (7.14) | 3 (21.43) | 11 (78.57) | 9 (64.29) | 5 (35.71) | |||
| Location | ||||||||||
| head | 60 (88.24) | 55 (91.67) | 5 (8.33) | 0.54 | 15 (25.00) | 45 (75.00) | >0.99 | 35 (58.33) | 25 (41.67) | >0.99 |
| Body-tail | 8 (11.76) | 7 (87.50) | 1 (12.50) | 2 (25.00) | 6 (75.00) | 5 (62.50) | 3 (37.50) | |||
| VI | ||||||||||
| Absent | 39 (72.22) | 33 (84.62) | 6 (15.38) | 0.17 | 13 (33.33) | 26 (66.67) | 0.51 | 18 (46.15) | 21 (53.85) | 0.23 |
| Present | 15 (27.78) | 15 (100.0) | 0 (0.0) | 3 (20.00) | 12 (80.00) | 10 (66.67) | 5 (33.33) | |||
| PNI Absent Present | 22 (34.38) | 17 (77.27) | 5 (22.73) | 0.02 | 6 (27.27) | 16 (72.73) | >0.99 | 10 (45.45) | 12 (54.55) | 0.11 |
| 42 (65.62) | 41 (97.62) | 1 (2.38) | 11 (26.19) | 31 (73.81) | 28 (66.67) | 14 (33.33) | ||||
| Variables | n (%) | CCNF Expression | p | RRM2 Expression | p | SPDL1 Expression | p | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Low n = 75 | High n = 102 | Low n = 115 | High n = 62 | Low n = 133 | High n = 44 | |||||
| Gender | ||||||||||
| Male | 97 (54.80) | 44 (45.36) | 53 (54.64) | 0.45 | 67 (69.07) | 30 (30.93) | 0.27 | 73 (75.26) | 24 (24.74) | >0.99 |
| Female | 80 (45.20) | 31 (38.75) | 49 (61.25) | 48 (60.00) | 32 (40.00) | 60 (75.00) | 20 (25.00) | |||
| Age | ||||||||||
| ≤60 | 59 (33.33) | 26 (44.07) | 33 (55.93) | 0.75 | 39 (66.10) | 20 (33.90) | 0.87 | 44 (74.58) | 15 (25.42) | >0.99 |
| >60 | 118 (66.67) | 49 (41.53) | 69 (58.47) | 76 (64.41) | 42 (35.59) | 89 (75.42) | 29 (24.58) | |||
| Grading | ||||||||||
| G1 | 31 (17.71) | 21 (67.74) | 10 (32.26) | 0.01 | 24 (77.42) | 7 (22.58) | 0.26 | 27 (87.10) | 4 (12.90) | 0.25 |
| G2 | 94 (53.71) | 36 (38.30) | 58 (61.70) | 60 (63.83) | 34 (36.17) | 69 (73.40) | 25 (26.60) | |||
| G3-G4 | 50 (28.57) | 17 (34.00) | 33 (66.00) | 30 (60.00) | 20 (40.00) | 36 (72.00) | 14 (28.00) | |||
| pT status | ||||||||||
| T1-T2 | 30 (17.14) | 18 (60.00) | 12 (40.00) | 0.04 | 22 (73.33) | 8 (26.67) | 0.30 | 26 (86.67) | 4 (13.33) | 0.11 |
| T3-T4 | 145 (82.86) | 55 (37.93) | 90 (62.07) | 91 (62.76) | 54 (37.24) | 105 (72.41) | 40 (27.59) | |||
| pN status | ||||||||||
| N0 | 49 (28.49) | 24 (48.98) | 25 (51.02) | 0.24 | 34 (69.39) | 15 (30.61) | 0.48 | 39 (79.59) | 10 (20.41) | 0.44 |
| N1 | 123 (71.51) | 48 (39.02) | 75 (60.98) | 77 (62.60) | 46 (37.40) | 90 (73.17) | 33 (26.83) | |||
| TNM stage | ||||||||||
| I | 21 (12) | 13 (61.90) | 8 (38.10) | 0.06 | 15 (71.43) | 6 (28.57) | 0.63 | 18 (85.71) | 3 (14.29) | 0.29 |
| II-IV | 154 (88) | 60 (38.96) | 94 (61.04) | 98 (63.64) | 56 (36.36) | 113 (73.38) | 41 (26.62) | |||
| Variable | Univariate Analysis | Multivariate Analysis: CCNF | Multivariate Analysis: RRM2 | Multivariate Analysis: SPDL1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | HR | 95% CI | p | HR | 95% CI | p | |||||
| Lower | Upper | Lower | Upper | Lower | Upper | Lower | Upper | |||||||||
| CCNF | 2.40 | 1.52 | 3.80 | 0.0002 | 1.66 | 1.04 | 2.67 | 0.03 | - | - | - | - | - | - | - | - |
| RRM2 | 2.39 | 1.58 | 3.63 | <0.0001 | - | - | - | - | 1.94 | 1.27 | 2.97 | 0.002 | - | - | - | - |
| SPDL1 | 2.80 | 1.81 | 4.31 | <0.0001 | - | - | - | - | - | - | - | - | 2.39 | 1.53 | 3.75 | 0.0001 |
| age | 1.41 | 0.90 | 2.21 | 0.13 | 1.27 | 0.80 | 2.03 | 0.32 | 1.24 | 0.78 | 1.99 | 0.36 | 1.24 | 0.78 | 1.98 | 0.37 |
| gender | 0.81 | 0.54 | 1.23 | 0.33 | 0.86 | 0.56 | 1.32 | 0.48 | 0.86 | 0.56 | 1.32 | 0.50 | 0.79 | 0.52 | 1.21 | 0.28 |
| grade | 2.18 | 1.15 | 4.13 | 0.02 | 1.49 | 0.77 | 2.89 | 0.24 | 1.69 | 0.88 | 3.24 | 0.12 | 1.90 | 0.98 | 3.69 | 0.06 |
| pN | 2.10 | 1.25 | 3.52 | 0.005 | 1.90 | 1.09 | 3.30 | 0.02 | 1.86 | 1.07 | 3.25 | 0.03 | 1.93 | 1.11 | 3.38 | 0.02 |
| pT | 2.21 | 1.14 | 4.28 | 0.02 | 1.37 | 0.67 | 2.81 | 0.39 | 1.44 | 0.70 | 2.97 | 0.32 | 1.20 | 0.58 | 2.50 | 0.62 |
| stage | 0.74 | 0.23 | 2.34 | 0.60 | - | - | - | - | - | - | - | - | - | - | - | |
| Variable | Univariate Analysis | Multivariate Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | |||
| Lower | Upper | Lower | Upper | |||||
| Cyclin F | 0.47 | 0.17 | 1.32 | 0.15 | 0.37 | 0.10 | 1.43 | 0.15 |
| RRM2 | 1.52 | 0.76 | 3.06 | 0.24 | 1.07 | 0.45 | 2.56 | 0.87 |
| SPDL1 | 0.45 | 0.24 | 0.83 | 0.01 | 0.36 | 0.16 | 0.80 | 0.01 |
| age | 1.38 | 0.76 | 2.48 | 0.29 | 2.30 | 1.06 | 5.02 | 0.04 |
| gender | 1.06 | 0.60 | 1.85 | 0.85 | 1.12 | 0.52 | 2.40 | 0.78 |
| grade | 1.43 | 0.44 | 4.61 | 0.55 | 1.11 | 0.20 | 6.07 | 0.91 |
| pN | 1.07 | 0.60 | 1.91 | 0.81 | - | - | - | - |
| pT | 0.91 | 0.41 | 2.03 | 0.81 | - | - | - | - |
| stage | 1.87 | 0.96 | 3.65 | 0.07 | 2.44 | 1.06 | 5.62 | 0.04 |
| PNI | 1.80 | 0.96 | 3.35 | 0.07 | 1.40 | 0.61 | 3.20 | 0.43 |
| VI | 2.44 | 1.24 | 4.81 | 0.01 | 2.32 | 0.97 | 5.54 | 0.06 |
| Variable | Univariate Analysis | Multivariate Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | |||
| Lower | Upper | Lower | Upper | |||||
| CCNF + RRM2 + SPDL1 | 4.64 | 2.61 | 8.27 | <0.0001 | 3.51 | 1.93 | 6.36 | <0.0001 |
| age | 1.62 | 0.87 | 3.03 | 0.13 | 1.34 | 0.70 | 2.55 | 0.37 |
| gender | 0.94 | 0.53 | 1.66 | 0.83 | 1.22 | 0.67 | 2.23 | 0.51 |
| grade | 3.66 | 1.51 | 8.86 | 0.004 | 2.89 | 1.14 | 7.34 | 0.03 |
| pN | 2.74 | 1.28 | 5.91 | 0.01 | 2.69 | 1.05 | 6.91 | 0.04 |
| pT | 3.20 | 1.25 | 8.18 | 0.02 | 1.26 | 0.38 | 4.16 | 0.70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klimaszewska-Wiśniewska, A.; Buchholz, K.; Neska-Długosz, I.; Durślewicz, J.; Grzanka, D.; Zabrzyński, J.; Sopońska, P.; Grzanka, A.; Gagat, M. Expression of Genomic Instability-Related Molecules: Cyclin F, RRM2 and SPDL1 and Their Prognostic Significance in Pancreatic Adenocarcinoma. Cancers 2021, 13, 859. https://doi.org/10.3390/cancers13040859
Klimaszewska-Wiśniewska A, Buchholz K, Neska-Długosz I, Durślewicz J, Grzanka D, Zabrzyński J, Sopońska P, Grzanka A, Gagat M. Expression of Genomic Instability-Related Molecules: Cyclin F, RRM2 and SPDL1 and Their Prognostic Significance in Pancreatic Adenocarcinoma. Cancers. 2021; 13(4):859. https://doi.org/10.3390/cancers13040859
Chicago/Turabian StyleKlimaszewska-Wiśniewska, Anna, Karolina Buchholz, Izabela Neska-Długosz, Justyna Durślewicz, Dariusz Grzanka, Jan Zabrzyński, Paulina Sopońska, Alina Grzanka, and Maciej Gagat. 2021. "Expression of Genomic Instability-Related Molecules: Cyclin F, RRM2 and SPDL1 and Their Prognostic Significance in Pancreatic Adenocarcinoma" Cancers 13, no. 4: 859. https://doi.org/10.3390/cancers13040859
APA StyleKlimaszewska-Wiśniewska, A., Buchholz, K., Neska-Długosz, I., Durślewicz, J., Grzanka, D., Zabrzyński, J., Sopońska, P., Grzanka, A., & Gagat, M. (2021). Expression of Genomic Instability-Related Molecules: Cyclin F, RRM2 and SPDL1 and Their Prognostic Significance in Pancreatic Adenocarcinoma. Cancers, 13(4), 859. https://doi.org/10.3390/cancers13040859








