New Trends in the Detection of Gynecological Precancerous Lesions and Early-Stage Cancers
Simple Summary
Abstract
1. Introduction
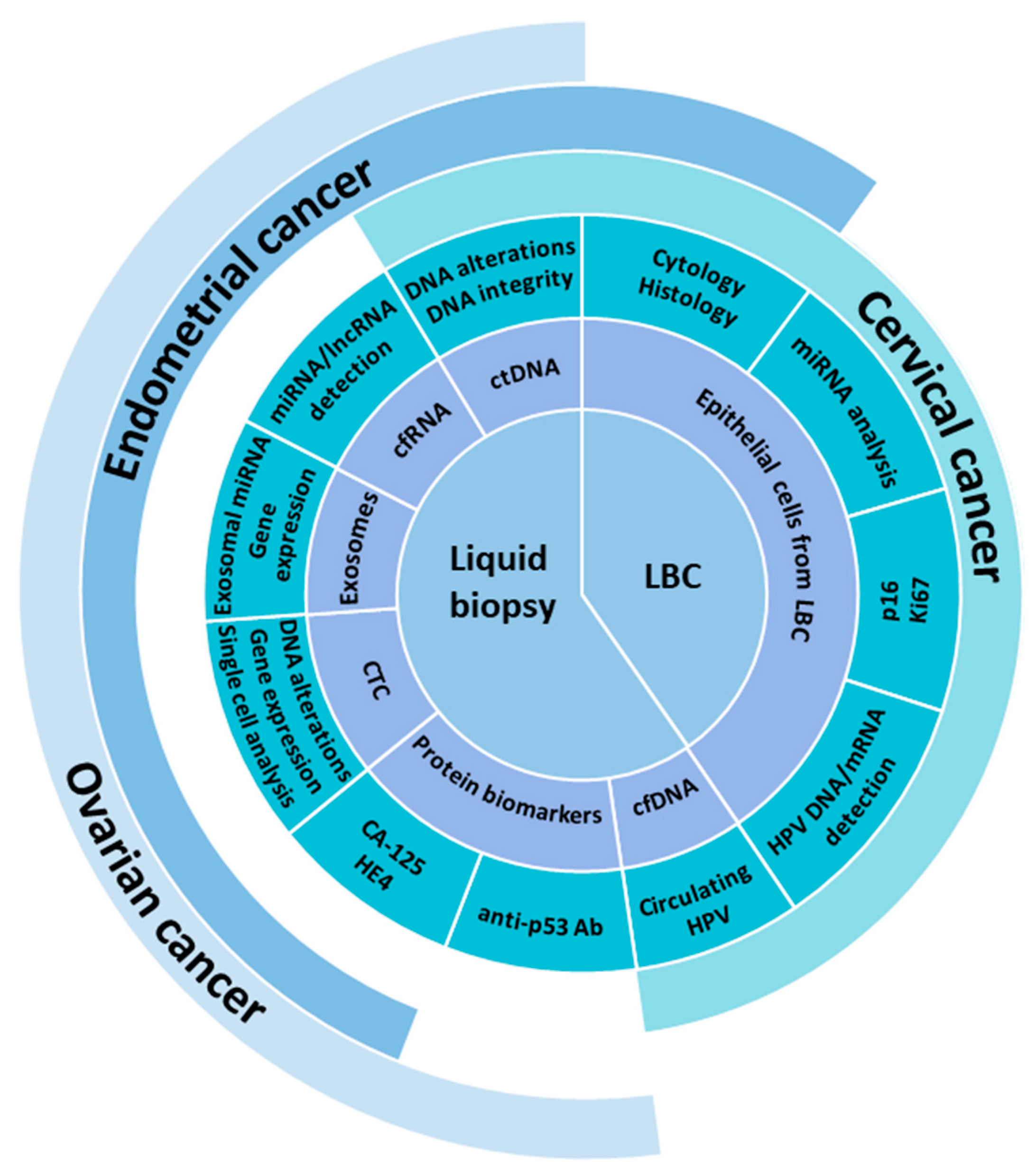
2. Overview of Circulating Biomarkers
2.1. Circulating Tumor Cells
2.2. Cell-Free DNA and Circulating Tumor DNA
2.3. Circulating Cell-Free RNA
2.4. Circulating Extracellular Vesicles
3. Cervical Precancerous Lesions
3.1. Current Diagnostics of Cervical (Pre)Cancer
3.2. Novel Biomarkers in Liquid-Based Cytology
3.3. The Analysis of Circulating DNA in Cervical Precancerous Lesions
4. Endometrial Precancerous Lesions and Early-Stage Endometrial Cancer
4.1. Endometrial Cancer
4.2. EC Development: Endometrial Precancer–Cancer Sequence
4.3. The Current State of the Diagnosis and Screening of Endometrial Precancer and/or Early EC
5. Strategies for the Early Detection of Epithelial Ovarian Cancer (EOC)
5.1. Current EOC Screening
5.2. Protein Biomarkers in Liquid Biopsies
5.3. Circulating Tumor Cells
5.4. Circulating Free DNA and Circulating Tumor DNA
5.5. Exosomes and Circulating Cell-Free MicroRNAs
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Costas, L.; Frias-Gomez, J.; Guardiola, M.; Benavente, Y.; Pineda, M.; Pavón, M.Á.; Martínez, J.M.; Climent, M.; Barahona, M.; Canet, J.; et al. New perspectives on screening and early detection of endometrial cancer. Int. J. Cancer 2019, 145, 3194–3206. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, I.J.; Menon, U.; Ryan, A.; Gentry-Maharaj, A.; Burnell, M.; Kalsi, J.K.; Amso, N.N.; Apostolidou, S.; Benjamin, E.; Cruickshank, D.; et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): A randomised controlled trial. Lancet 2016, 387, 945–956. [Google Scholar] [CrossRef]
- Morice, P.; Leary, A.; Creutzberg, C.; Abu-Rustum, N.; Darai, E. Endometrial cancer. Lancet 2016, 387, 1094–1108. [Google Scholar] [CrossRef]
- Hyun, K.-A.; Gwak, H.; Lee, J.; Kwak, B.; Jung, H.-I. Salivary Exosome and Cell-Free DNA for Cancer Detection. Micromachines 2018, 9, 340. [Google Scholar] [CrossRef] [PubMed]
- Martins, I.; Ribeiro, I.P.; Jorge, J.; Gonçalves, A.C.; Sarmento-Ribeiro, A.B.; Melo, J.B.; Carreira, I.M. Liquid Biopsies: Applications for Cancer Diagnosis and Monitoring. Genes 2021, 12, 349. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, J. A Pap-Based Test to Detect Endometrial and Ovarian Cancers Early. JAMA J. Am. Med. Assoc. 2018, 319, 1853. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Li, L.; Douville, C.; Cohen, J.D.; Yen, T.-T.; Kinde, I.; Sundfelt, K.; Kjær, S.K.; Hruban, R.H.; Shih, I.-M.; et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian can-cers. Sci. Transl. Med. 2018, 10, 433. [Google Scholar] [CrossRef] [PubMed]
- Nair, N.; Camacho-Vanegas, O.; Rykunov, D.; Dashkoff, M.; Camacho, S.C.; Schumacher, C.A.; Irish, J.C.; Harkins, T.T.; Freeman, E.; Garcia, I.; et al. Genomic Analysis of Uterine Lavage Fluid Detects Early Endometrial Cancers and Reveals a Prevalent Landscape of Driver Mutations in Women without Histopathologic Evidence of Cancer: A Prospective Cross-Sectional Study. PLoS Med. 2016, 13, e1002206. [Google Scholar] [CrossRef]
- Ghezelayagh, T.; Fredrickson, J.; Manhardt, E.; Radke, M.; Kohrn, B.; Gray, H.; Urban, R.; Pennington, K.; Liao, J.; Doll, K.; et al. Uterine lavage for the detection of ovarian cancer using an expanded gene panel. Gynecol. Oncol. 2021, 162, S49. [Google Scholar] [CrossRef]
- Stockley, J.; Akhand, R.; Kennedy, A.; Nyberg, C.; Crosbie, E.J.; Edmondson, R.J. Detection of MCM5 as a novel non-invasive aid for the diagnosis of endometrial and ovarian tumours. BMC Cancer 2020, 20, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Donkers, H.; Hirschfeld, M.; Weiß, D.; Erbes, T.; Jäger, M.; Pijnenborg, J.; Bekkers, R.; Galaal, K. Detection of microRNA in urine to identify patients with endometrial cancer: A feasibility study. Int. J. Gynecol. Cancer 2021, 31, 868–874. [Google Scholar] [CrossRef]
- Qu, W.L.; Gao, Q.S.; Chen, H.B.; Tang, Z.Q.; Zhu, X.Q.; Jiang, S.-W. HE4-test of urine and body fluids for diagnosis of gynecologic cancer. Expert Rev. Mol. Diagn. 2017, 17, 239–244. [Google Scholar] [CrossRef] [PubMed]
- O’Flynn, H.; Ryan, N.A.J.; Narine, N.; Shelton, D.; Rana, D.; Crosbie, E.J. Diagnostic accuracy of cytology for the detection of endometrial cancer in urine and vaginal samples. Nat. Commun. 2021, 12, 952. [Google Scholar] [CrossRef]
- Cheng, S.C.; Chen, K.; Chiu, C.Y.; Lu, K.Y.; Lu, H.Y.; Chiang, M.H.; Tsai, C.K.; Lo, C.J.; Cheng, M.L.; Chang, T.C.; et al. Metabolomic biomarkers in cervicovaginal fluid for detecting endometrial cancer through nuclear magnetic resonance spectroscopy. Metabolomics 2019, 15, 1–11. [Google Scholar] [CrossRef]
- Van Raemdonck, G.A.A.; Tjalma, W.A.A.; Coen, E.P.; Depuydt, C.E.; Van Ostade, X.W.M. Identification of Protein Biomarkers for Cervical Cancer Using Human Cervicovaginal Fluid. PLoS ONE 2014, 9, e106488. [Google Scholar] [CrossRef]
- Calis, P.; Yüce, K.; Basaran, D.; Salman, C. Assessment of Cervicovaginal Cancer Antigen 125 Levels: A Preliminary Study for Endometrial Cancer Screening. Gynecol. Obstet. Investig. 2016, 81, 518–522. [Google Scholar] [CrossRef]
- Qiu, J.H.; Xu, J.X.; Zhang, K.; Gu, W.; Nie, L.M.; Wang, G.X.; Luo, Y. Refining Cancer Management Using Integrated Liquid Biopsy. Theranostics 2020, 10, 2374–2384. [Google Scholar] [CrossRef]
- Nahar, F.; Hossain, M.A.; Paul, S.K.; Ahmed, M.U.; Khatun, S.; Bhuiyan, G.R.; Nasreen, S.A.; Haque, N.; Ahmed, S.; Kobayashi, N.; et al. Molecular Diagnosis of Human Papilloma Virus by PCR. Mymensingh Med. J. 2019, 28, 175–181. [Google Scholar] [PubMed]
- Cho, H.-W.; Ouh, Y.-T.; Hong, J.H.; Min, K.-J.; So, K.A.; Kim, T.J.; Paik, E.S.; Lee, J.W.; Moon, J.H.; Lee, J.K. Comparison of urine, self-collected vaginal swab, and cervical swab samples for detecting human papillomavirus (HPV) with Roche Cobas HPV, Anyplex II HPV, and RealTime HR-S HPV assay. J. Virol. Methods 2019, 269, 77–82. [Google Scholar] [CrossRef]
- Coorevits, L.; Traen, A.; Bingé, L.; Van Dorpe, J.; Praet, M.; Boelens, J.; Padalko, E. Are vaginal swabs comparable to cervical smears for human papillomavirus DNA testing? J. Gynecol. Oncol. 2018, 29, e8. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Circulating Tumor Cells: Liquid Biopsy of Cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef]
- Nguyen, D.X.; Bos, P.D.; Massague, J. Metastasis: From dissemination to organ-specific colonization. Nat. Rev. Cancer 2009, 9, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Vasseur, A.; Kiavue, N.; Bidard, F.-C.; Pierga, J.-Y.; Cabel, L. Clinical utility of circulating tumor cells: An update. Mol. Oncol. 2021, 15, 1647–1666. [Google Scholar] [CrossRef]
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011, 192, 373–382. [Google Scholar] [CrossRef]
- Lowe, A.C. Circulating Tumor Cells: Applications in Cytopathology. Surg. Pathol. Clin. 2018, 11, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Kitz, J.; Lowes, L.E.; Goodale, D.; Allan, A.L. Circulating Tumor Cell Analysis in Preclinical Mouse Models of Metastasis. Diagnostics 2018, 8, 30. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Cell lines from circulating tumor cells. Oncoscience 2015, 2, 815–816. [Google Scholar] [CrossRef] [PubMed]
- Cayrefourcq, L.; Mazard, T.; Joosse, S.; Solassol, J.; Ramos, J.; Assenat, E.; Schumacher, U.; Costes, V.; Maudelonde, T.; Pantel, K.; et al. Establishment and Characterization of a Cell Line from Human Circulating Colon Cancer Cells. Cancer Res. 2015, 75, 892–901. [Google Scholar] [CrossRef]
- Hu, C.-L.; Zhang, Y.-J.; Zhang, X.-F.; Fei, X.; Zhang, H.; Li, C.-G.; Sun, B. 3D Culture of Circulating Tumor Cells for Evaluating Early Recurrence and Metastasis in Patients with Hepatocellular Carcinoma. OncoTargets Ther. 2021, 14, 2673–2688. [Google Scholar] [CrossRef]
- Esposito, A.; Bardelli, A.; Criscitiello, C.; Colombo, N.; Gelao, L.; Fumagalli, L.; Minchella, I.; Locatelli, M.; Goldhirsch, A.; Curigliano, G. Monitoring tumor-derived cell-free DNA in patients with solid tumors: Clinical perspectives and research opportunities. Cancer Treat. Rev. 2014, 40, 648–655. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Salehi, M.; Sharifi, M. Exosomal miRNAs as novel cancer biomarkers: Challenges and opportunities. J. Cell. Physiol. 2018, 233, 6370–6380. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Pan, J.; Fang, S.; Zhou, C.; Han, Y.; Chen, J.; Meng, X.; Jin, X.; Gong, Z. Liquid biopsy: Circulating exosomal long noncoding RNAs in cancer. Clin. Chim. Acta 2019, 495, 331–337. [Google Scholar] [CrossRef]
- Norouzi-Barough, L.; Shahi, A.A.K.; Mohebzadeh, F.; Masoumi, L.; Haddadi, M.R.; Shirian, S. Early diagnosis of breast and ovarian cancers by body fluids circulating tumor-derived exosomes. Cancer Cell Int. 2020, 20, 187. [Google Scholar] [CrossRef] [PubMed]
- Sumrin, A.; Moazzam, S.; Khan, A.A.; Ramzan, I.; Batool, Z.; Kaleem, S.; Ali, M.; Bashir, H.; Bilal, M. Exosomes as Biomarker of Cancer. Braz. Arch. Biol. Technol. 2018, 61, e18160730. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, J.-Q.; Liu, J.-L.; Tian, L. Exosomes in tumor microenvironment: Novel transporters and biomarkers. J. Transl. Med. 2016, 14, 1–9. [Google Scholar] [CrossRef]
- Tang, M.K.; Wong, A.S. Exosomes: Emerging biomarkers and targets for ovarian cancer. Cancer Lett. 2015, 367, 26–33. [Google Scholar] [CrossRef]
- Halvaei, S.; Daryani, S.; Eslami-S, Z.; Samadi, T.; Jafarbeik-Iravani, N.; Bakhshayesh, T.O.; Majidzadeh-A, K.; Esmaeili, R. Exosomes in Cancer Liquid Biopsy: A Focus on Breast Cancer. Mol. Ther. Nucleic Acids 2018, 10, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Pospíchalová, V.; Svoboda, J.; Dave, Z.; Kotrbová, A.; Kaiser, K.; Klemova, D.; Ilkovics, L.; Hampl, A.; Crha, I.; Jandakova, E.; et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J. Extracell. Vesicles 2015, 4, 25530. [Google Scholar] [CrossRef]
- Singh, U.; Anjum, Q.S.; Negi, N.; Singh, N.; Goel, M.; Srivastava, K. Comparative study between liquid-based cytology & conventional Pap smear for cytological follow up of treated patients of cancer cervix. Indian J. Med. Res. 2018, 147, 263–267. [Google Scholar] [CrossRef]
- Ronco, G.; Cuzick, J.; Pierotti, P.; Cariaggi, M.P.; Palma, P.D.; Naldoni, C.; Ghiringhello, B.; Rossi, P.G.; Minucci, D.; Parisio, F.; et al. Accuracy of liquid based versus conventional cytology: Overall results of new technologies for cervical cancer screening: Randomised controlled trial. BMJ 2007, 335, 28. [Google Scholar] [CrossRef]
- Wright, T.C., Jr.; Stoler, M.H.; Behrens, C.M.; Apple, R.; Derion, T.; Wright, T.L. The ATHENA human papillomavirus study: Design, methods, and baseline results. Am. J. Obstet. Gynecol. 2012, 206, 46.e1–46.e11. [Google Scholar] [CrossRef]
- Luttmer, R.; Dijkstra, M.G.; Snijders, P.J.F.; Berkhof, J.; Van Kemenade, F.J.; Rozendaal, L.; Helmerhorst, T.J.M.; Verheijen, R.H.M.; Ter Harmsel, W.A.; Van Baal, W.M.; et al. p16/Ki-67 dual-stained cytology for detecting cervical (pre)cancer in a HPV-positive gynecologic outpatient population. Mod. Pathol. 2016, 29, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Liu, J.; Gong, L.; Sun, X.W.; Long, W.B. Combining HPV DNA load with p16/Ki-67 staining to detect cervical precancerous lesions and predict the progression of CIN1-2 lesions. Virol. J. 2019, 16, 117–119. [Google Scholar] [CrossRef]
- Shi, Q.; Xu, L.; Yang, R.; Meng, Y.P.; Qiu, L.H. Ki-67 and P16 proteins in cervical cancer and precancerous lesions of young women and the diagnostic value for cervical cancer and precancerous lesions. Oncol. Lett. 2019, 18, 1351–1355. [Google Scholar] [CrossRef]
- Ziemke, P. p16/Ki-67 Immunocytochemistry in Gynecological Cytology: Limitations in Practice. Acta Cytol. 2017, 61, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Meites, E.; Szilagyi, P.G.; Chesson, H.W.; Unger, E.R.; Romero, J.R.; Markowitz, L.E. Human Papillomavirus Vaccination for Adults: Updated Recommendations of the Advisory Committee on Immunization Practices. MMWR Morb. Mortal. Wkly. Rep. 2019, 68, 698–702. [Google Scholar] [CrossRef]
- Schiller, J.T.; Castellsagué, X.; Garland, S.M. A Review of Clinical Trials of Human Papillomavirus Prophylactic Vaccines. Vaccine 2012, 30, F123–F138. [Google Scholar] [CrossRef] [PubMed]
- Kreimer, A.R.; González, P.; Katki, H.A.; Porras, C.; Schiffman, M.; Rodriguez, A.C.; Solomon, D.; Jiménez, S.; Schiller, J.T.; Lowy, D.R.; et al. Efficacy of a bivalent HPV 16/18 vaccine against anal HPV 16/18 infection among young women: A nested analysis within the Costa Rica Vaccine Trial. Lancet Oncol. 2011, 12, 862–870. [Google Scholar] [CrossRef]
- Chaturvedi, A.K.; Graubard, B.I.; Broutian, T.; Pickard, R.K.L.; Tong, Z.-Y.; Xiao, W.; Kahle, L.; Gillison, M.L. Effect of Prophylactic Human Papillomavirus (HPV) Vaccination on Oral HPV Infections Among Young Adults in the United States. J. Clin. Oncol. 2018, 36, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A. The next generation of HPV vaccines: Nonavalent vaccine V503 on the horizon. Expert Rev. Vaccines 2014, 13, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Prendiville, W.; Sankaranarayanan, R. Colposcopy and Treatment of Cervical Precancer; IARC Technical Publications: Lyon, France, 2017. [Google Scholar]
- World Health Organization. WHO Guidelines for Screening and Treatment of Precancerous Lesions for Cervical Cancer Prevention; World Health Organization: Geneva, Switzerland, 2013. [Google Scholar]
- Munkhdelger, J.; Kim, G.; Wang, H.-Y.; Lee, D.; Kim, S.; Choi, Y.; Choi, E.; Park, S.; Jin, H.; Park, K.H.; et al. Performance of HPV E6/E7 mRNA RT-qPCR for screening and diagnosis of cervical cancer with ThinPrep® Pap test samples. Exp. Mol. Pathol. 2014, 97, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Pisarska, J.; Baldy-Chudzik, K. MicroRNA-Based Fingerprinting of Cervical Lesions and Cancer. J. Clin. Med. 2020, 9, 3668. [Google Scholar] [CrossRef]
- Bhat, A.A.; Younes, S.N.; Raza, S.S.; Zarif, L.; Nisar, S.; Ahmed, I.; Mir, R.; Kumar, S.; Sharawat, S.K.; Hashem, S.; et al. Correction to: Role of non-coding RNA networks in leukemia progression, metastasis and drug resistance. Mol. Cancer 2020, 19, 1. [Google Scholar] [CrossRef]
- Liu, K.S.; Gao, L.; Ma, X.S.; Huang, J.-J.; Chen, J.; Zeng, L.; Ashby, C.R., Jr.; Zou, C.; Chen, Z.-S. Long non-coding RNAs regulate drug resistance in cancer. Mol. Cancer 2020, 19, 54. [Google Scholar] [CrossRef]
- Barwal, T.S.; Sharma, U.; Vasquez, K.M.; Prakash, H.; Jain, A. A panel of circulating long non-coding RNAs as liquid biopsy biomarkers for breast and cervical cancers. Biochimie 2020, 176, 62–70. [Google Scholar] [CrossRef]
- Zhu, H.; Zhu, H.; Tian, M.; Wang, D.; He, J.; Xu, T. DNA Methylation and Hydroxymethylation in Cervical Cancer: Diagnosis, Prognosis and Treatment. Front. Genet. 2020, 11, 347. [Google Scholar] [CrossRef]
- Martisova, A.; Holcakova, J.; Izadi, N.; Sebuyoya, R.; Hrstka, R.; Bartosik, M. DNA Methylation in Solid Tumors: Functions and Methods of Detection. Int. J. Mol. Sci. 2021, 22, 4247. [Google Scholar] [CrossRef]
- Wentzensen, N.; Sherman, M.E.; Schiffman, M.; Wang, S.S. Utility of methylation markers in cervical cancer early detection: Appraisal of the state-of-the-science. Gynecol. Oncol. 2009, 112, 293–299. [Google Scholar] [CrossRef]
- Yang, H.-J. Aberrant DNA methylation in cervical carcinogenesis. Chin. J. Cancer 2013, 32, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.-T.; Sytwu, H.-K.; Yan, M.-D.; Shih, Y.-L.; Chang, C.-C.; Yu, M.-H.; Chu, T.-Y.; Lai, H.-C.; Lin, Y.-W. Promoter methylation of SFRPs gene family in cervical cancer. Gynecol. Oncol. 2009, 112, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Carestiato, F.N.; Amaro-Filho, S.M.; Moreira, M.A.M.; Cavalcanti, S.M.B. Methylation of p16 ink4a promoter is independent of human papillomavirus DNA physical state: A comparison between cervical pre-neoplastic and neoplastic samples. Memórias Inst. Oswaldo Cruz 2018, 114, e180456. [Google Scholar] [CrossRef]
- Lim, E.H.; Ng, S.L.; Li, J.; Chang, A.R.; Ng, J.; Ilancheran, A.; Low, J.; Quek, S.C.; Tay, E.H. Cervical dysplasia: Assessing methylation status (Methylight) of CCNA1, DAPK1, HS3ST2, PAX1 and TFPI2 to improve diagnostic accuracy. Gynecol. Oncol. 2010, 119, 225–231. [Google Scholar] [CrossRef]
- Del Pino, M.; Sierra, A.; Marimon, L.; Delgado, C.M.; Rodriguez-Trujillo, A.; Barnadas, E.; Saco, A.; Torné, A.; Ordi, J. CADM1, MAL, and miR124 Promoter Methylation as Biomarkers of Transforming Cervical Intrapithelial Lesions. Int. J. Mol. Sci. 2019, 20, 2262. [Google Scholar] [CrossRef] [PubMed]
- Dankai, W.; Khunamornpong, S.; Siriaunkgul, S.; Soongkhaw, A.; Janpanao, A.; Utaipat, U.; Kitkumthorn, N.; Mutirangura, A.; Srisomboon, J.; Lekawanvijit, S. Role of genomic DNA methylation in detection of cytologic and histologic abnormalities in high risk HPV-infected women. PLoS ONE 2019, 14, e0210289. [Google Scholar] [CrossRef]
- Yang, N.; Nijhuis, E.R.; Volders, H.H.; Eijsink, J.J.; Lendvai, Á.; Zhang, B.; Hollema, H.; Schuuring, E.; Wisman, G.B.A.; van der Zee, A.G. Gene promoter methylation patterns throughout the process of cervical carcinogenesis. Cell. Oncol. 2010, 32, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Gao, H.; Tan, H.-Z. SOX1 Promoter Hypermethylation as a Potential Biomarker for High-Grade Squamous Intraepithelial Neoplasia Lesion and Cervical Carcinoma: A Meta-Analysis with Trial Sequential Analysis. Front. Genet. 2020, 11, 633. [Google Scholar] [CrossRef]
- Bowden, S.J.; Kalliala, I.; Veroniki, A.A.; Arbyn, M.; Mitra, A.; Lathouras, K.; Mirabello, L.; Chadeau-Hyam, M.; Paraskevaidis, E.; Flanagan, J.M.; et al. The use of human papillomavirus DNA methylation in cervical intraepithelial neoplasia: A systematic review and meta-analysis. EBioMedicine 2019, 50, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Hublarova, P.; Hrstka, R.; Rotterova, P.; Rotter, L.; Coupkova, M.; Badal, V.; Nenutil, R.; Vojtesek, B. Prediction of Human Papillomavirus 16 E6 Gene Expression and Cervical Intraepithelial Neoplasia Progression by Methylation Status. Int. J. Gynecol. Cancer 2009, 19, 321–325. [Google Scholar] [CrossRef]
- Heitmann, E.R.; Lankachandra, K.M.; Wall, J.; Harris, G.D.; McKinney, H.J.; Jalali, G.R.; Verma, Y.; Kershnar, E.; Kilpatrick, M.W.; Tsipouras, P.; et al. 3q26 Amplification Is an Effective Negative Triage Test for LSIL: A Historical Prospective Study. PLoS ONE 2012, 7, e39101. [Google Scholar] [CrossRef]
- Stoler, M.H.; Schiffman, M. Interobserver Reproducibility of Cervical Cytologic and Histologic InterpretationsRealistic Estimates From the ASCUS-LSIL Triage Study. JAMA 2001, 285, 1500–1505. [Google Scholar] [CrossRef] [PubMed]
- Solomon, D.; Schiffman, M.; Tarone, R.; Grp, A. Comparison of Three Management Strategies for Patients with Atypical Squamous Cells of Undetermined Significance: Baseline Results from a Randomized Trial. J. Natl. Cancer Inst. 2001, 93, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Kinney, W.K.; Manos, M.; Hurley, L.B.; Ransley, J.E. Where’s the high-grade cervical neoplasia? The importance of minimally abnormal Papanicolaou diagnoses. Obstet. Gynecol. 1998, 91, 973–976. [Google Scholar] [CrossRef]
- Cox, J.T.; Schiffman, M.; Solomon, D.; Grp, A. Prospective follow-up suggests similar risk of subsequent cervical intraepithelial neoplasia grade 2 or 3 among women with cervical intraepithelial neoplasia grade 1 or negative colposcopy and directed biopsy. Am. J. Obstet. Gynecol. 2003, 188, 1406–1412. [Google Scholar] [CrossRef]
- Kitchener, H.C.; Almonte, M.; Thomson, C.; Wheeler, P.; Sargent, A.; Stoykova, B.; Gilham, C.; Baysson, H.; Roberts, C.; Dowie, R.; et al. HPV testing in combination with liquid-based cytology in primary cervical screening (ARTISTIC): A randomised controlled trial. Lancet Oncol. 2009, 10, 672–682. [Google Scholar] [CrossRef]
- Pao, C.C.; Hor, J.J.; Yang, F.P.; Lin, C.Y.; Tseng, C.J. Detection of human papillomavirus mRNA and cervical cancer cells in peripheral blood of cervical cancer patients with metastasis. J. Clin. Oncol. 1997, 15, 1008–1012. [Google Scholar] [CrossRef]
- Pornthanakasem, W.; Shotelersuk, K.; Termrungruanglert, W.; Voravud, N.; Niruthisard, S.; Mutirangura, A. Human papillomavirus DNA in plasma of patients with cervical cancer. BMC Cancer 2001, 1, 2. [Google Scholar] [CrossRef]
- Widschwendter, A.; Blassnig, A.; Wiedemair, A.; Müller-Holzner, E.; Müller, H.M.; Marth, C. Human papillomavirus DNA in sera of cervical cancer patients as tumor marker. Cancer Lett. 2003, 202, 231–239. [Google Scholar] [CrossRef]
- Sathish, N.; Abraham, P.; Peedicayil, A.; Sridharan, G.; John, S.; Shaji, R.; Chandy, G. HPV DNA in plasma of patients with cervical carcinoma. J. Clin. Virol. 2004, 31, 204–209. [Google Scholar] [CrossRef]
- Jeannot, E.; Becette, V.; Campitelli, M.; Calméjane, M.; Lappartient, E.; Ruff, E.; Saada, S.; Holmes, A.; Bellet, D.; Sastre-Garau, X. Circulating human papillomavirus DNA detected using droplet digital PCR in the serum of patients diagnosed with early stage human papillomavirus-associated invasive carcinoma. J. Pathol. Clin. Res. 2016, 2, 201–209. [Google Scholar] [CrossRef]
- Gu, Y.; Wan, C.; Qiu, J.; Cui, Y.; Jiang, T.; Zhuang, Z. Circulating HPV cDNA in the blood as a reliable biomarker for cervical cancer: A meta-analysis. PLoS ONE 2020, 15, e0224001. [Google Scholar] [CrossRef]
- Cocuzza, C.E.; Martinelli, M.; Sina, F.; Piana, A.; Sotgiu, G.; Dell’Anna, T.; Musumeci, R. Human papillomavirus DNA detection in plasma and cervical samples of women with a recent history of low grade or precancerous cervical dysplasia. PLoS ONE 2017, 12, e0188592. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-Y.; Chae, D.-K.; Lee, S.-H.; Lim, Y.; An, J.; Chae, C.H.; Kim, B.C.; Bhak, J.; Bolser, D.; Cho, D.-H. Efficient mutation screening for cervical cancers from circulating tumor DNA in blood. BMC Cancer 2020, 20, 694. [Google Scholar] [CrossRef] [PubMed]
- Charo, L.M.; Eskander, R.N.; Okamura, R.; Patel, S.P.; Nikanjam, M.; Lanman, R.B.; Piccioni, D.E.; Kato, S.; McHale, M.T.; Kurzrock, R. Clinical implications of plasma circulating tumor DNA in gynecologic cancer patients. Mol. Oncol. 2021, 15, 67–79. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Lortet-Tieulent, J.; Ferlay, J.; Bray, F.; Jemal, A. International Patterns and Trends in Endometrial Cancer Incidence, 1978–2013. J. Natl. Cancer Inst. 2018, 110, 354–361. [Google Scholar] [CrossRef]
- Schlosshauer, P.W.; Ellenson, L.H.; Soslow, R.A. β-Catenin and E-Cadherin Expression Patterns in High-Grade Endometrial Carcinoma Are Associated with Histological Subtype. Mod. Pathol. 2002, 15, 1032–1037. [Google Scholar] [CrossRef]
- Murali, R.; Davidson, B.; Fadare, O.; Carlson, J.; Crum, C.P.; Gilks, C.B.; Irving, J.A.; Malpica, A.; Matias-Guiu, X.; McCluggage, W.G.; et al. High-grade Endometrial Carcinomas. Int. J. Gynecol. Pathol. 2019, 38, S40–S63. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef]
- Concin, N.; Matias-Guiu, X.; Vergote, I.; Cibula, D.; Mirza, M.R.; Marnitz, S.; Ledermann, J.; Bosse, T.; Chargari, C.; Fagotti, A.; et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. Int. J. Gynecol. Cancer 2021, 31, 12–39. [Google Scholar] [CrossRef] [PubMed]
- Murali, R.; Soslow, R.; Weigelt, B. Classification of endometrial carcinoma: More than two types. Lancet Oncol. 2014, 15, e268–e278. [Google Scholar] [CrossRef]
- Kaitu’U-Lino, T.J.; Ye, L.; Gargett, C.E. Reepithelialization of the Uterine Surface Arises from Endometrial Glands: Evidence from a Functional Mouse Model of Breakdown and Repair. Endocrinology 2010, 151, 3386–3395. [Google Scholar] [CrossRef]
- Lac, V.; Nazeran, T.M.; Tessier-Cloutier, B.; Aguirre-Hernandez, R.; Albert, A.; Lum, A.; Khattra, J.; Praetorius, T.; Mason, M.; Chiu, D.; et al. Oncogenic mutations in histologically normal endometrium: The new normal? J. Pathol. 2019, 249, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.; Leongamornlert, D.; Coorens, T.H.H.; Sanders, M.A.; Ellis, P.; Dentro, S.C.; Dawson, K.J.; Butler, T.; Rahbari, R.; Mitchell, T.J.; et al. The mutational landscape of normal human endometrial epithelium. Nature 2020, 580, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Kyo, S.; Sato, S.; Nakayama, K. Cancer-associated mutations in normal human endometrium: Surprise or expected? Cancer Sci. 2020, 111, 3458–3467. [Google Scholar] [CrossRef]
- Temko, D.; Van Gool, I.C.; Rayner, E.; Glaire, M.; Makino, S.; Brown, M.; Chegwidden, L.; Palles, C.; Depreeuw, J.; Beggs, A.; et al. Somatic POLE exonuclease domain mutations are early events in sporadic endometrial and colorectal carcinogenesis, determining driver mutational landscape, clonal neoantigen burden and immune response. J. Pathol. 2018, 245, 283–296. [Google Scholar] [CrossRef]
- Aguilar, M.; Zhang, H.; Zhang, M.S.; Cantarell, B.; Sahoo, S.S.; Li, H.D.; Cuevas, I.C.; Lea, J.; Miller, D.S.; Chen, H.; et al. Serial genomic analysis of endometrium supports the existence of histologically indistinct endometrial cancer precursors. J. Pathol. 2021, 254, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Martignetti, J.A.; Pandya, D.; Nagarsheth, N.; Chen, Y.; Camacho, O.; Tomita, S.; Brodman, M.; Ascher-Walsh, C.; Kolev, V.; Cohen, S.; et al. Detection of endometrial precancer by a targeted gynecologic cancer liquid biopsy. Mol. Case Stud. 2018, 4, a003269. [Google Scholar] [CrossRef] [PubMed]
- Huvila, J.; Pors, J.; Thompson, E.F.; Gilks, C.B. Endometrial carcinoma: Molecular subtypes, precursors and the role of pathology in early diagnosis. J. Pathol. 2021, 253, 355–365. [Google Scholar] [CrossRef]
- Lacey, J.V., Jr.; Ioffe, O.B.; Ronnett, B.M.; Rush, B.B.; Richesson, D.A.; Chatterjee, N.; Langholz, B.; Glass, A.G.; Sherman, M.E. Endometrial carcinoma risk among women diagnosed with endometrial hyperplasia: The 34-year experience in a large health plan. Br. J. Cancer 2007, 98, 45–53. [Google Scholar] [CrossRef]
- Trimble, C.L.; Kauderer, J.; Zaino, R.J.; Silverberg, S.G.; Lim, P.C.; Burke, J.J., 2nd; Alberts, D.S.; Curtin, J.P. Concurrent endometrial carcinoma in women with a biopsy diagnosis of atypical endometrial hyperplasia: A Gynecologic Oncology Group study. Cancer 2006, 106, 812–819. [Google Scholar] [CrossRef]
- Riethdorf, L.; Begemann, C.; Riethdorf, S.; Milde-Langosch, K.; Loning, T. Comparison of benign and malignant endometrial lesions for their p53 state, using immunohistochemistry and temperature-gradient gel electrophoresis. Virchows Arch. 1996, 428, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, M.; Katoh, T.; Hori, S.; Suzuki, K.; Ohno, K.; Maruyama, M.; Matsui, N.; Miyazaki, S.; Ogane, N.; Kameda, Y. Endometrial intraepithelial carcinoma in association with polyp: Review of eight cases. Diagn. Pathol. 2013, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Maksem, J.A.; Meiers, I.; Robboy, S.J. A primer of endometrial cytology with histological correlation. Diagn. Cytopathol. 2007, 35, 817–844. [Google Scholar] [CrossRef]
- Bergman, L.; Beelen, M.L.; Gallee, M.P.; Hollema, H.; Benraadt, J.; van Leeuwen, F.E.; Comprehensive Cancer Centres’ ALERT Group. Risk and prognosis of endometrial cancer after tamoxifen for breast cancer. Lancet 2000, 356, 881–887. [Google Scholar] [CrossRef]
- Hrstka, R.; Podhorec, J.; Nenutil, R.; Sommerova, L.; Obacz, J.; Durech, M.; Faktor, J.; Bouchal, P.; Skoupilova, H.; Vojtesek, B. Tamoxifen-Dependent Induction of AGR2 Is Associated with Increased Aggressiveness of Endometrial Cancer Cells. Cancer Investig. 2017, 35, 313–324. [Google Scholar] [CrossRef]
- Ryan, N.A.J.; Glaire, M.A.; Blake, D.; Cabrera-Dandy, M.; Evans, D.G.; Crosbie, E.J. The proportion of endometrial cancers associated with Lynch syndrome: A systematic review of the literature and meta-analysis. Genet. Med. 2019, 21, 2167–2180. [Google Scholar] [CrossRef]
- Gammon, A.; Jasperson, K.; Champine, M. Genetic basis of Cowden syndrome and its implications for clinical practice and risk management. Appl. Clin. Genet. 2016, 9, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Hampel, H.; Frankel, W.; Panescu, J.; Lockman, J.; Sotamaa, K.; Fix, D.; Comeras, I.; La Jeunesse, J.; Nakagawa, H.; Westman, J.A.; et al. Screening for Lynch Syndrome (Hereditary Nonpolyposis Colorectal Cancer) among Endometrial Cancer Patients. Cancer Res. 2006, 66, 7810–7817. [Google Scholar] [CrossRef] [PubMed]
- Bafligil, C.; Thompson, D.J.; Lophatananon, A.; Smith, M.J.; Ryan, N.A.J.; Naqvi, A.; Evans, D.G.; Crosbie, E.J. Association between genetic polymorphisms and endometrial cancer risk: A systematic review. J. Med. Genet. 2020, 57, 591–600. [Google Scholar] [CrossRef] [PubMed]
- O’Mara, T.A.; Crosbie, E.J. Polygenic risk score opportunities for early detection and prevention strategies in endometrial cancer. Br. J. Cancer 2020, 123, 1045–1046. [Google Scholar] [CrossRef] [PubMed]
- Frias-Gomez, J.; Benavente, Y.; Ponce, J.; Brunet, J.; Ibáñez, R.; Peremiquel-Trillas, P.; Baixeras, N.; Zanca, A.; Piulats, J.M.; Aytés, Á.; et al. Sensitivity of cervico-vaginal cytology in endometrial carcinoma: A systematic review and meta-analysis. Cancer Cytopathol. 2020, 128, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Frias-Gomez, J.; Tovar, E.; Vidal, A.; Murgui, L.; Ibáñez, R.; Peremiquel-Trillas, P.; Paytubi, S.; Baixeras, N.; Zanca, A.; Ponce, J.; et al. Sensitivity of cervical cytology in endometrial cancer detection in a tertiary hospital in Spain. Cancer Med. 2021, 10, 6762–6766. [Google Scholar] [CrossRef]
- Jones, E.R.; Carter, S.; O’Flynn, H.; Njoku, K.; Barr, C.E.; Narine, N.; Shelton, D.; Rana, D.; Crosbie, E.J. Developing Tests for Endometrial Cancer Detection (DETECT): Protocol for a diagnostic accuracy study of urine and vaginal samples for the detection of endometrial cancer by cytology in women with postmenopausal bleeding. BMJ Open 2021, 11, e050755. [Google Scholar] [CrossRef] [PubMed]
- Njoku, K.; Chiasserini, D.; Whetton, A.D.; Crosbie, E.J. Proteomic Biomarkers for the Detection of Endometrial Cancer. Cancers 2019, 11, 1572. [Google Scholar] [CrossRef]
- Mu, A.K.-W.; Lim, B.-K.; Hashim, O.H.; Shuib, A.S. Detection of Differential Levels of Proteins in the Urine of Patients with Endometrial Cancer: Analysis Using Two-Dimensional Gel Electrophoresis and O-Glycan Binding Lectin. Int. J. Mol. Sci. 2012, 13, 9489–9501. [Google Scholar] [CrossRef]
- Kurnit, K.C.; Westin, S.N.; Coleman, R.L. Microsatellite instability in endometrial cancer: New purpose for an old test. Cancer 2019, 125, 2154–2163. [Google Scholar] [CrossRef]
- Bolivar, A.M.; Luthra, R.; Mehrotra, M.; Chen, W.; Barkoh, B.A.; Hu, P.; Zhang, W.; Broaddus, R.R. Targeted next-generation sequencing of endometrial cancer and matched circulating tumor DNA: Identification of plasma-based, tumor-associated mutations in early stage patients. Mod. Pathol. 2019, 32, 405–414. [Google Scholar] [CrossRef]
- Casas-Arozamena, C.; Díaz, E.; Moiola, C.P.; Alonso-Alconada, L.; Ferreiros, A.; Abalo, A.; Gil, C.L.; Oltra, S.S.; De Santiago, J.; Cabrera, S.; et al. Genomic Profiling of Uterine Aspirates and cfDNA as an Integrative Liquid Biopsy Strategy in Endometrial Cancer. J. Clin. Med. 2020, 9, 585. [Google Scholar] [CrossRef]
- Tsukamoto, O.; Miura, K.; Mishima, H.; Abe, S.; Kaneuchi, M.; Higashijima, A.; Miura, S.; Kinoshita, A.; Yoshiura, K.-I.; Masuzaki, H. Identification of endometrioid endometrial carcinoma-associated microRNAs in tissue and plasma. Gynecol. Oncol. 2014, 132, 715–721. [Google Scholar] [CrossRef]
- Srivastava, A.; Moxley, K.; Ruskin, R.; Dhanasekaran, D.N.; Zhao, Y.D.; Ramesh, R. A Non-invasive Liquid Biopsy Screening of Urine-Derived Exosomes for miRNAs as Biomarkers in Endometrial Cancer Patients. AAPS J. 2018, 20, 82. [Google Scholar] [CrossRef] [PubMed]
- Roman-Canal, B.; Moiola, C.P.; Gatius, S.; Bonnin, S.; Ruiz-Miró, M.; González, E.; González-Tallada, X.; Llordella, I.; Hernández, I.; Porcel, J.M.; et al. EV-Associated miRNAs from Peritoneal Lavage are a Source of Biomarkers in Endometrial Cancer. Cancers 2019, 11, 839. [Google Scholar] [CrossRef]
- Fan, X.; Zou, X.; Liu, C.; Cheng, W.; Zhang, S.; Geng, X.; Zhu, W. MicroRNA expression profile in serum reveals novel diagnostic biomarkers for endometrial cancer. Biosci. Rep. 2021, 41, BSR20210111. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, W.; Wang, F.; Yang, S.; Hu, J.; Lu, B.; Pan, Z.; Ma, Y.; Zheng, M.; Zhou, L.; et al. Plasma-derived exosomal miR-15a-5p as a promising diagnostic biomarker for early detection of endometrial carcinoma. Mol. Cancer 2021, 20, 57. [Google Scholar] [CrossRef]
- Njoku, K.; Sutton, C.J.; Whetton, A.D.; Crosbie, E.J. Metabolomic Biomarkers for Detection, Prognosis and Identifying Recurrence in Endometrial Cancer. Metabolites 2020, 10, 314. [Google Scholar] [CrossRef]
- Bahado-Singh, R.O.; Lugade, A.; Field, J.; Al-Wahab, Z.; Han, B.; Mandal, R.; Bjorndahl, T.C.; Turkoglu, O.; Graham, S.F.; Wishart, D.; et al. Metabolomic prediction of endometrial cancer. Metabolomics 2017, 14, 6. [Google Scholar] [CrossRef]
- Raffone, A.; Troisi, J.; Boccia, D.; Travaglino, A.; Capuano, G.; Insabato, L.; Mollo, A.; Guida, M.; Zullo, F. Metabolomics in endometrial cancer diagnosis: A systematic review. Acta Obstet. Gynecol. Scand. 2020, 99, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Trousil, S.; Lee, P.; Pinato, D.J.; Ellis, J.K.; Dina, R.; Aboagye, E.O.; Keun, H.C.; Sharma, R. Alterations of Choline Phospholipid Metabolism in Endometrial Cancer Are Caused by Choline Kinase Alpha Overexpression and a Hyperactivated Deacylation Pathway. Cancer Res. 2014, 74, 6867–6877. [Google Scholar] [CrossRef]
- Njoku, K.; Campbell, A.E.; Geary, B.; MacKintosh, M.L.; Derbyshire, A.E.; Kitson, S.J.; Sivalingam, V.N.; Pierce, A.; Whetton, A.D.; Crosbie, E.J. Metabolomic Biomarkers for the Detection of Obesity-Driven Endometrial Cancer. Cancers 2021, 13, 718. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2017. CA A Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Bast, R.C., Jr.; Lu, Z.; Han, C.Y.; Lu, K.H.; Anderson, K.S.; Drescher, C.W.; Skates, S.J. Biomarkers and Strategies for Early Detection of Ovarian Cancer. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2504–2512. [Google Scholar] [CrossRef]
- Buys, S.S.; Partridge, E.; Greene, M.H.; Prorok, P.C.; Reding, D.; Riley, T.L.; Hartge, P.; Fagerstrom, R.M.; Ragard, L.R.; Chia, D.; et al. Ovarian cancer screening in the Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening trial: Findings from the initial screen of a randomized trial. Am. J. Obstet. Gynecol. 2005, 193, 1630–1639. [Google Scholar] [CrossRef]
- Kobayashi, H.; Yamada, Y.; Sado, T.; Sakata, M.; Yoshida, S.; Kawaguchi, R.; Kanayama, S.; Shigetomi, H.; Haruta, S.; Tsuji, Y.; et al. A randomized study of screening for ovarian cancer: A multicenter study in Japan. Int. J. Gynecol. Cancer 2008, 18, 414–420. [Google Scholar] [CrossRef]
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef]
- Jervis, S.; Song, H.; Lee, A.; Dicks, E.; Harrington, P.; Baynes, C.; Manchanda, R.; Easton, D.F.; Jacobs, I.; Pharoah, P.P.D.; et al. A risk prediction algorithm for ovarian cancer incorporating BRCA1, BRCA2, common alleles and other familial effects. J. Med. Genet. 2015, 52, 465–475. [Google Scholar] [CrossRef]
- Yang, X.; Leslie, G.; Gentry-Maharaj, A.; Ryan, A.; Intermaggio, M.; Lee, A.; Kalsi, J.K.; Tyrer, J.; Gaba, F.; Manchanda, R.; et al. Evaluation of polygenic risk scores for ovarian cancer risk prediction in a prospective cohort study. J. Med. Genet. 2018, 55, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Phelan, C.M.; Kuchenbaecker, K.B.; Tyrer, J.P.; Kar, S.P.; Lawrenson, K.; Winham, S.J.; Dennis, J.; Pirie, A.; Riggan, M.J.; Chornokur, G.; et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat. Genet. 2017, 49, 680–691. [Google Scholar] [CrossRef] [PubMed]
- Kuchenbaecker, K.B.; Ramus, S.J.; Tyrer, J.; Lee, A.; Shen, H.C.; Beesley, J.; Lawrenson, K.; McGuffog, L.; Healey, S.; Lee, J.M.; et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat. Genet. 2015, 47, 164–171. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Hurson, A.N.; Zhang, H.; Choudhury, P.P.; Easton, D.F.; Milne, R.L.; Simard, J.; Hall, P.; Michailidou, K.; Dennis, J.; et al. Assessment of polygenic architecture and risk prediction based on common variants across fourteen cancers. Nat. Commun. 2020, 11, 3353. [Google Scholar] [CrossRef]
- Mehra, K.K.; Chang, M.C.; Folkins, A.K.; Raho, C.J.; Lima, J.F.; Yuan, L.; Mehrad, M.; Tworoger, S.S.; Crum, C.P.; Saleemuddin, A. The impact of tissue block sampling on the detection of p53 signatures in fallopian tubes from women with BRCA 1 or 2 mutations (BRCA+) and controls. Mod. Pathol. 2011, 24, 152–156. [Google Scholar] [CrossRef]
- Dorigo, O.; Berek, J.S. Personalizing CA125 Levels for Ovarian Cancer Screening. Cancer Prev. Res. 2011, 4, 1356–1359. [Google Scholar] [CrossRef] [PubMed]
- Drescher, C.W.; Hawley, S.; Thorpe, J.D.; Marticke, S.; McIntosh, M.; Gambhir, S.S.; Urban, N. Impact of Screening Test Performance and Cost on Mortality Reduction and Cost-effectiveness of Multimodal Ovarian Cancer Screening. Cancer Prev. Res. 2012, 5, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Blyuss, O.; Gentry-Maharaj, A.; Fourkala, E.-O.; Ryan, A.; Zaikin, A.; Menon, U.; Jacobs, I.; Timms, J.F. Serial Patterns of Ovarian Cancer Biomarkers in a Prediagnosis Longitudinal Dataset. BioMed Res. Int. 2015, 2015, 681416. [Google Scholar] [CrossRef] [PubMed]
- Russell, M.R.; Graham, C.; D’Amato, A.; Gentry-Maharaj, A.; Ryan, A.; Kalsi, J.K.; Whetton, A.D.; Menon, U.; Jacobs, I.; Graham, R.L.J. Diagnosis of epithelial ovarian cancer using a combined protein biomarker panel. Br. J. Cancer 2019, 121, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.G.; Jabre-Raughley, M.; Brown, A.K.; Robison, K.M.; Miller, M.C.; Allard, W.J.; Kurman, R.J.; Bast, R.C.; Skates, S.J. Comparison of a novel multiple marker assay vs the Risk of Malignancy Index for the prediction of epithelial ovarian cancer in patients with a pelvic mass. Am. J. Obstet. Gynecol. 2010, 203, 228.e1–228.e6. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S. Serum Biomarker Based Algorithms in Diagnosis of Ovarian Cancer: A Review. Indian J. Clin. Biochem. 2018, 33, 382–386. [Google Scholar] [CrossRef]
- Lee, Y.-K.; Park, N.-H. Prognostic value and clinicopathological significance of p53 and PTEN in epithelial ovarian cancers. Gynecol. Oncol. 2009, 112, 475–480. [Google Scholar] [CrossRef]
- Yang, W.-L.; Gentry-Maharaj, A.; Simmons, A.R.; Ryan, A.; Fourkala, E.O.; Lu, Z.; Baggerly, K.A.; Zhao, Y.; Lu, K.H.; Bowtell, D.D.; et al. Elevation of TP53 Autoantibody Before CA125 in Preclinical Invasive Epithelial Ovarian Cancer. Clin. Cancer Res. 2017, 23, 5912–5922. [Google Scholar] [CrossRef]
- Anderson, K.S.; Cramer, D.W.; Sibani, S.; Wallstrom, G.; Wong, J.; Park, J.; Qiu, J.; Vitonis, A.; LaBaer, J. Autoantibody Signature for the Serologic Detection of Ovarian Cancer. J. Proteome Res. 2015, 14, 578–586. [Google Scholar] [CrossRef]
- Hurley, L.C.; Levin, N.K.; Chatterjee, M.; Coles, J.; Muszkat, S.; Howarth, Z.; Dyson, G.; Tainsky, M.A. Evaluation of paraneoplastic antigens reveals TRIM21 autoantibodies as biomarker for early detection of ovarian cancer in combination with autoantibodies to NY-ESO-1 and TP53. Cancer Biomark. 2020, 27, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Yang, W.-L.; Pak, D.; Celestino, J.; Lu, K.H.; Ning, J.; Lokshin, A.E.; Cheng, Z.; Lu, Z.; Bast, R.C., Jr. Osteopontin, Macrophage Migration Inhibitory Factor and Anti-Interleukin-8 Autoantibodies Complement CA125 for Detection of Early Stage Ovarian Cancer. Cancers 2019, 11, 596. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Lu, Z.; Guo, J.; Fellman, B.M.; Ning, J.; Lu, K.H.; Menon, U.; Kobayashi, M.; Hanash, S.M.; Celestino, J.; et al. Human epididymis protein 4 antigen-autoantibody complexes complement cancer antigen 125 for detecting early-stage ovarian cancer. Cancer 2020, 126, 725–736. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, X.; Qiu, C.P.; Qin, J.J.; Wang, K.Y.; Sun, G.Y.; Jiang, D.; Li, J.T.; Wang, L.; Shi, J.X.; et al. Using protein microarray to identify and evaluate autoantibodies to tumor-associated antigens in ovarian cancer. Cancer Sci. 2021, 112, 537–549. [Google Scholar] [CrossRef]
- Van Berckelaer, C.; Brouwers, A.J.; Peeters, D.J.; Tjalma, W.; Trinh, X.B.; van Dam, P.A. Current and future role of circulating tumor cells in patients with epithelial ovarian cancer. Eur. J. Surg. Oncol. 2016, 42, 1772–1779. [Google Scholar] [CrossRef]
- Deng, G.; Herrler, M.; Burgess, D.; Manna, E.; Krag, D.; Burke, J.F. Enrichment with anti-cytokeratin alone or combined with anti-EpCAM antibodies significantly increases the sensitivity for circulating tumor cell detection in metastatic breast cancer patients. Breast Cancer Res. 2008, 10, R69. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.F.; Kindelberger, D.; Doyle, C.; Lowe, A.; Barry, W.T.; Matulonis, U.A. Predictive value of circulating tumor cells (CTCs) in newly-diagnosed and recurrent ovarian cancer patients. Gynecol. Oncol. 2013, 131, 352–356. [Google Scholar] [CrossRef]
- Cheng, X.; Zhang, L.; Chen, Y.; Qing, C. Circulating cell-free DNA and circulating tumor cells, the “liquid biopsies” in ovarian cancer. J. Ovarian Res. 2017, 10, 1–10. [Google Scholar] [CrossRef]
- Guo, Y.-X.; Neoh, K.H.; Chang, X.-H.; Sun, Y.; Cheng, H.-Y.; Ye, X.; Ma, R.-Q.; Han, R.P.S.; Cui, H. Diagnostic value of HE4+ circulating tumor cells in patients with suspicious ovarian cancer. Oncotarget 2018, 9, 7522–7533. [Google Scholar] [CrossRef] [PubMed]
- Asante, D.-B.; Calapre, L.; Ziman, M.; Meniawy, T.M.; Gray, E.S. Liquid biopsy in ovarian cancer using circulating tumor DNA and cells: Ready for prime time? Cancer Lett. 2020, 468, 59–71. [Google Scholar] [CrossRef]
- Giannopoulou, L.; Kasimir-Bauer, S.; Lianidou, E.S. Liquid biopsy in ovarian cancer: Recent advances on circulating tumor cells and circulating tumor DNA. Clin. Chem. Lab. Med. 2018, 56, 186–197. [Google Scholar] [CrossRef]
- Thusgaard, C.F.; Korsholm, M.; Koldby, K.M.; Kruse, T.A.; Thomassen, M.; Jochumsen, K.M. Epithelial ovarian cancer and the use of circulating tumor DNA: A systematic review. Gynecol. Oncol. 2021, 161, 884–895. [Google Scholar] [CrossRef] [PubMed]
- Widschwendter, M.; Zikan, M.; Wahl, B.; Lempiäinen, H.; Paprotka, T.; Evans, I.; Jones, A.; Ghazali, S.; Reisel, D.; Eichner, J.; et al. The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer. Genome Med. 2017, 9, 116. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.M.; Miller, H.; Matsuo, K.; Roman, L.D.; Salhia, B. Circulating Cell-Free DNA Methylation Profiles in the Early Detection of Ovarian Cancer: A Scoping Review of the Literature. Cancers 2021, 13, 838. [Google Scholar] [CrossRef]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef]
- Zhao, Z.; Yang, Y.; Zeng, Y.; He, M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip 2016, 16, 489–496. [Google Scholar] [CrossRef]
- Liu, S.-L.; Sun, P.; Li, Y.; Liu, S.-S.; Lu, Y. Exosomes as critical mediators of cell-to-cell communication in cancer pathogenesis and their potential clinical application. Transl. Cancer Res. 2019, 8, 298–311. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef]
- Yoshimura, A.; Sawada, K.; Nakamura, K.; Kinose, Y.; Nakatsuka, E.; Kobayashi, M.; Miyamoto, M.; Ishida, K.; Matsumoto, Y.; Kodama, M.; et al. Exosomal miR-99a-5p is elevated in sera of ovarian cancer patients and promotes cancer cell invasion by increasing fibronectin and vitronectin expression in neighboring peritoneal mesothelial cells. BMC Cancer 2018, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Szajnik, M.; Derbis, M.; Lach, M.; Patalas, P.; Michalak, M.; Drzewiecka, H.; Szpurek, D.; Nowakowski, A.; Spaczynski, M.; Baranowski, W.; et al. Exosomes in Plasma of Patients with Ovarian Carcinoma: Potential Biomarkers of Tumor Progression and Response to Therapy. Gynecol. Obstet. 2013, 4, 3. [Google Scholar] [CrossRef]
- Li, J.; Sherman-Baust, C.A.; Tsai-Turton, M.; Bristow, R.E.; Roden, R.B.; Morin, P.J. Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer 2009, 9, 244. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; König, A.-K.; Marmé, F.; Runz, S.; Wolterink, S.; Koensgen, D.; Mustea, A.; Sehouli, J.; Altevogt, P. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett. 2009, 278, 73–81. [Google Scholar] [CrossRef]
- Zhou, J.; Gong, G.; Tan, H.; Dai, F.; Zhu, X.; Chen, Y.; Wang, J.; Liu, Y.; Chen, P.; Wu, X.; et al. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol. Rep. 2015, 33, 2915–2923. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhou, X.; Zeng, Y. Multiplexed immunophenotyping of circulating exosomes on nano-engineered ExoProfile chip towards early diagnosis of cancer. Chem. Sci. 2019, 10, 5495–5504. [Google Scholar] [CrossRef]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef]
- Jaffee, E.M.; Van Dang, C.; Agus, D.B.; Alexander, B.M.; Anderson, K.C.; Ashworth, A.; Barker, A.D.; Bastani, R.; Bhatia, S.; Bluestone, J.A.; et al. Future cancer research priorities in the USA: A Lancet Oncology Commission. Lancet Oncol. 2017, 18, e653–e706. [Google Scholar] [CrossRef]
- ACOG Committee Opinion No. 734: The Role of Transvaginal Ultrasonography in Evaluating the Endometrium of Women with Postmenopausal Bleeding. Obstet. Gynecol. 2018, 131, e124–e129. [CrossRef] [PubMed]
- Doubeni, C.A.; Doubeni, A.R.; Myers, A.E. Diagnosis and Management of Ovarian Cancer. Am. Fam. Physician 2016, 93, 937–944. [Google Scholar]
- Bernard, L.; Kwon, J.S.; Simpson, A.N.; Ferguson, S.E.; Sinasac, S.; Pina, A.; Reade, C.J. The levonorgestrel intrauterine system for prevention of endometrial cancer in women with obesity: A cost-effectiveness study. Gynecol. Oncol. 2021, 161, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Yasin, H.K.; Taylor, A.H.; Ayakannu, T. A Narrative Review of the Role of Diet and Lifestyle Factors in the Development and Prevention of Endometrial Cancer. Cancers 2021, 13, 2149. [Google Scholar] [CrossRef]
- Dilley, S.E.; Havrilesky, L.J.; Bakkum-Gamez, J.; Cohn, D.E.; Straughn, J.M., Jr.; Caughey, A.B.; Rodriguez, M.I. Cost-effectiveness of opportunistic salpingectomy for ovarian cancer prevention. Gynecol. Oncol. 2017, 146, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.L.; Powell, C.B.; Chen, L.-M.; Carter, J.; Jump, V.L.B.; Parker, L.P.; Borowsky, M.E.; Gibb, R.K. Society of Gynecologic Oncology recommendations for the prevention of ovarian cancer. Cancer 2015, 121, 2108–2120. [Google Scholar] [CrossRef] [PubMed]
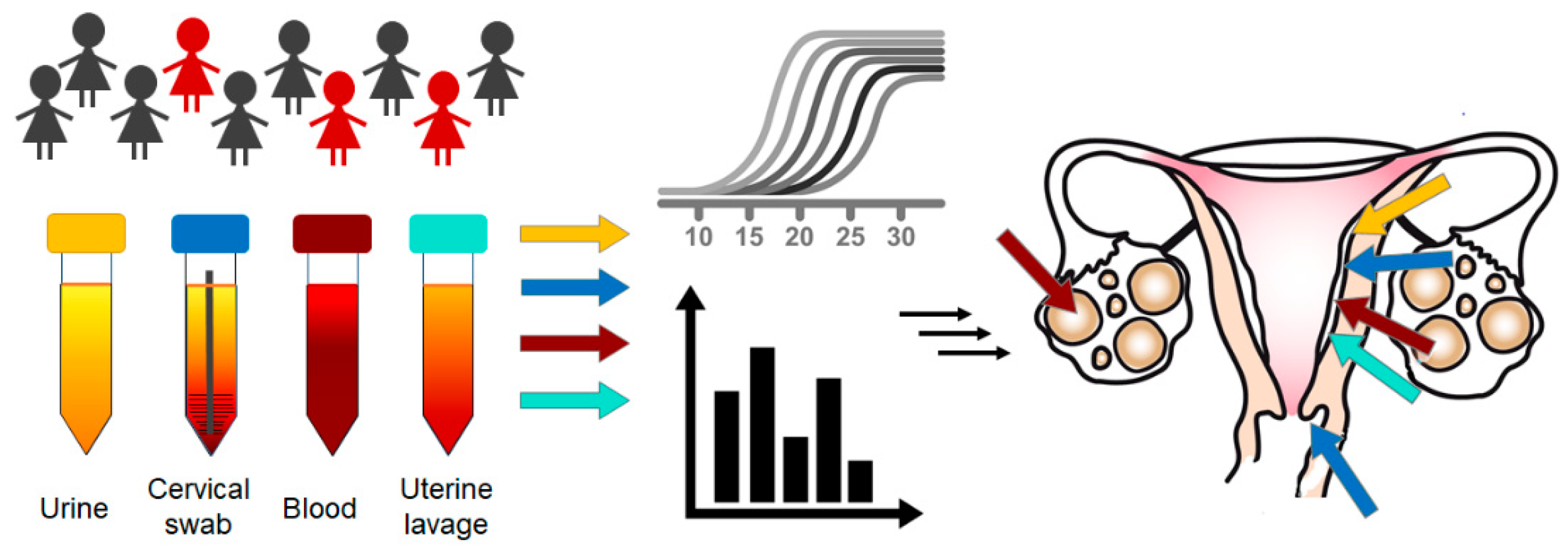
| Source | Biomarker | (Pre)Cancer | Application and References |
|---|---|---|---|
| Pap smear/Pap test/PapSEEK test | DNA | CC EC OEC | Pap test as a screening method for early-stage cervical, ovarian, and endometrial cancers [7,8] |
| Uterine lavage | DNA | EC OEC | Uterine lavage fluid is used to detect early endometrial carcinomas by genomic analysis [9]; DNA sequencing from uterine lavage samples with a focus on a panel of candidate ovarian cancer driver genes, including TP53 [10] and MCM5 [11]. |
| Urine | Protein biomarkers Cytology miRNA HPV DNA | OEC EC CC | MicroRNA (miR-223, let7-i, miR-34a, and miR-200c) expression levels in urine as a non-invasive diagnostic test for endometrial cancer [12]; summary of studies testing the level of HE4 in urine and body fluids for the diagnosis of gynecological cancer [13]; cytology of urine and vagina fluids as a sensitive and specific tool for the detection of gynecological tumors [14] |
| Cervicovaginal secretions | Metabolomic biomarkers Protein biomarkers Cytology | OEC EC CC | Phosphocholine, asparagine, and malate from cervicovaginal fluid have been identified by nuclear magnetic resonance spectroscopy as promising metabolomics biomarkers for EC detection [15]; alpha-actinin-4 as a promising biomarker for the detection of precancerous state of cervical cancer [16]; CA-125 in cervicovaginal secretion as a potential biomarker for EC detection [17]; cytology of urine and vagina fluids as a sensitive and specific tool for the detection of gynecological tumors [14] |
| Endocervical swabs | Protein biomarkers Enzymatic activity HPV DNA | EC CC | Proprotein convertase activity [11,18]; HPV DNA testing from cervical swabs as an alternative mechanism to routine cytological screening [19] |
| Tampons or vaginal swabs | Protein Biomarkers HPV DNA | CC | Comparison of vaginal swabs and urine samples with cervical smears for HPV testing in cervical cancer screening strategies [20,21] |
| Test | Vendor | Application |
|---|---|---|
| Cervical cancer | ||
| ThinPrep® Pap Test | Hologic (Marlborough, MA, USA) | Cervical smear taken into a liquid medium followed by computer evaluation of the specimen |
| SurePath Pap Test | Becton Dickinson (Franklin Lakes, NJ, USA) | A liquid-based Pap test used in the screening and detection of cervical cancer, pre-cancerous lesions, atypical cells, and all other cytological categories |
| Roche Cobas® HPV | Roche (Basel, Switzerland) | A qualitative in vitro test for the detection of HPV in patient specimens by amplification of target DNA and its hybridization for the detection of 14 high-risk HPV types |
| Cervista HPV16/18 assay | Hologic (Marlborough, MA, USA) | A qualitative, in vitro diagnostic test for the detection of DNA from two high-risk HPV types: 16 and 18 |
| Hybrid Capture 2 | Qiagen (Hilden, Germany) | The platform for the nucleic acid hybridization assay for the detection of HPV, Chlamydia trachomatis, and Neisseria gonorrhoeae |
| Linear Array HPV | Roche (Basel, Switzerland) | Test for genotyping HPV in cervical biopsies and other formalin-fixed, paraffin-embedded specimens |
| INNO-LiPA® HPV Genotyping Extra II | Fujirebio (Tokyo, Japan) | Line probe assay, based on the reverse hybridization principle, designed for the identification of 32 different genotypes of HPV |
| Endometrial cancer | ||
| None available | ||
| Ovarian cancer | ||
| OVA1® and OVERA® tests | Aspira Women’s Health Inc(Austin, TX, USA) | In vitro diagnostic multivariate index assay that analyzes the serum levels of proteomic biomarkers |
| Elecsys HE4 assay | Roche (Basel, Switzerland) | Sandwich electrochemiluminescent immunoassay, which measures the amount of HE4 in a patient sample against a calibration curve |
| Elecsys® CA 125 II | Roche (Basel, Switzerland) | Biomarker test to determine the amount of CA 125 protein in a blood sample |
| Bard1 Life Sciences test | BARD1 Life Sciences (Notting Hill, Australia) | Autoantibody test for early detection of ovarian, breast, and lung cancers |
| (Pre)Cancer | Prevention 1 | Screening | Diagnostics | Treatment | References |
|---|---|---|---|---|---|
| Cervical | Vaccination | Pap test HPV triage | Colposcopy + histology | Conization | [53] |
| Endometrial | None 2 | None | Transvaginal sonography Endometrial biopsy Hysteroscopy | Hysterectomy | [118] |
| Ovarian | None 3 | None | Histology in advanced stages (>75% of cases) Transvaginal sonography ROMA (CA125 + HE4) | Radical surgery Systemic therapy Targeted therapy | [134] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holcakova, J.; Bartosik, M.; Anton, M.; Minar, L.; Hausnerova, J.; Bednarikova, M.; Weinberger, V.; Hrstka, R. New Trends in the Detection of Gynecological Precancerous Lesions and Early-Stage Cancers. Cancers 2021, 13, 6339. https://doi.org/10.3390/cancers13246339
Holcakova J, Bartosik M, Anton M, Minar L, Hausnerova J, Bednarikova M, Weinberger V, Hrstka R. New Trends in the Detection of Gynecological Precancerous Lesions and Early-Stage Cancers. Cancers. 2021; 13(24):6339. https://doi.org/10.3390/cancers13246339
Chicago/Turabian StyleHolcakova, Jitka, Martin Bartosik, Milan Anton, Lubos Minar, Jitka Hausnerova, Marketa Bednarikova, Vit Weinberger, and Roman Hrstka. 2021. "New Trends in the Detection of Gynecological Precancerous Lesions and Early-Stage Cancers" Cancers 13, no. 24: 6339. https://doi.org/10.3390/cancers13246339
APA StyleHolcakova, J., Bartosik, M., Anton, M., Minar, L., Hausnerova, J., Bednarikova, M., Weinberger, V., & Hrstka, R. (2021). New Trends in the Detection of Gynecological Precancerous Lesions and Early-Stage Cancers. Cancers, 13(24), 6339. https://doi.org/10.3390/cancers13246339






