miR-582-5p Is a Tumor Suppressor microRNA Targeting the Hippo-YAP/TAZ Signaling Pathway in Non-Small Cell Lung Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
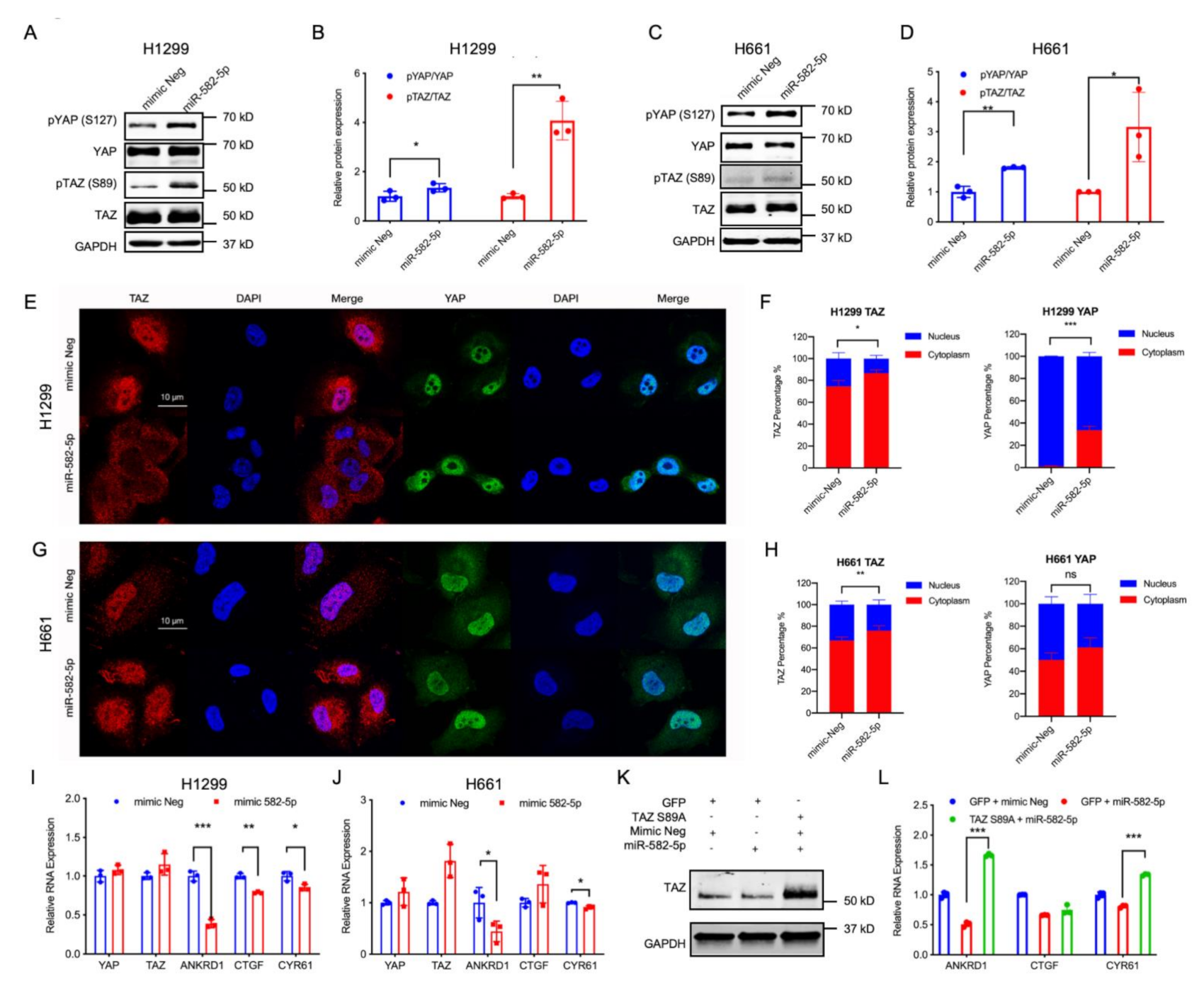
2.1. MiR-582-5p Expression Positively Correlates with the Phosphorylation Rate of YAP/TAZ in NSCLC Cell Lines
2.2. miR-582-5p Overexpression Suppresses YAP/TAZ Transcriptional Activity
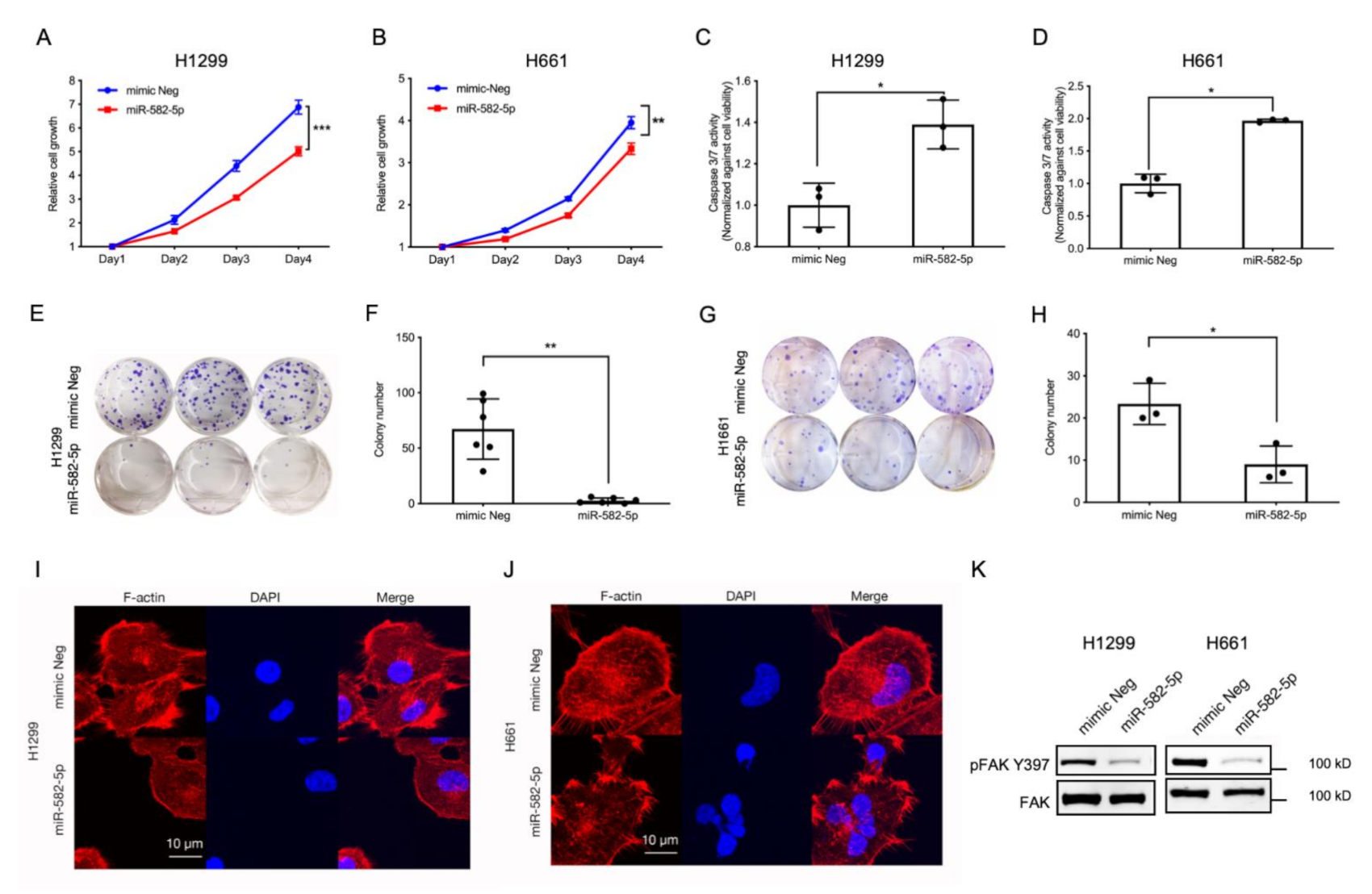
2.3. Overexpression of miR-582-5p Inhibits Cell Growth, Induces Cell Apoptosis, and Compromises F-Actin Cytoskeleton
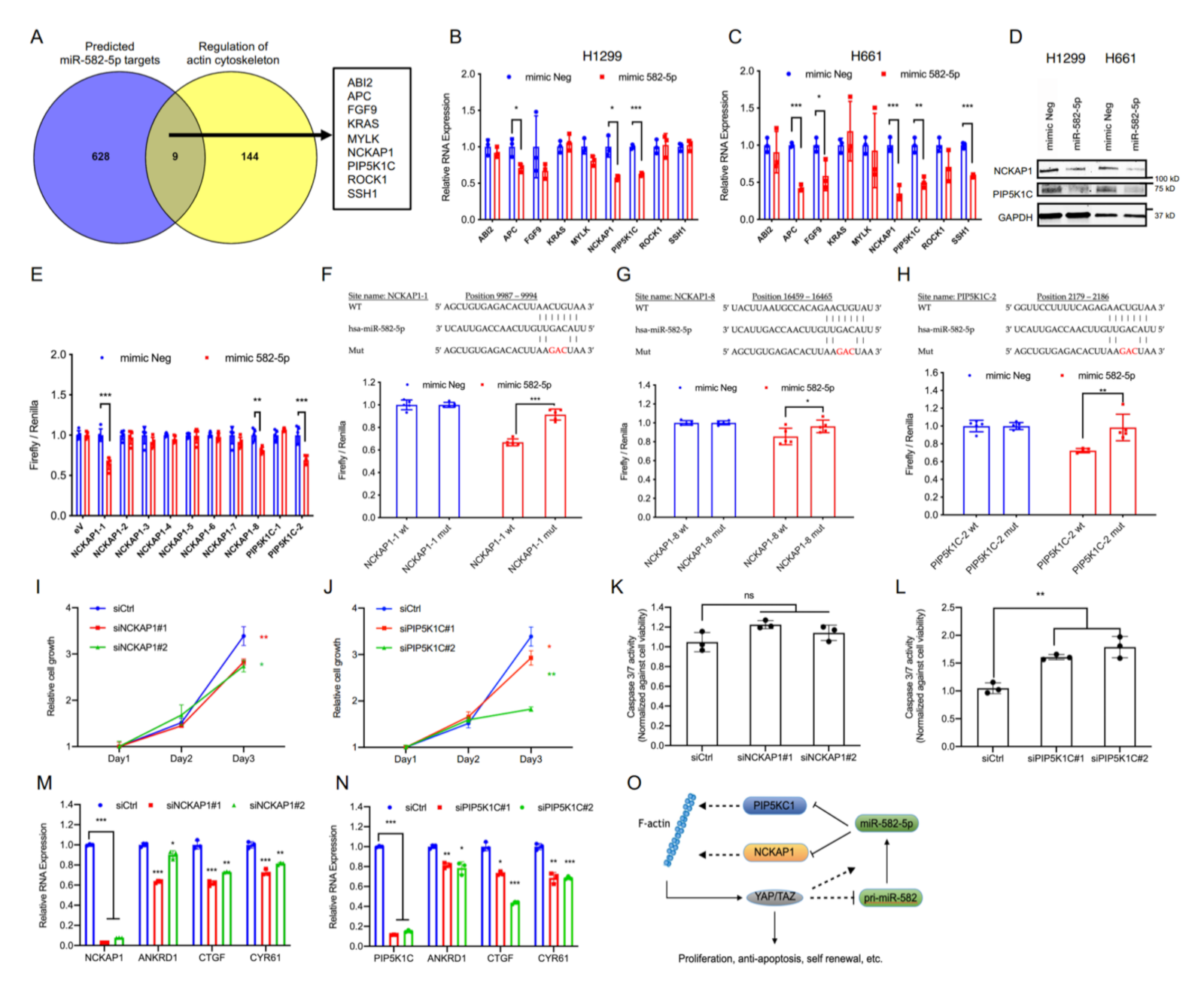
2.4. miR-582-5p Suppresses Hippo-YAP/TAZ Signaling by Targeting Actin Cytoskeleton Regulators, PIP5K1C and NCKAP1
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Transfection
4.3. RNA Isolation and Quantitative Real-Time PCR (RT-qPCR)
4.4. Quantification of miR-582-5p Expression
4.5. Western Blot Analysis
4.6. Immunofluorescence (IF) Staining and Confocal Microscopy
4.7. Cell Proliferation Analysis
4.8. Cell Apoptosis Analysis
4.9. Colony Formation Analysis
4.10. Database Analysis
4.11. Plasmid Construction
4.12. Lentiviral Packaging and Viral Transduction
4.13. Luciferase Assays
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Cancer Fact Sheets 2018. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 3 December 2020).
- Kanodra, N.M.; Silvestri, G.A.; Tanner, N.T. Screening and early detection efforts in lung cancer. Cancer 2015, 121, 1347–1356. [Google Scholar] [CrossRef]
- Morgensztern, D.; Ng, S.H.; Gao, F.; Govindan, R. Trends in stage distribution for patients with non-small cell lung cancer: A National Cancer Database survey. J. Thorac. Oncol. 2010, 5, 29–33. [Google Scholar] [CrossRef]
- Gyoba, J.; Shan, S.; Roa, W.; Bedard, E.L. Diagnosing Lung Cancers through Examination of Micro-RNA Biomarkers in Blood, Plasma, Serum and Sputum: A Review and Summary of Current Literature. Int. J. Mol. Sci. 2016, 17, 494. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.X.; Zhao, B.; Panupinthu, N.; Jewell, J.L.; Lian, I.; Wang, L.H.; Zhao, J.; Yuan, H.; Tumaneng, K.; Li, H.; et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 2012, 150, 780–791. [Google Scholar] [CrossRef]
- Pan, D. The hippo signaling pathway in development and cancer. Dev. Cell 2010, 19, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Moroishi, T.; Hansen, C.G.; Guan, K.L. The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 2015, 15, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Piccolo, S.; Dupont, S.; Cordenonsi, M. The biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev. 2014, 94, 1287–1312. [Google Scholar] [CrossRef] [PubMed]
- Low, B.C.; Pan, C.Q.; Shivashankar, G.V.; Bershadsky, A.; Sudol, M.; Sheetz, M. YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett. 2014, 588, 2663–2670. [Google Scholar] [CrossRef]
- Moya, I.M.; Halder, G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 2019, 20, 211–226. [Google Scholar] [CrossRef]
- Varelas, X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development 2014, 141, 1614–1626. [Google Scholar] [CrossRef]
- Lei, Q.Y.; Zhang, H.; Zhao, B.; Zha, Z.Y.; Bai, F.; Pei, X.H.; Zhao, S.; Xiong, Y.; Guan, K.L. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell Biol. 2008, 28, 2426–2436. [Google Scholar] [CrossRef]
- Zhao, B.; Ye, X.; Yu, J.; Li, L.; Li, W.; Li, S.; Yu, J.; Lin, J.D.; Wang, C.Y.; Chinnaiyan, A.M.; et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008, 22, 1962–1971. [Google Scholar] [CrossRef]
- Dong, J.; Feldmann, G.; Huang, J.; Wu, S.; Zhang, N.; Comerford, S.A.; Gayyed, M.F.; Anders, R.A.; Maitra, A.; Pan, D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 2007, 130, 1120–1133. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Kim, J. Regulation of Hippo signaling by actin remodeling. BMB Rep. 2018, 51, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Aragona, M.; Panciera, T.; Manfrin, A.; Giulitti, S.; Michielin, F.; Elvassore, N.; Dupont, S.; Piccolo, S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 2013, 154, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Degese, M.S.; Iglesias-Bartolome, R.; Vaque, J.P.; Molinolo, A.A.; Rodrigues, M.; Zaidi, M.R.; Ksander, B.R.; Merlino, G.; Sodhi, A.; et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell 2014, 25, 831–845. [Google Scholar] [CrossRef]
- Plouffe, S.W.; Lin, K.C.; Moore, J.L., 3rd; Tan, F.E.; Ma, S.; Ye, Z.; Qiu, Y.; Ren, B.; Guan, K.L. The Hippo pathway effector proteins YAP and TAZ have both distinct and overlapping functions in the cell. J. Biol. Chem. 2018, 293, 11230–11240. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Nakahara, K.; Pham, J.W.; Kim, K.; He, Z.; Sontheimer, E.J.; Carthew, R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 2004, 117, 69–81. [Google Scholar] [CrossRef]
- Gregory, R.I.; Chendrimada, T.P.; Shiekhattar, R. MicroRNA biogenesis: Isolation and characterization of the microprocessor complex. Methods Mol. Biol. 2006, 342, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.W.; Men, T.; Feng, R.C.; Li, Y.C.; Zhou, D.; Teng, C.B. miR-375 inhibits proliferation of mouse pancreatic progenitor cells by targeting YAP1. Cell Physiol. Biochem. 2013, 32, 1808–1817. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.M.; Poon, R.T.; Luk, J.M. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem. Biophys. Res. Commun 2010, 394, 623–627. [Google Scholar] [CrossRef]
- Kang, W.; Huang, T.; Zhou, Y.; Zhang, J.; Lung, R.W.M.; Tong, J.H.M.; Chan, A.W.H.; Zhang, B.; Wong, C.C.; Wu, F.; et al. miR-375 is involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in gastric carcinogenesis. Cell Death Dis. 2018, 9, 92. [Google Scholar] [CrossRef] [PubMed]
- Higashi, T.; Hayashi, H.; Ishimoto, T.; Takeyama, H.; Kaida, T.; Arima, K.; Taki, K.; Sakamoto, K.; Kuroki, H.; Okabe, H.; et al. miR-9-3p plays a tumour-suppressor role by targeting TAZ (WWTR1) in hepatocellular carcinoma cells. Br. J. Cancer 2015, 113, 252–258. [Google Scholar] [CrossRef]
- Liu, J.; Liu, S.; Deng, X.; Rao, J.; Huang, K.; Xu, G.; Wang, X. MicroRNA-582-5p suppresses non-small cell lung cancer cells growth and invasion via downregulating NOTCH1. PLoS ONE 2019, 14, e0217652. [Google Scholar] [CrossRef] [PubMed]
- Galluzzo, P.; Bocchetta, M. Notch signaling in lung cancer. Expert Rev. Anticancer Ther. 2011, 11, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, L. Upregulation of miR-582-5p regulates cell proliferation and apoptosis by targeting AKT3 in human endometrial carcinoma. Saudi J. Biol. Sci. 2018, 25, 965–970. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Tao, L.P.; Yao, S.C.; Huang, Q.K.; Chen, Z.F.; Sun, Y.J.; Jin, S.Q. MicroRNA-582-5p suppressed gastric cancer cell proliferation via targeting AKT3. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 5112–5120. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Yang, J.; Li, S.; Chen, J. Upregulation of miR-582-5p inhibits cell proliferation, cell cycle progression and invasion by targeting Rab27a in human colorectal carcinoma. Cancer Gene Ther. 2015, 22, 475–480. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, W.; Ran, Y.; Xiong, Y.; Zhong, Z.; Fan, X.; Wang, Z.; Ye, Q. miR-582-5p inhibits proliferation of hepatocellular carcinoma by targeting CDK1 and AKT3. Tumour. Biol. 2015, 36, 8309–8316. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zou, C.; Tang, Y.; Wa, Q.; Peng, X.; Chen, X.; Yang, C.; Ren, D.; Huang, Y.; Liao, Z.; et al. miR-582-3p and miR-582-5p Suppress Prostate Cancer Metastasis to Bone by Repressing TGF-beta Signaling. Mol. Ther. Nucleic Acids 2019, 16, 91–104. [Google Scholar] [CrossRef]
- Mori, M.; Triboulet, R.; Mohseni, M.; Schlegelmilch, K.; Shrestha, K.; Camargo, F.D.; Gregory, R.I. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell 2014, 156, 893–906. [Google Scholar] [CrossRef]
- Chaulk, S.G.; Lattanzi, V.J.; Hiemer, S.E.; Fahlman, R.P.; Varelas, X. The Hippo pathway effectors TAZ/YAP regulate dicer expression and microRNA biogenesis through Let-7. J. Biol. Chem. 2014, 289, 1886–1891. [Google Scholar] [CrossRef]
- Lee, Y.; Finch-Edmondson, M.; Cognart, H.; Zhu, B.; Song, H.; Chuan, L.B.; Sudol, M. Common and Unique Transcription Signatures of YAP and TAZ in Gastric Cancer Cells. Cancers (Basel) 2020, 12, 3667. [Google Scholar] [CrossRef]
- Zhao, B.; Wei, X.; Li, W.; Udan, R.S.; Yang, Q.; Kim, J.; Xie, J.; Ikenoue, T.; Yu, J.; Li, L.; et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007, 21, 2747–2761. [Google Scholar] [CrossRef]
- Martens, M.; Ammar, A.; Riutta, A.; Waagmeester, A.; Slenter, D.N.; Hanspers, K.; Ryan, A.M.; Digles, D.; Lopes, E.N.; Ehrhart, F.; et al. WikiPathways: Connecting communities. Nucleic Acids Res. 2020, 49, D613–D621. [Google Scholar] [CrossRef] [PubMed]
- Kruger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Hu, J.K.; Du, W.; Shelton, S.J.; Oldham, M.C.; DiPersio, C.M.; Klein, O.D. An FAK-YAP-mTOR Signaling Axis Regulates Stem Cell-Based Tissue Renewal in Mice. Cell Stem Cell 2017, 21, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Lachowski, D.; Cortes, E.; Robinson, B.; Rice, A.; Rombouts, K.; Del Rio Hernandez, A.E. FAK controls the mechanical activation of YAP, a transcriptional regulator required for durotaxis. FASEB J. 2018, 32, 1099–1107. [Google Scholar] [CrossRef]
- Wu, Z.; Li, X.; Sunkara, M.; Spearman, H.; Morris, A.J.; Huang, C. PIPKIgamma regulates focal adhesion dynamics and colon cancer cell invasion. PLoS ONE 2011, 6, e24775. [Google Scholar] [CrossRef]
- Teng, Y.; Qin, H.; Bahassan, A.; Bendzunas, N.G.; Kennedy, E.J.; Cowell, J.K. The WASF3-NCKAP1-CYFIP1 Complex Is Essential for Breast Cancer Metastasis. Cancer Res. 2016, 76, 5133–5142. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Ren, M.Q.; Cheney, R.; Sharma, S.; Cowell, J.K. Inactivation of the WASF3 gene in prostate cancer cells leads to suppression of tumorigenicity and metastases. Br. J. Cancer 2010, 103, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; He, L.; Shay, C.; Lang, L.; Loveless, J.; Yu, J.; Chemmalakuzhy, R.; Jiang, H.; Liu, M.; Teng, Y. Nck-associated protein 1 associates with HSP90 to drive metastasis in human non-small-cell lung cancer. J. Exp. Clin. Cancer Res. 2019, 38, 122. [Google Scholar] [CrossRef]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing [Internet]. Vienna, Austria; 2013. Available online: https://www.r-project.org/ (accessed on 3 December 2020).
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]

| Gene | Forward | Reverse |
|---|---|---|
| GAPDH | 5′-TGGACTCCACGACGTACTCA-3′ | 5′-AATCCCATCACCATCTTCCA-3′ |
| 18SrRNA | 5′-ACCCGTGGTCACCATGGTA-3′ | 5′-CGAACGTCTGCCCTATCAACTT-3′ |
| YAP1 | 5′-AAGCTCAACTGAAGGCATGTCA-3′ | 5′-GGATCTCTAATGCAATGATAG-3′ |
| TAZ (WWTR1) | 5′-AACCCCAAGACTGAGGTGTG-3′ | 5′-TGACATCTCTCCGCTTCCTT-3′ |
| ANKRD1 | 5′-TGCTGAATGCCTTCTCCCA-3′ | 5′-GCCTGCTGCCCTATCACA-3′ |
| CTGF | 5′-TGCAGTTCCTGACCCCTTAATG-3′ | 5′-AGCCAATTCCTGTAATGAACCAA-3′ |
| KRAS | 5′-GAGTACAGTGCAATGAGGGAC-3′ | 5′-CCTGAGCCTGTTTTGTGTCTAC-3′ |
| PIP5K1C | 5′-AGGCCATCGAATCGGATGAC-3′ | 5′-CCGAAAGACCGTGTTGCTCA-3′ |
| NCKAP1 | 5′-TCCTAAATACTGACGCTACAGCA-3′ | 5′-GCCTCCTTGCATTCTCTTATGTC-3′ |
| ITGB8 | 5′-ACCAGGAGAAGTGTCTATCCAG-3′ | 5′-CCAAGACGAAAGTCACGGGA-3′ |
| SSH1 | 5′-ACCTTCTGCGTTGCGAAGAC-3′ | 5′-AGGTGGATTTTCGTGTCGCTC-3′ |
| ABI2 | 5′-CAAAGCCTACACCACCCAATC-3′ | 5′-AGGTTGGCTGGAGCAATAATC-3′ |
| ROCK1 | 5′-AAGTGAGGTTAGGGCGAAATG-3′ | 5′-AAGGTAGTTGATTGCCAACGAA-3′ |
| MYLK | 5′-CCCGAGGTTGTCTGGTTCAAA-3′ | 5′-GCAGGTGTACTTGGCATCGT-3′ |
| FGF9 | 5′-GGCCTGGTCAGCATTCGAG-3′ | 5′-GTATCGCCTTCCAGTGTCCAC-3′ |
| APC | 5′-AAGCATGAAACCGGCTCACAT-3′ | 5′-CATTCGTGTAGTTGAACCCTGA-3′ |
| pri-miR-582 | 5′-GTCATTCATGCACACATTGAAGAG-3′ | 5′-TCTACTAGAGAGAGATTTGCTAGTGGTGTT-3′ |
| Gene | Sequences |
|---|---|
| 18SrRNA | Forward primer: 5′-CGAACGTCTGCCCTATCAACTT-3′ |
| Reverse primer: 5’-ACCCGTGGTCACCATGGTA-3′ | |
| Probe: 5′-FAM-TCGGAAGCTAAGCAGGGTCGGGC-Tamra-3′ |
| Antibody | Antigen | Species | Dilution | Manufacturer | Cat # |
|---|---|---|---|---|---|
| Primary | YAP | Mouse | 1:500 | Santa Cruz | sc-101199 |
| TAZ | Rabbit | 1:2000 | Cell Signaling | 4883S | |
| p-YAP (S127) | Rabbit | 1:1000 | Cell Signaling | 4911S | |
| p-TAZ (S89) | Rabbit | 1:1000 | Cell Signaling | 59971S | |
| CYR61 | Rabbit | 1:3000 | Cell Signaling | 14479S | |
| GAPDH | Rabbit | 1:2000 | Santa Cruz | sc-25778 | |
| FAK | Rabbit | 1:3000 | Cell Signaling | 13009S | |
| p-FAK Y397 | Rabbit | 1:1000 | Cell Signaling | 3283S | |
| NOTCH1 | Rabbit | 1:1000 | Cell Signaling | 4380S | |
| NOTCH2 | Rabbit | 1:1000 | Cell Signaling | 4530S | |
| NOTCH3 | Rabbit | 1:1000 | Cell Signaling | 5276S | |
| Pan-AKT | Rabbit | 1:1000 | Abcam | AB8805 | |
| pLATS1(S909) | Rabbit | 1:1000 | Cell Signaling | 9157S | |
| LATS1 | Rabbit | 1:1000 | Protein Tech | 17049-1-AP |
| Gene | Forward | Reverse |
|---|---|---|
| NCKAP1-1 | 5′-AATTCGACAGGAGCTGTGAGACACTTAACTGTAATCTTACC-3′ | 5′-TCGAGGTAAGATTACAGTTAAGTGTCTCACAGCTCCTGTCG-3′ |
| NCKAP1-2 | 5′-AATTCCTTGTTTATTTCTGAAAAAGAACTGTATTTAGC-3′ | 5′-TCGAGCTAAATACAGTTCTTTTTCAGAAATAAACAAGG-3′ |
| NCKAP1-3 | 5′-AATTCGAAACATTTGCCAAACTAAATACTGTAACACTGC-3′ | 5′-TCGAGCAGTGTTACAGTATTTAGTTTGGCAAATGTTTCG-3′ |
| NCKAP1-4 | 5′-AATTCCAATGGCCTGATCTCGGCTCACTGTAACCTCCC-3′ | 5′-TCGAGGGAGGTTACAGTGAGCCGAGATCAGGCCATTGG-3′ |
| NCKAP1-5 | 5′-AATTCGGAAATCATATTTAAAATTTACTGTAATTTTAC-3′ | 5′-TCGAGTAAAATTACAGTAAATTTTAAATATGATTTCCG-3′ |
| NCKAP1-6 | 5′-AATTCGACTTTACATGTTTGATCTTGACTGTAAAACTAC-3′ | 5′-TCGAGTAGTTTTACAGTCAAGATCAAACATGTAAAGTCG-3′ |
| NCKAP1-7 | 5′-AATTCGGCTTTCCTACTCAATCAAAAACTGTAGCTTGC-3′ | 5′-TCGAGCAAGCTACAGTTTTTGATTGAGTAGGAAAGCCG-3′ |
| NCKAP1-8 | 5′-AATTCGAATGTACTTAATGCCACAGAACTGTATGCTTC-3′ | 5′-TCGAGAAGCATACAGTTCTGTGGCATTAAGTACATTCG-3′ |
| NCKAP1-1mut | 5′-GACAGGAGCTGTGAGACACTTAAGACTAATCTTACC-3′ | 5′-TCGAGGTAAGATTAGTCTTAAGTGTCTCACAGCTCCTGTCG-3′ |
| NCKAP1-8mut | 5′-AATTCGAATGTACTTAATGCCACAGAAGACTATGCTTC-3′ | 5′-TCGAGAAGCATAGTCTTCTGTGGCATTAAGTACATTCG-3′ |
| PIP5K1C-1 | 5′-AATTCGCAGTGTCCAAATTCCTGTACTGTAAAGACTC-3′ | 5′-TCGAGAGTCTTTACAGTACAGGAATTTGGACACTGG-3′ |
| PIP5K1C-2 | 5′-AATTCTTTGGTTCCTTTTCAGAGAACTGTAAACCG C-3′ | 5′-TCGAGCGGTTTACAGTTCTCTGAAAAGGAACCAAAG-3′ |
| PIP5K1C-2mut | 5′-AATTCTTTGGTTCCTTTTCAGAGAAGACTAAACCG C-3′ | 5′-TCGAGCGGTTTAGTCTTCTCTGAAAAGGAACCAAAG-3′ |
| TAZ Subcloning | 5′-CGTGGATCCGCCACCATGGACTACAAGACGATGACGACAAGAATCCGGCCTCGGCGCCCCCTC-3′ | 5′-CGTCTCGAGACGCGTTTACAGCCAGGTTAGAAAGG-3′ |
| TAZ mutant | 5′-CCGCTCGCACCGGTCGCCCGCG-3′ | 5′-ACATGCTGGGCACCCCCA-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, B.; V, M.; Finch-Edmondson, M.; Lee, Y.; Wan, Y.; Sudol, M.; DasGupta, R. miR-582-5p Is a Tumor Suppressor microRNA Targeting the Hippo-YAP/TAZ Signaling Pathway in Non-Small Cell Lung Cancer. Cancers 2021, 13, 756. https://doi.org/10.3390/cancers13040756
Zhu B, V M, Finch-Edmondson M, Lee Y, Wan Y, Sudol M, DasGupta R. miR-582-5p Is a Tumor Suppressor microRNA Targeting the Hippo-YAP/TAZ Signaling Pathway in Non-Small Cell Lung Cancer. Cancers. 2021; 13(4):756. https://doi.org/10.3390/cancers13040756
Chicago/Turabian StyleZhu, Bowen, Mitheera V, Megan Finch-Edmondson, Yaelim Lee, Yue Wan, Marius Sudol, and Ramanuj DasGupta. 2021. "miR-582-5p Is a Tumor Suppressor microRNA Targeting the Hippo-YAP/TAZ Signaling Pathway in Non-Small Cell Lung Cancer" Cancers 13, no. 4: 756. https://doi.org/10.3390/cancers13040756
APA StyleZhu, B., V, M., Finch-Edmondson, M., Lee, Y., Wan, Y., Sudol, M., & DasGupta, R. (2021). miR-582-5p Is a Tumor Suppressor microRNA Targeting the Hippo-YAP/TAZ Signaling Pathway in Non-Small Cell Lung Cancer. Cancers, 13(4), 756. https://doi.org/10.3390/cancers13040756









