1. Introduction
Ovarian cancer is a deadly gynecologic malignancy responsible for over 13,000 deaths annually in the US [
1]. Among ovarian cancer diagnoses, the most common subtype is high-grade serous ovarian carcinomas (HGSOC) [
2]. Standard therapy for HGSOC includes surgical resection of the tumor followed by platinum-based chemotherapy with carboplatin [
3]. Patients with advanced-stage disease not amenable to frontline cytoreduction are often first treated with chemotherapy (neoadjuvant chemotherapy) to reduce overall tumor volume, followed by an interval debulking surgery [
4]. While many patients achieve remission with initial treatment, approximately 80% of patients with advanced-stage disease experience tumor relapse [
3] which is associated with the development of platinum-resistant disease. Ultimately, it is platinum resistance that claims the lives of women diagnosed with HGSOC. Effective targeting of these tumor cells and prevention of disease relapse remain a clinical challenge.
While alterations in p53 are found in about half of all human tumors [
5], mutations in this protein are especially prevalent in HGSOCs, observed in >90% of cases [
6]. According to a recent study mapping the evolutionary history of many cancers, mutations in p53 may be a very early event in HGSOC development and can be detected many years before diagnosis [
7]. Further, driver mutations in p53 may be an initiating event in HGSOC [
8,
9,
10], and high levels of p53 immunostaining serve as a biomarker of early-stage disease in serous tubal intraepithelial carcinoma lesions. p53 plays an important role in maintaining the stability of the cell’s genetic information, guarding against genomic chaos [
11]. Wildtype function of p53 is crucial as a tumor suppressor by acting as a cellular stress sensor, inducing cell-cycle arrest, and promoting DNA repair upon cellular injury or genotoxic damage. If a cell accumulates DNA damage too severe for repair, p53 activates an apoptotic program, which leads to the elimination of the damaged cells [
5,
12,
13]. This function prevents genomically unstable cells from replicating and becoming cancerous. Because mutations in p53 are associated with chemotherapy resistance and potentially worse clinical outcomes [
12,
13], therapeutic targeting of this protein is a major goal of many researchers globally.
The clinical challenge of disease recurrence despite the administration of platinum-based chemotherapy has necessitated the exploration of alternative options for the treatment of ovarian cancers. Primarily, these alternatives include the addition of targeted agents to standard chemotherapy. Because p53 is a regulator of cellular apoptosis, restoration of p53 function may enhance platinum-mediated apoptosis. Such approaches are currently being explored clinically. For example, APR-246 has been tested in clinical trials in combination with cytotoxic drugs, such as carboplatin and doxorubicin (NCT:02098343), in treating patients with recurrent HGSOC. Further, the development of agents that target p53 is a growing field in cancer therapeutics. One such peptide, called ReACp53, has been shown to target multiple cancer models in vitro and in vivo [
14,
15]. While the exact mechanism of ReACp53 induced cytotoxicity remains unknown [
16,
17], it was designed to inhibit mutant p53 aggregation restoring its wildtype function (reviewed in [
18]). The aim of the present study is to test the efficacy of ReACp53 in combination with carboplatin chemotherapy in targeting human ovarian cancers.
2. Materials and Methods
2.1. Cell Lines and Primary Patient Samples
Human ovarian cancer cell lines were obtained from ATCC or the National Institutes of Health (NIH), and frequently STR verified during experiments. All cell lines were maintained in recommended media (RPMI/10% FBS or DMEM/10% FBS) at 5% CO2 and 37 °C. This study was approved by the UCLA Office of the Human Research Protection Program and (IRB# 20-001762, IRB# 10-000727) and the VA Greater Los Angeles Institutional Review Board (IRB# 2019-020090). Clinical information from consenting patients was obtained from medical records. This heterogeneous population of patients consisted of tumors from patients who were chemo naïve, neoadjuvant chemotherapy-treated (both platinum-sensitive and platinum-resistant), and recurrent platinum-resistant. Platinum resistance was defined as tumor relapse <6 months from the last infusion of platinum-based chemotherapy. Platinum sensitivity was defined as tumor relapse >6 months after the final administration of platinum drugs. Solid tumor samples from consenting patients were obtained fresh from the operating room, brought back to the lab, and dissociated mechanically and enzymatically (collagenase 1 mg/mL and dispase 1 mg/mL, Gibco). Effusion samples were harvested by centrifugation and filtered through a 100 μM filter. All patient tumor samples were cryopreserved in FBS/10% DMSO in multiple aliquots to facilitate experimental repeats.
2.2. Drug Preparation
ReACp53 peptide (amino acid sequence: RRRRRRRRRRPILTRITLE) used for the experiments outlined was either purchased from the Chinese Peptide Company or generously provided by Dr. Alice Soragni (UCLA). In either case, lyophilized ReACp53 was reconstituted in PBS (pH 8.5) at 5 mM and then sterile filtered for final use. 5-Carboxyfluorescein tagged peptide (5-FAM-ReACp53) was used to confirm cellular penetrance (
Supplementary Figure S1A,B). The specificity of the peptide was tested in vitro using three cell lines (OVCAR3, SKOV3, and MCF7) using a previously reported high throughput in vitro organoid drug assay (
Supplementary Figure S1C) [
19,
20]. Carboplatin (Tocris) was reconstituted in ddH2O at a concentration of 10 mM and sterile filtered for final use.
2.3. High Throughput In Vitro 3D Organoid Drug Assay
The efficacy of the ReACp53 and carboplatin combination was tested using a previously reported high throughput in vitro organoid drug assay [
19,
20] using human ovarian cancer cell lines and primary patient HGSOC tumor cells. In this assay, 5000 cells per well were suspended in a mixture of Matrigel matrix (Corning, Corning, NY, USA) and MammoCult medium (STEMCELL Technologies, Vancouver, BC, Canada) and plated around the rim of the wells of a 96-well plate. Following organoid establishment for two days, cells were treated with drugs for three days, replenished daily. Dose combinations were administered in triplicate wells for each plate (5 independent plates per cell line and 2 independent plates for each primary patient tumor sample). Cell viability was assessed using an ATP luminescence assay (CellTiter-Glo 3D Viability Assay, Promega, Madison, WI, USA) [
19,
20]. Viability percentages were calculated by normalizing each luminescence value to control wells (0 µM carboplatin, 0 µM ReACp53) using GraphPad Prism 8. Each cell line or primary tumor sample was plated by two independent investigators.
2.4. Measurement of Apoptosis Markers in Response to ReACp53 and Carboplatin Combination
To assess apoptosis in ovarian cancer cells treated with ReACp53 and carboplatin, 40,000 OVCAR3 cells per well were plated in 24-well tissue cultures dishes embedded in Matrigel (Corning) to grow 3D organoids. Carboplatin (50 μM), ReACp53 (4 μM), or the combination of the two agents was administered daily for 72 h. Treated organoids from some wells were harvested from Matrigel using 5 mg/mL dispase (Gibco, Waltham, MA, USA) for 30 min at 37 °C. The level of extracellular annexin V was assessed using a FITC Annexin V Apoptosis Detection Kit (BD Pharmagen, Franklin Lakes, NJ, USA). Staurosporine or DMSO-treated organoids were used as gating controls. Stained cells were analyzed using a BD LSRII flow cytometer and FACSDiva software (BD Biosciences, Franklin Lakes, NJ, USA).
Other wells of similarly treated OVCAR3 organoids were harvested, lysed using RIPA buffer supplemented with protease inhibitor cocktail (ThermoFisher Scientific, Waltham, MA, USA), and used for Western blot analysis to measure the level of apoptosis biomarkers, cleaved PARP, and cleaved caspase 3. Total protein concentration of the lysate was measured using a BCA protein assay kit (ThermoFisher Scientific). Thirty micrograms of lysate per lane were loaded and run on 4–12% Bis-Tris polyacrylamide gels (Invitrogen, Waltham, MA, USA) and transferred to nitrocellulose membranes (Millipore, Burlington, MA, USA). Nonspecific antibody binding on membranes was blocked using phosphate-buffered saline containing 0.1% Tween and 5% non-fat dried milk (PBST+5% NFDM) and incubated for 1 h at room temperature. Blocked membranes were incubated with primary antibody diluted 1:1000 in PBST+5% NFDM overnight at 4 °C. Membranes were then washed in PBST and incubated in secondary antibody diluted 1:1000 in PBST+5% NFDM for 1 h at room temperature. Membranes were washed a final time using PBST and treated with Immobilon chemiluminescence substrate (Millipore, WBKLS0500). Protein was detected using a Biorad ChemiDoc Imaging System. Primary antibodies were the following: anti-PARP (Cell Signaling Technology, 9532, Topsfield, MA, USA); anti-Caspase 3 (Cell Signaling Technology, 9662), and anti-GAPDH (Cell Signaling Technology, 5174). The secondary antibody was HRP-linked anti-rabbit IgG (Cell Signaling Technology, 7074). The relative quantification of protein bands was done by densitometric analysis using ImageJ software.
2.5. Animals
All animal experiments were approved by the UCLA Animal Research Committee (protocol 2008-153), the West Los Angeles Veterans Administration Medical Center IACUC (protocol 2019-020090) and performed under the oversight of the UCLA Division of Laboratory Animal Medicine. Female NSG mice (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ, Jackson Laboratory) ages 6–8 weeks were used for all in vivo experiments. All mice were housed in specific pathogen-free (SPF) facilities, in autoclaved cages with sterile bedding and food.
2.6. Establishment of Intraperitoneal Xenografts
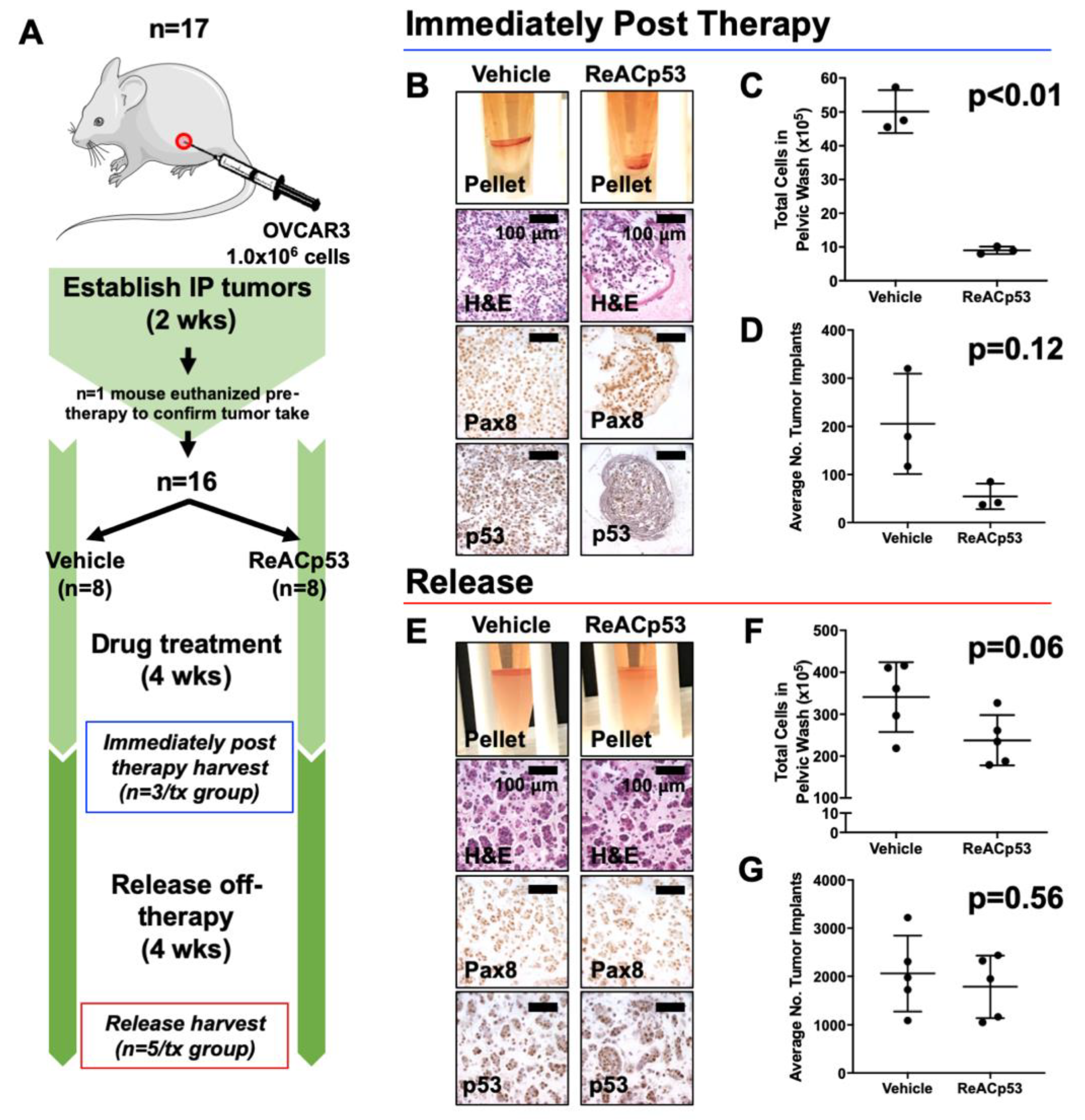
Intraperitoneal xenografts were established in female NSG mice using OVCAR3, SKOV3, and OVCAR8 ovarian cancer cell lines. Xenografts were established by injecting a 50/50 Matrigel/tumor cell suspension (100 μL total) into the IP space of mice using a 20-gauge needle. Two weeks after tumor establishment, mice were allocated into experimental groups by simple randomization, with one mouse randomly selected from each experimental cohort and euthanized to confirm tumor take.
2.7. Quantification of Tumor Burden In Vivo
Intraperitoneal disease burden based on peritoneal lavage: Pelvic washes were performed using RPMI medium (Gibco) on mice following euthanasia. Cells obtained from pelvic washings were harvested and enzymatically digested with collagenase and dispase (1 mg/mL each, Gibco) for 45–60 min at 37 °C. Digested cell pellets were resuspended in RPMI and quantified either by manual counting on a hemocytometer by two independent investigators or using an automated cell counter (Countess II, ThermoFisher Scientific). Values reported reflect the total number of cells recovered from peritoneal washings.
Intraperitoneal disease burden based on tumor implant counts: The number of tumor implants was quantified by two independent investigators using histologic sections of harvested mouse organs immunohistochemically stained for either p53 or Pax8, at five levels throughout the depth of the tissue block (
Supplementary Figure S2A). The number of tumor implants per level was summed to give the total number of tumor implants per mouse and then averaged across treatment groups. A tumor implant was defined as a continuous region of tumor cells that exhibited positive staining for p53 (for OVCAR3 tumors with aggregating p53 mutations) or Pax8 (for SKOV3 tumors known to be p53-null). Non-continuous regions of positive staining that were near one another were counted as separate tumor implants (
Supplementary Figure S2B).
2.8. Flow Cytometry to Determine Percentage of Tumor Cells from Peritoneal Lavage
Immunostaining of mouse pelvic wash cells for Trop1 expression was performed by incubating cells in anti-CD326 PE-Cyanine7 conjugated antibody (Invitrogen) at a concentration of 5 μL per million cells for 30 min. Stained cells were analyzed using a BD FACSCelesta and FACSDiva software (BD Biosciences, Franklin Lakes, NJ, USA).
2.9. Immunohistochemistry to Detect Tumor Cells
The primary antibodies used to detect tumor cells were anti-p53 (sc-126, clone DO-1, 1:250; Santa Cruz Biotechnology, Dallas, TX, USA) and anti-Pax8 (MRQ-50, 1:500; Cell Marque). The secondary antibodies used were biotinylated rabbit anti-mouse (Jackson Immunoresearch, 1:1000), and the tertiary antibody used was streptavidin-conjugated horseradish peroxidase (Jackson Immunoresearch, 1:1000). Detection was performed using 3,3′-diaminobenzidine (DAB) chromagen (HK130-5K, Biogenex).
2.10. Determination of p53 Mutation Status in Patient Samples and Cell Lines
p53 mutation status for each patient sample was determined by clinical sequencing of the primary tumor (FoundationOne, Cambridge, MA, USA) or whole-exome sequencing. For whole-exome sequencing: the Kapa Hyper library kit was used to make the genomic DNA library. The workflow consisted of fragmentation of gDNA, end repair to generate blunt ends, A-tailing, adaptor ligation, and PCR amplification. Different indices were used for multiplexing samples in one lane. Whole-exome DNA was captured from total genomic DNA using the SeqCap EZ System from NimbleGen according to the manufacturer’s instructions. Briefly, the gDNA library was incubated with SeqCap biotinylated DNA baits, and the hybrids were purified using streptavidin-coated magnetic beads, then followed by PCR. Sequencing was performed on an Illumina HiSeq3000 for a pair-end 150 bp run.
Raw reads were mapped to the GRch38 human genome reference assembly (GCA_000001405.15) using BWA-MEM. Variants were called using GATK v4.0.10.0 [
21] according to GATK’s best practices for somatic mutation calling using the Mutect2 tool. Variant calling in cancer cells was aided by using a panel of normal variants (PON) generated from patient-matched PBMCs and the af-only-gnomad.hg38.vcf germline resources and then the called somatic variants were filtered according to GATK recommendations. TP53 mutation status for each cell line was obtained from the International Agency for Research on Cancer (IARC) p53 database (Version R20) [
22].
2.11. Synergy Analysis
Synergy scores using four separate reference models (the Highest Single Agent (HSA) model, the Loewe Additivity model, the Bliss Independence model, and the Zero Interaction Potency (ZIP) model) [
23] were calculated for each cell line and patient sample tested in the high throughput in vitro organoid drug assay using the computational tool SynergyFinder 2.0, which has an R-package as well as a web application [
24]. The synergistic or antagonistic effect of the pairwise combination of doses was visualized as two-dimensional synergy/antagonism heatmaps and then summarized over the full dose–response matrix using a synergy score calculated for each reference model. Experimental results were run in triplicate, and results were averaged for analytical purposes. SynergyFinder 2.0 allows for inputting data from independent replicate experiments in order to calculate a 95% confidence interval for synergy scoring [
24]. Based on these reference models, a summary synergy score value >10 is considered synergistic, between −10 and +10 is considered additive, and a synergy score <−10 is considered antagonistic [
24]. We considered each sample as synergistic, additive, or antagonistic based upon the majority results from the four models available in SynergyFinder (e.g., if 3 or more models agreed, the combination was synergistic).
2.12. Statistics
Results are reported as means ± standard deviation. The p-values for comparing means were computed using one-way analysis of variance, allowing for non-constant variance (variance heterogeneity). The Shapiro–Wilkes test on the residual errors was computed to confirm normality. When data was not normal, p-values were computed with the non-parametric Kruskal–Wallis test. Calculations were carried out using R version 4.0.5 (R Foundation for Statistical Computing, Vienna, Austria). Survival curves were estimated with the Kaplan–Meier method and were compared between groups using the log-rank test. p < 0.05 were considered statistically significant. IC50 values were calculated using the four-parameter logistic model and GraphPad Prism 8.
4. Discussion
Carboplatin is the frontline treatment for patients diagnosed with HGSOC [
3]. While the initial response to this chemotherapy is favorable, the majority of patients experience tumor recurrence associated with the development of platinum-resistant disease. The underlying biologic cause for platinum resistance has been explored for decades and is likely a multifactorial process mediated by drug transport, tolerance of cancer cells to DNA damage, loss of p53 function, and evasion of apoptosis, among others [
29]. Mutations in p53 are also associated with resistance to chemotherapy in many cancers and worse potential outcomes [
12,
13]. In fact, alterations in p53 are frequently found in castration-resistant prostate cancer (CRPC) and may be enriched in advanced, metastatic disease compared to primary prostate cancer [
30].
The efficacy of ReACp53 in targeting tumor cells has been evaluated by other researchers. For example, ReACp53 was effective in targeting prostate cancer cells with aggregating mutations in p53 by increasing mitochondrial cell death and inhibiting DNA synthesis [
14]. This peptide also inhibited xenograft growth of prostate cancer tumors in vivo [
14]. ReACp53 could also prohibit the growth of pancreatic cancer xenografts harboring aggregating mutations in p53 [
15]. Recent work suggests that ReACp53 can also restore sensitivity to cisplatin in a human lung cancer cell line expressing an exogenous p53R282W mutation in vitro [
31].
Our study focused on the analysis of epithelial ovarian cancer samples known to frequently carry mutations in p53 [
6]. Given the prevalence of p53 mutations observed in HGSOCs, these tumors are an ideal model for further testing ReACp53 peptide alone and in combination with standard carboplatin chemotherapy. Synergy analysis was used to determine relative sensitivity to the ReACp53 and carboplatin combination. Notably, for 17/18 (94%) total samples tested, including both cell lines and patient tumor samples, results from at least three out of four synergy models agreed. In the majority of primary HGSOC tumor samples tested, an additive effect for the combination was observed, indicating that tumor targeting may be enhanced when adding ReACp53 to carboplatin. These samples included patients with clinically-determined platinum-sensitive and -resistant disease. Importantly, in 5/6 samples from patients with platinum-resistant disease, the addition of ReACp53 enhanced the efficacy of carboplatin when administered in combination, suggesting this approach may be applicable in targeting therapy-resistant tumors.
In the four platinum-resistant ovarian cancer cell lines tested, the combination of ReACp53 and carboplatin yielded additive effects. In OVCAR4 wells with intermediate sensitivity to platinum drugs, synergy was observed. Within the tested cell lines, the highest level of synergy was seen when ReACp53 and carboplatin were combined in targeting OVCAR3 cells. In these cells, reduced cell viability may be mediated through enhanced apoptosis. Potential synergy in carboplatin and ReACp53 was validated using OVCAR3 cells in an in vivo survival analysis. Given that an additive effect was seen when ReACp53 was added to carboplatin in OVCAR8 cells, the in vivo efficacy of this combination was tested using this ovarian cancer cell line as well. Here, an improvement in survival was not observed when the combination was administered compared to carboplatin treatment alone. A challenge with this in vivo survival study was the aggressive nature of OVCAR8 cells that required euthanizing a subset of the mice prior to, or soon after, administration of ReACp53 and carboplatin. Some mice in this cohort could not complete the four-week course of treatment; hence, OVCAR8 cells may not be an optimal model for survival analysis. Collectively, our data suggest that ReACp53 in combination with carboplatin may provide a potential strategy for targeting human epithelial ovarian cancers. Given the tolerability of ReACp53 and carboplatin observed in vivo, further analyses into the potential of this combinatorial strategy may be warranted.
While ReACp53 is designed to specifically target mutant aggregating p53 protein, there is emerging data that it may have cytotoxic effects through other mechanisms, including targeting p53 interactions with p63, p73, or the wildtype protein [
17,
32]. There is some evidence that in malignant hematopoietic cell lines, ReACp53 can target cell lines both with and without aggregating p53 mutations in a p73-dependent manner [
32]. Additionally, it is also suggested that ReACp53 may impact cell cycle transition in malignant cells [
14]. Work from others suggests that ReACp53 and other peptides designed to target p53 aggregation may have p53-dependent and independent effects [
17]. In this study, we observed that ReACp53 could potentially target cell lines and tumors with p53 mutations not known to result in aggregation (
Supplementary Figure S11), suggesting that other potential mechanisms of ReACp53 action may be present in targeting tumors as reported by other investigators [
17,
32].
ReACp53 is a peptide. Therapeutic peptides have some advantages, including potentially less toxicity, increased specificity, and reduced side effects, compared to other small molecules and cytotoxic drugs [
25]. However, the effective use of peptides for cancer therapy is hindered by many challenges, including a metabolic breakdown and fast clearance in vivo [
25]. Such inherent challenges in the administration of peptide-based therapeutics must be addressed in order to further test the viability of ReACp53 for clinical use. Thus far, we have tested a frequent intraperitoneal route of administration for ReACp53, a strategy that is feasible but not performed frequently in patients. Other well-tolerated p53 targeting peptides administered intraperitoneally, such as ADH-6, have similarly demonstrated efficacy in targeting p53-mutated cancers [
15]. In future work, consideration can be given to other routes of delivery, including intravenous infusion or strategies being tested for nanoparticle delivery of peptides [
33].
Overall, the findings presented here demonstrated that the addition of ReACp53 to carboplatin may enhance targeting in a subset of HGSOC tumors. The ReACp53 and carboplatin combination demonstrated synergistic effects in some ovarian cancer cell lines and additive effects in primary HGSOC tumor samples tested.