Polycythemia Vera and Essential Thrombocythemia Patients Exhibit Unique Serum Metabolic Profiles Compared to Healthy Individuals and Secondary Thrombocytosis Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
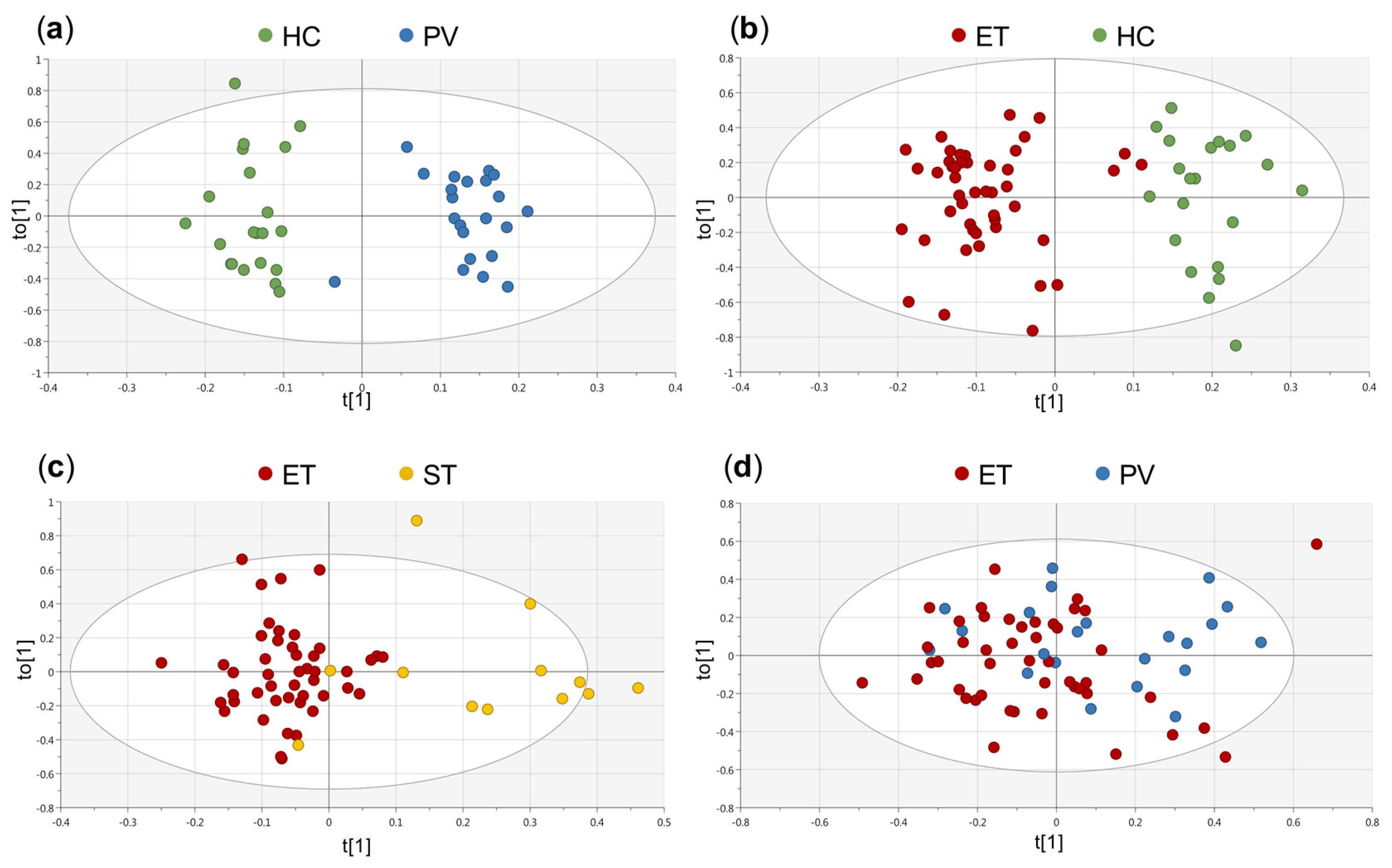
2.1. Untargeted Analysis of Serum Metabolic Profile
2.2. GSEA of a Transcriptomic Study
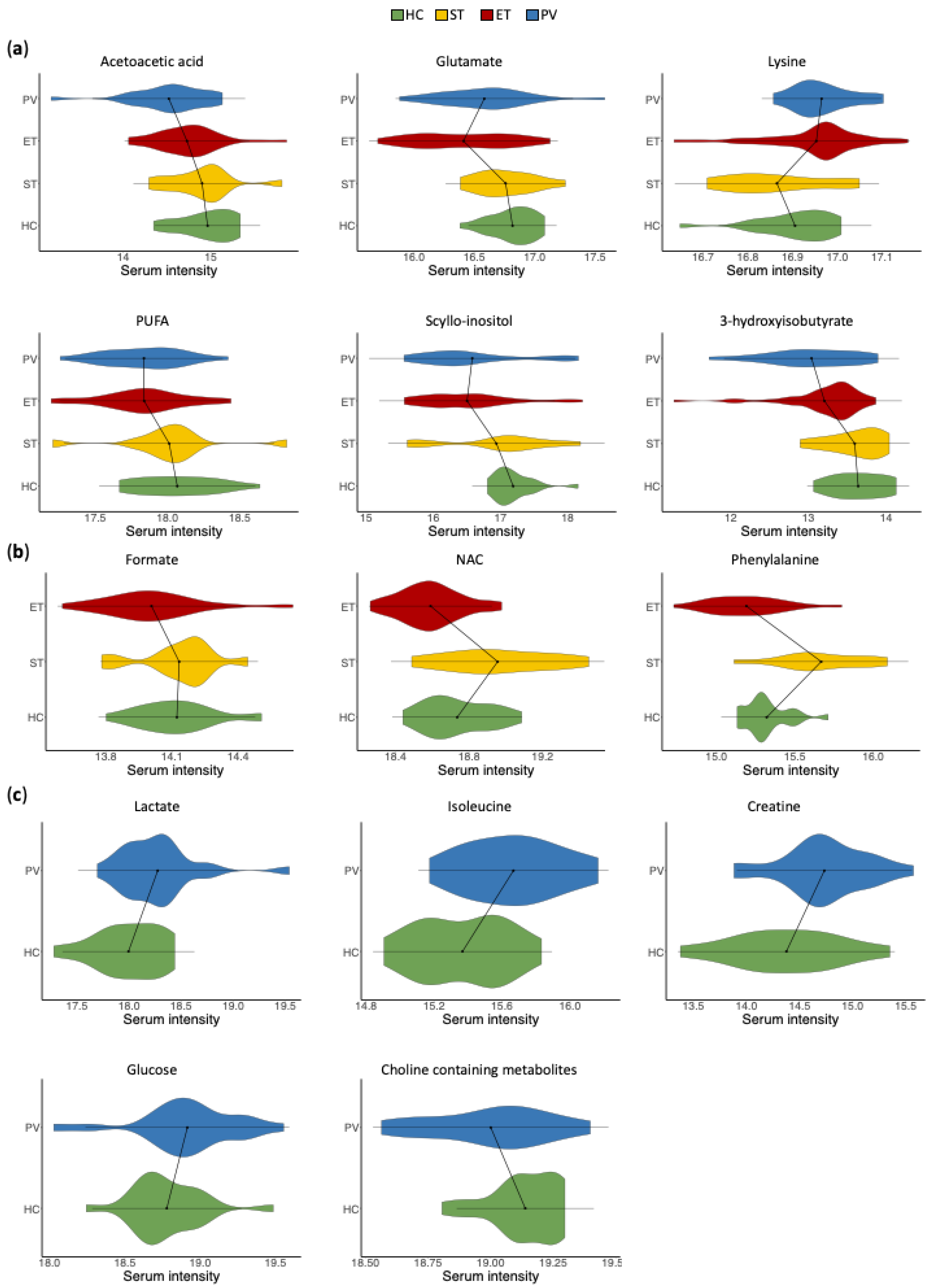
2.3. Univariate Analysis of Serum Metabolic Profiles
3. Discussion
4. Materials and Methods
4.1. Study Cohort
4.2. Serum Sample Collection
4.3. NMR Sample Preparation
4.4. NMR Sample Measurements
4.5. NMR Data Processing
4.6. Multivariate Statistical Analysis
4.7. Transcriptomic Analysis
4.8. Metabolite Quantification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rollison, D.E.; Howlader, N.; Smith, M.T.; Strom, S.S.; Merritt, W.D.; Ries, L.A.; Edwards, B.K.; List, A.F. Epidemiology of Myelodysplastic Syndromes and Chronic Myeloproliferative Disorders in the United States, 2001–2004, Using Data from the NAACCR and SEER Programs. Blood 2008, 112, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Moulard, O.; Mehta, J.; Fryzek, J.; Olivares, R.; Iqbal, U.; Mesa, R.A. Epidemiology of Myelofibrosis, Essential Thrombocythemia, and Polycythemia Vera in the European Union. Eur. J. Haematol. 2014, 92, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Tonkin, J.; Francis, Y.; Pattinson, A.; Peters, T.; Taylor, M.; Thompson, R.; Wallis, L. Myeloproliferative Neoplasms: Diagnosis, Management and Treatment. Nurs. Stand. 2012, 26, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Tefferi, A.; Barbui, T. Polycythemia Vera and Essential Thrombocythemia: 2019 Update on Diagnosis, Risk-Stratification and Management. Am. J. Hematol. 2019, 94, 133–143. [Google Scholar] [CrossRef]
- Nangalia, J.; Green, T.R. The Evolving Genomic Landscape of Myeloproliferative Neoplasms. Hematol. Am. Soc. Hematol. Educ. Program. 2014, 2014, 287–296. [Google Scholar] [CrossRef]
- Fowlkes, S.; Murray, C.; Fulford, A.; De Gelder, T.; Siddiq, N. Myeloproliferative Neoplasms (MPNs)—Part 1: An Overview of the Diagnosis and Treatment of the “Classical” MPNs. Can. Oncol. Nurs. J. 2018, 28, 262–268. [Google Scholar] [CrossRef]
- Barbui, T.; Thiele, J.; Gisslinger, H.; Kvasnicka, H.M.; Vannucchi, A.M.; Guglielmelli, P.; Orazi, A.; Tefferi, A. The 2016 WHO Classification and Diagnostic Criteria for Myeloproliferative Neoplasms: Document Summary and in-Depth Discussion. Blood Cancer J. 2018, 8, 15. [Google Scholar] [CrossRef]
- Kutti, J.; Wadenvik, H. Diagnostic and Differential Criteria of Essential Thrombocythemia and Reactive Thrombocytosis. Leuk. Lymphoma 1996, 22, 41–45. [Google Scholar] [CrossRef]
- Lindon, J.C.; Nicholson, J.K.; Holmes, E.; Everett, J.R. Metabonomics: Metabolic Processes Studied by NMR Spectroscopy of Biofluids. Concepts Magn. Reson. 2000, 12, 289–320. [Google Scholar] [CrossRef]
- Puchades-Carrasco, L.; Lecumberri, R.; Martínez-López, J.; Lahuerta, J.-J.; Mateos, M.-V.; Prósper, F.; San-Miguel, J.F.; Pineda-Lucena, A. Multiple Myeloma Patients Have a Specific Serum Metabolomic Profile That Changes after Achieving Complete Remission. Clin. Cancer Res. 2013, 19, 4770–4779. [Google Scholar] [CrossRef] [PubMed]
- Puchades-Carrasco, L.; Jantus-Lewintre, E.; Pérez-Rambla, C.; García-García, F.; Lucas, R.; Calabuig, S.; Blasco, A.; Dopazo, J.; Camps, C.; Pineda-Lucena, A. Serum Metabolomic Profiling Facilitates the Non-Invasive Identification of Metabolic Biomarkers Associated with the Onset and Progression of Non-Small Cell Lung Cancer. Oncotarget 2016, 7, 12904–12916. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Cebrián, N.; García-Flores, M.; Rubio-Briones, J.; López-Guerrero, J.A.; Pineda-Lucena, A.; Puchades-Carrasco, L. Targeted Metabolomics Analyses Reveal Specific Metabolic Alterations in High-Grade Prostate Cancer Patients. J. Proteome Res. 2020, 19, 4082–4092. [Google Scholar] [CrossRef] [PubMed]
- MacIntyre, D.A.; Jiménez, B.; Lewintre, E.J.; Martín, C.R.; Schäfer, H.; Ballesteros, C.G.; Mayans, J.R.; Spraul, M.; García-Conde, J.; Pineda-Lucena, A. Serum Metabolome Analysis by 1H-NMR Reveals Differences between Chronic Lymphocytic Leukaemia Molecular Subgroups. Leukemia 2010, 24, 788–797. [Google Scholar] [CrossRef] [PubMed]
- Hasselbalch, H.C.; Thomassen, M.; Hasselbalch Riley, C.; Kjær, L.; Stauffer Larsen, T.; Jensen, M.K.; Bjerrum, O.W.; Kruse, T.A.; Skov, V. Whole Blood Transcriptional Profiling Reveals Deregulation of Oxidative and Antioxidative Defence Genes in Myelofibrosis and Related Neoplasms. Potential Implications of Downregulation of Nrf2 for Genomic Instability and Disease Progression. PLoS ONE 2014, 9, e112786. [Google Scholar] [CrossRef]
- Allegra, A.; Innao, V.; Gerace, D.; Bianco, O.; Musolino, C. The Metabolomic Signature of Hematologic Malignancies. Leuk. Res. 2016, 49, 22–35. [Google Scholar] [CrossRef]
- Steiner, N.; Müller, U.; Hajek, R.; Sevcikova, S.; Borjan, B.; Jöhrer, K.; Göbel, G.; Pircher, A.; Gunsilius, E. The Metabolomic Plasma Profile of Myeloma Patients Is Considerably Different from Healthy Subjects and Reveals Potential New Therapeutic Targets. PLoS ONE 2018, 13, e0202045. [Google Scholar] [CrossRef]
- Du, H.; Wang, L.; Liu, B.; Wang, J.; Su, H.; Zhang, T.; Huang, Z. Analysis of the Metabolic Characteristics of Serum Samples in Patients With Multiple Myeloma. Front. Pharmacol. 2018, 9, 884. [Google Scholar] [CrossRef]
- Barberini, L.; Noto, A.; Fattuoni, C.; Satta, G.; Zucca, M.; Cabras, M.G.; Mura, E.; Cocco, P. The Metabolomic Profile of Lymphoma Subtypes: A Pilot Study. Molecules 2019, 24, 2367. [Google Scholar] [CrossRef]
- Wiklund, S.; Johansson, E.; Sjöström, L.; Mellerowicz, E.J.; Edlund, U.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-Based Metabolomics Data for Identification of Biochemically Interesting Compounds Using OPLS Class Models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef]
- Jiang, L.; Si, Z.-H.; Li, M.-H.; Zhao, H.; Fu, Y.-H.; Xing, Y.-X.; Hong, W.; Ruan, L.-Y.; Li, P.-M.; Wang, J.-S. 1H NMR-Based Metabolomics Study of Liver Damage Induced by Ginkgolic Acid (15:1) in Mice. J. Pharm. Biomed. Anal. 2017, 136, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Skov, V.; Burton, M.; Thomassen, M.; Stauffer Larsen, T.; Riley, C.H.; Brinch Madelung, A.; Kjær, L.; Bondo, H.; Stamp, I.; Ehinger, M.; et al. A 7-Gene Signature Depicts the Biochemical Profile of Early Prefibrotic Myelofibrosis. PLoS ONE 2016, 11, e0161570. [Google Scholar] [CrossRef] [PubMed]
- Cluntun, A.A.; Lukey, M.J.; Cerione, R.A.; Locasale, J.W. Glutamine Metabolism in Cancer: Understanding the Heterogeneity. Trends Cancer 2017, 3, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Miyagi, Y.; Higashiyama, M.; Gochi, A.; Akaike, M.; Ishikawa, T.; Miura, T.; Saruki, N.; Bando, E.; Kimura, H.; Imamura, F.; et al. Plasma Free Amino Acid Profiling of Five Types of Cancer Patients and Its Application for Early Detection. PLoS ONE 2011, 6, e24143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, L.; Shen, J.; Cao, B.; Cheng, T.; Zhao, T.; Liu, X.; Zhang, H. Metabolic Signatures of Esophageal Cancer: NMR-Based Metabolomics and UHPLC-Based Focused Metabolomics of Blood Serum. Biochim. Biophys. Acta Mol. Basis Dis. 2013, 1832, 1207–1216. [Google Scholar] [CrossRef]
- Mika, A.; Kobiela, J.; Pakiet, A.; Czumaj, A.; Sokołowska, E.; Makarewicz, W.; Chmielewski, M.; Stepnowski, P.; Marino-Gammazza, A.; Sledzinski, T. Preferential Uptake of Polyunsaturated Fatty Acids by Colorectal Cancer Cells. Sci. Rep. 2020, 10, 1954. [Google Scholar] [CrossRef]
- Nitter, M.; Norgård, B.; de Vogel, S.; Eussen, S.J.P.M.; Meyer, K.; Ulvik, A.; Ueland, P.M.; Nygård, O.; Vollset, S.E.; Bjørge, T.; et al. Plasma Methionine, Choline, Betaine, and Dimethylglycine in Relation to Colorectal Cancer Risk in the European Prospective Investigation into Cancer and Nutrition (EPIC). Ann. Oncol. 2014, 25, 1609–1615. [Google Scholar] [CrossRef]
- Pietzke, M.; Arroyo, S.F.; Sumpton, D.; Mackay, G.M.; Martin-Castillo, B.; Camps, J.; Joven, J.; Menendez, J.A.; Vazquez, A. Stratification of Cancer and Diabetes Based on Circulating Levels of Formate and Glucose. Cancer Metab. 2019, 7, 3. [Google Scholar] [CrossRef]
- Villa, E.; Ali, E.; Sahu, U.; Ben-Sahra, I. Cancer Cells Tune the Signaling Pathways to Empower de Novo Synthesis of Nucleotides. Cancers 2019, 11, 688. [Google Scholar] [CrossRef]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Wojtowicz, W.; Chachaj, A.; Olczak, A.; Ząbek, A.; Piątkowska, E.; Rybka, J.; Butrym, A.; Biedroń, M.; Mazur, G.; Wróbel, T.; et al. Serum NMR Metabolomics to Differentiate Haematologic Malignancies. Oncotarget 2018, 9, 24414–24427. [Google Scholar] [CrossRef] [PubMed]
- Sillos, E.M.; Shenep, J.L.; Burghen, G.A.; Pui, C.-H.; Behm, F.G.; Sandlund, J.T. Lactic Acidosis: A Metabolic Complication of Hematologic Malignancies. Cancers 2001, 92, 2237–2246. [Google Scholar] [CrossRef]
- Matschinsky, F.M.; Wilson, D.F. The Central Role of Glucokinase in Glucose Homeostasis: A Perspective 50 Years After Demonstrating the Presence of the Enzyme in Islets of Langerhans. Front. Physiol. 2019, 10, 148. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Li, H.; Xu, H.; Woo, S.; Dong, H.; Lu, F.; Lange, A.J.; Wu, C. Glycolysis in the Control of Blood Glucose Homeostasis. Acta Pharma. Sin. B 2012, 2, 358–367. [Google Scholar] [CrossRef]
- Mayers, J.R.; Wu, C.; Clish, C.B.; Kraft, P.; Torrence, M.E.; Fiske, B.P.; Yuan, C.; Bao, Y.; Townsend, M.K.; Tworoger, S.S.; et al. Elevation of Circulating Branched-Chain Amino Acids Is an Early Event in Human Pancreatic Adenocarcinoma Development. Nat. Med. 2014, 20, 1193–1198. [Google Scholar] [CrossRef]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.D.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic Profiling, Metabolomic and Metabonomic Procedures for NMR Spectroscopy of Urine, Plasma, Serum and Tissue Extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef]
- Ghini, V.; Quaglio, D.; Luchinat, C.; Turano, P. NMR for Sample Quality Assessment in Metabolomics. New Biotechnol. 2019, 52, 25–34. [Google Scholar] [CrossRef]
- Dona, A.C.; Jiménez, B.; Schäfer, H.; Humpfer, E.; Spraul, M.; Lewis, M.R.; Pearce, J.T.M.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Precision High-Throughput Proton NMR Spectroscopy of Human Urine, Serum, and Plasma for Large-Scale Metabolic Phenotyping. Anal. Chem. 2014, 86, 9887–9894. [Google Scholar] [CrossRef]
- Vicente-Muñoz, S.; Morcillo, I.; Puchades-Carrasco, L.; Payá, V.; Pellicer, A.; Pineda-Lucena, A. Pathophysiologic Processes Have an Impact on the Plasma Metabolomic Signature of Endometriosis Patients. Fertil. Steril. 2016, 106, 1733–1741. [Google Scholar] [CrossRef]
- Pérez-Rambla, C.; Puchades-Carrasco, L.; García-Flores, M.; Rubio-Briones, J.; López-Guerrero, J.A.; Pineda-Lucena, A. Non-Invasive Urinary Metabolomic Profiling Discriminates Prostate Cancer from Benign Prostatic Hyperplasia. Metabolomics 2017, 13, 52. [Google Scholar] [CrossRef]
- Meiboom, S.; Gill, D. Modified Spin-Echo Method for Measuring Nuclear Relaxation Times. Rev. Sci. Instrum. 1958, 29, 688–691. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Foxall, P.J.; Spraul, M.; Farrant, R.D.; Lindon, J.C. 750 MHz 1H and 1H-13C NMR Spectroscopy of Human Blood Plasma. Anal. Chem. 1995, 67, 793–811. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://www.R-project.org (accessed on 25 June 2019).
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy—Analysis of Affymetrix GeneChip Data at the Probe Level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Montaner, D.; Dopazo, J. Multidimensional Gene Set Analysis of Genomic Data. PLoS ONE 2010, 5, e10348. [Google Scholar] [CrossRef]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New Approach for Understanding Genome Variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. Toward Understanding the Origin and Evolution of Cellular Organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Yekutieli, D.; Benjamini, Y. The Control of the False Discovery Rate in Multiple Testing under Dependency. Ann. Statist. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The Human Metabolome Database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Markley, J.L.; Ulrich, E.L.; Berman, H.M.; Henrick, K.; Nakamura, H.; Akutsu, H. BioMagResBank (BMRB) as a Partner in the Worldwide Protein Data Bank (WwPDB): New Policies Affecting Biomolecular NMR Depositions. J. Biomol. NMR 2008, 40, 153–155. [Google Scholar] [CrossRef]
- Jacob, D.; Deborde, C.; Lefebvre, M.; Maucourt, M.; Moing, A. NMRProcFlow: A Graphical and Interactive Tool Dedicated to 1D Spectra Processing for NMR-Based Metabolomics. Metabolomics 2017, 13, 36. [Google Scholar] [CrossRef]

| Comparison | Pathway Name | KEGG ID | LOR | p-Value | BY Adjusted |
|---|---|---|---|---|---|
| ET vs. HC | Oxidative phosphorylation | hsa00190 | −0.4946 | 7.57 × 10−8 | 3.24 × 10−5 |
| Citrate cycle (TCA cycle) | hsa00020 | −0.6648 | 1.87 × 10−4 | 4.00 × 10−2 | |
| alpha-linolenic acid metabolism | hsa00592 | 0.7378 | 3.18 × 10−4 | 4.53 × 10−2 | |
| PV vs. HC | Citrate cycle (TCA cycle) | hsa00020 | −0.9519 | 1.85 × 10−8 | 7.88 × 10−6 |
| Oxidative phosphorylation | hsa00190 | −0.4006 | 1.14 × 10−5 | 1.76 × 10−3 | |
| Carbon metabolism | hsa01200 | −0.4138 | 1.24 × 10−5 | 1.76 × 10−3 | |
| Valine, leucine and isoleucine degradation | hsa00280 | −0.6365 | 2.36 × 10−5 | 2.52 × 10−3 | |
| Starch and sucrose metabolism | hsa00500 | 0.6765 | 1.05 × 10−4 | 8.95 × 10−3 | |
| Propanoate metabolism | hsa00640 | −0.6678 | 1.48 × 10−4 | 1.06 × 10−2 | |
| Pyruvate metabolism | hsa00620 | −0.5943 | 4.15 × 10−4 | 2.53 × 10−2 | |
| PV vs. ET | − | − | − | − | − |
| Group | n (%) | Age (Mean ± SD) | Sex (m/f) | JAK2V617 (Mut/WT) | CALR (Mut/WT/NA) |
|---|---|---|---|---|---|
| HC | 21 (20.8%) | 58 ± 5.78 | 12/9 | −/21/− | −/21/− |
| PV | 22 (21.8%) | 68.86 ± 14.57 | 14/8 | 22/−/− | −/1/21 |
| ET | 46 (45.5%) | 61.89 ± 16.53 | 18/28 | 24/22 | 9/23/14 |
| ST | 12 (11.9% | 60.17 ± 16.07 | 4/8 | −/12/− | −/12/− |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Cebrián, N.; Rojas-Benedicto, A.; Albors-Vaquer, A.; Bellosillo, B.; Besses, C.; Martínez-López, J.; Pineda-Lucena, A.; Puchades-Carrasco, L. Polycythemia Vera and Essential Thrombocythemia Patients Exhibit Unique Serum Metabolic Profiles Compared to Healthy Individuals and Secondary Thrombocytosis Patients. Cancers 2021, 13, 482. https://doi.org/10.3390/cancers13030482
Gómez-Cebrián N, Rojas-Benedicto A, Albors-Vaquer A, Bellosillo B, Besses C, Martínez-López J, Pineda-Lucena A, Puchades-Carrasco L. Polycythemia Vera and Essential Thrombocythemia Patients Exhibit Unique Serum Metabolic Profiles Compared to Healthy Individuals and Secondary Thrombocytosis Patients. Cancers. 2021; 13(3):482. https://doi.org/10.3390/cancers13030482
Chicago/Turabian StyleGómez-Cebrián, Nuria, Ayelén Rojas-Benedicto, Arturo Albors-Vaquer, Beatriz Bellosillo, Carlos Besses, Joaquín Martínez-López, Antonio Pineda-Lucena, and Leonor Puchades-Carrasco. 2021. "Polycythemia Vera and Essential Thrombocythemia Patients Exhibit Unique Serum Metabolic Profiles Compared to Healthy Individuals and Secondary Thrombocytosis Patients" Cancers 13, no. 3: 482. https://doi.org/10.3390/cancers13030482
APA StyleGómez-Cebrián, N., Rojas-Benedicto, A., Albors-Vaquer, A., Bellosillo, B., Besses, C., Martínez-López, J., Pineda-Lucena, A., & Puchades-Carrasco, L. (2021). Polycythemia Vera and Essential Thrombocythemia Patients Exhibit Unique Serum Metabolic Profiles Compared to Healthy Individuals and Secondary Thrombocytosis Patients. Cancers, 13(3), 482. https://doi.org/10.3390/cancers13030482







