miR-125b Promotes Colorectal Cancer Migration and Invasion by Dual-Targeting CFTR and CGN
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
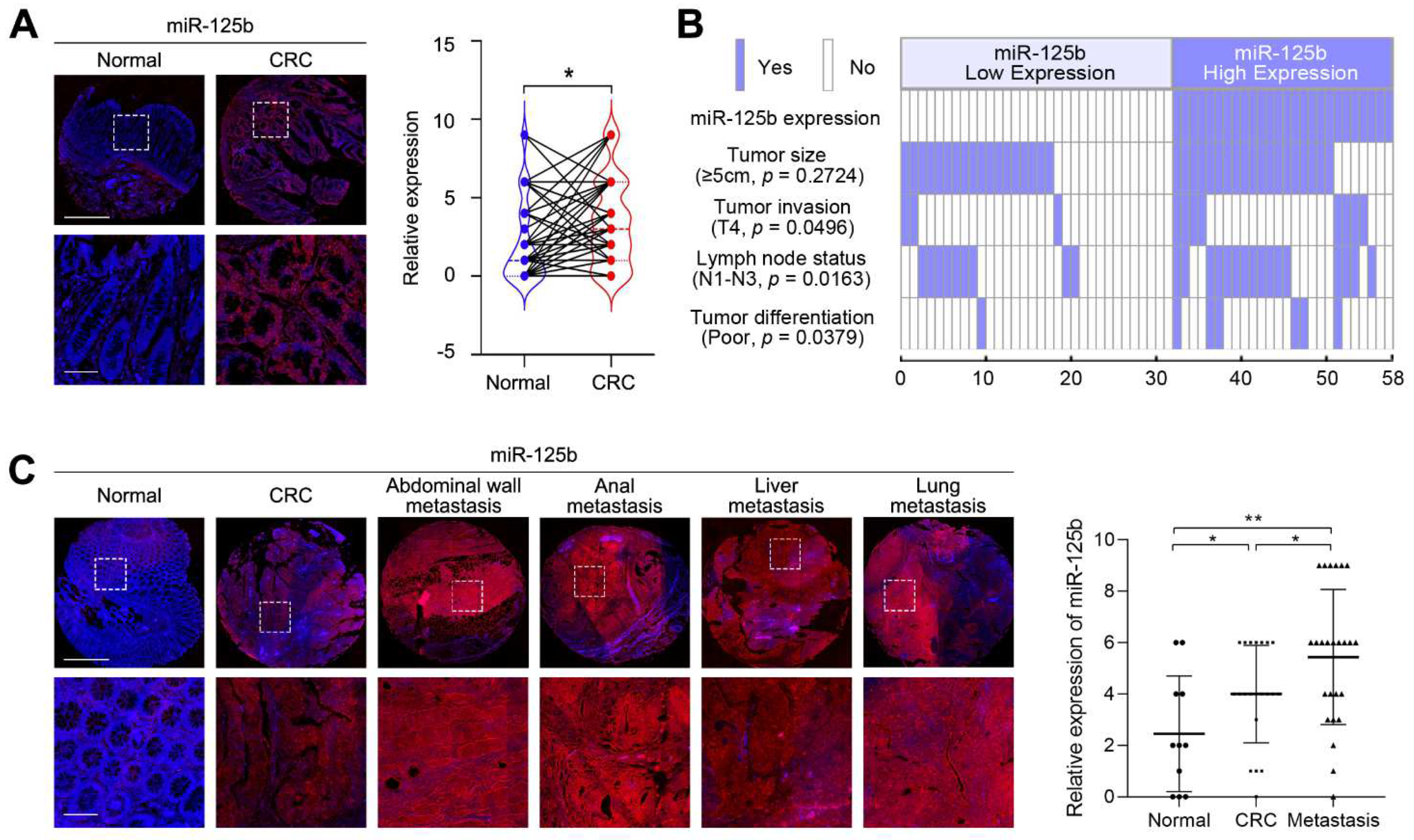
2.1. miR-125b Expression Is Upregulated in Metastatic CRC Tissues
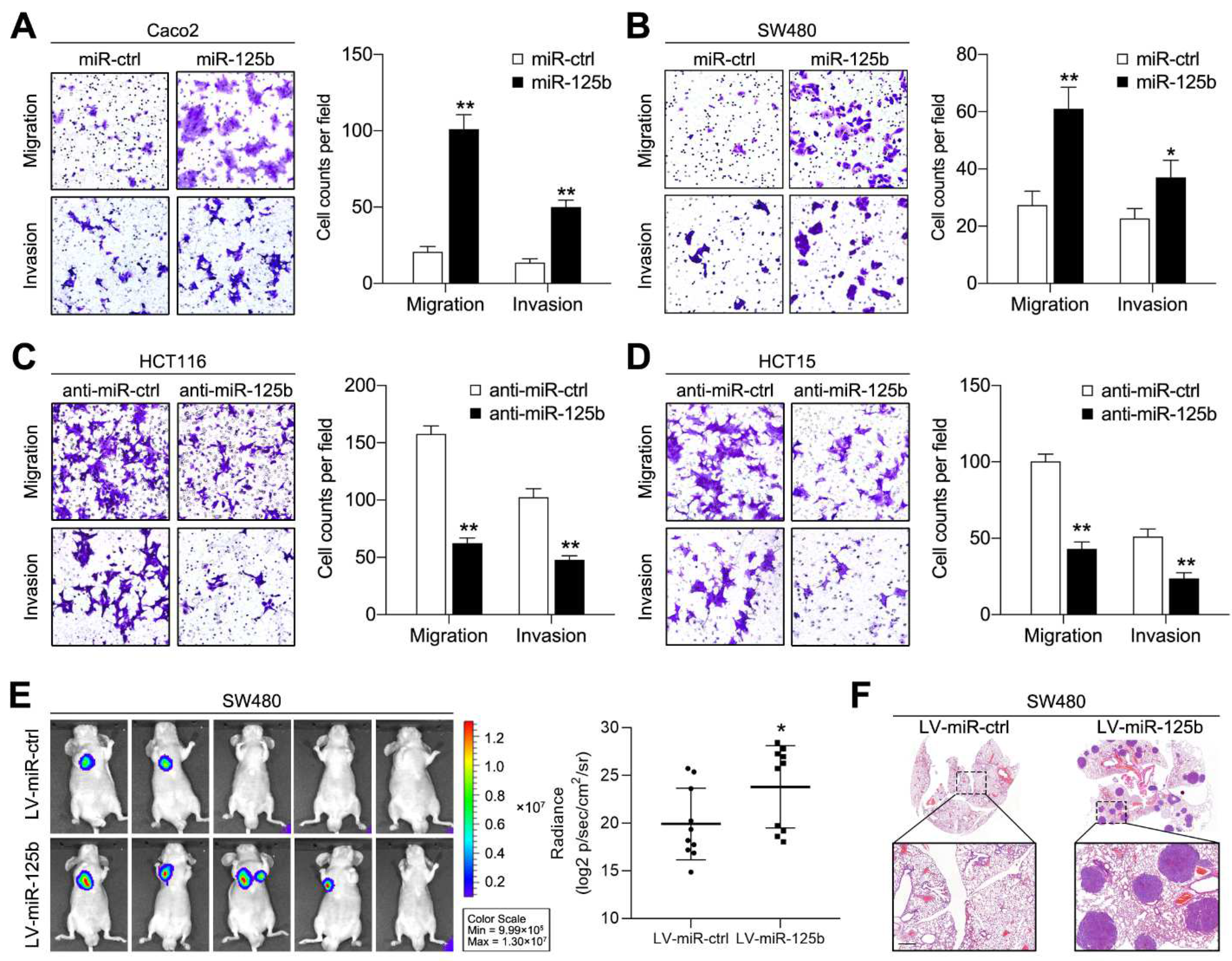
2.2. miR-125b Promotes CRC Cell Migration, Invasion and Metastasis
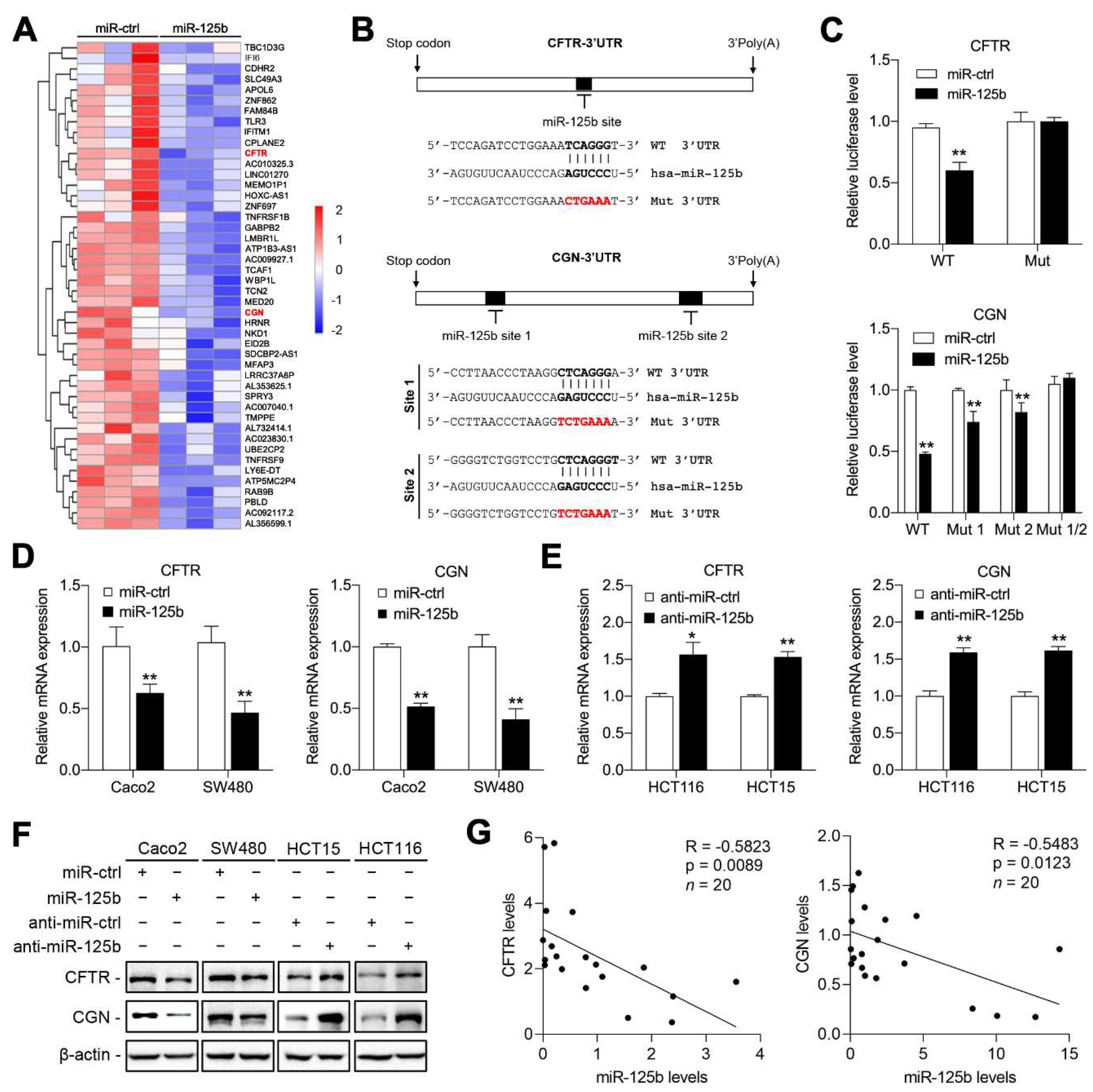
2.3. CFTR and CGN Are Direct Targets of miR-125b in CRC
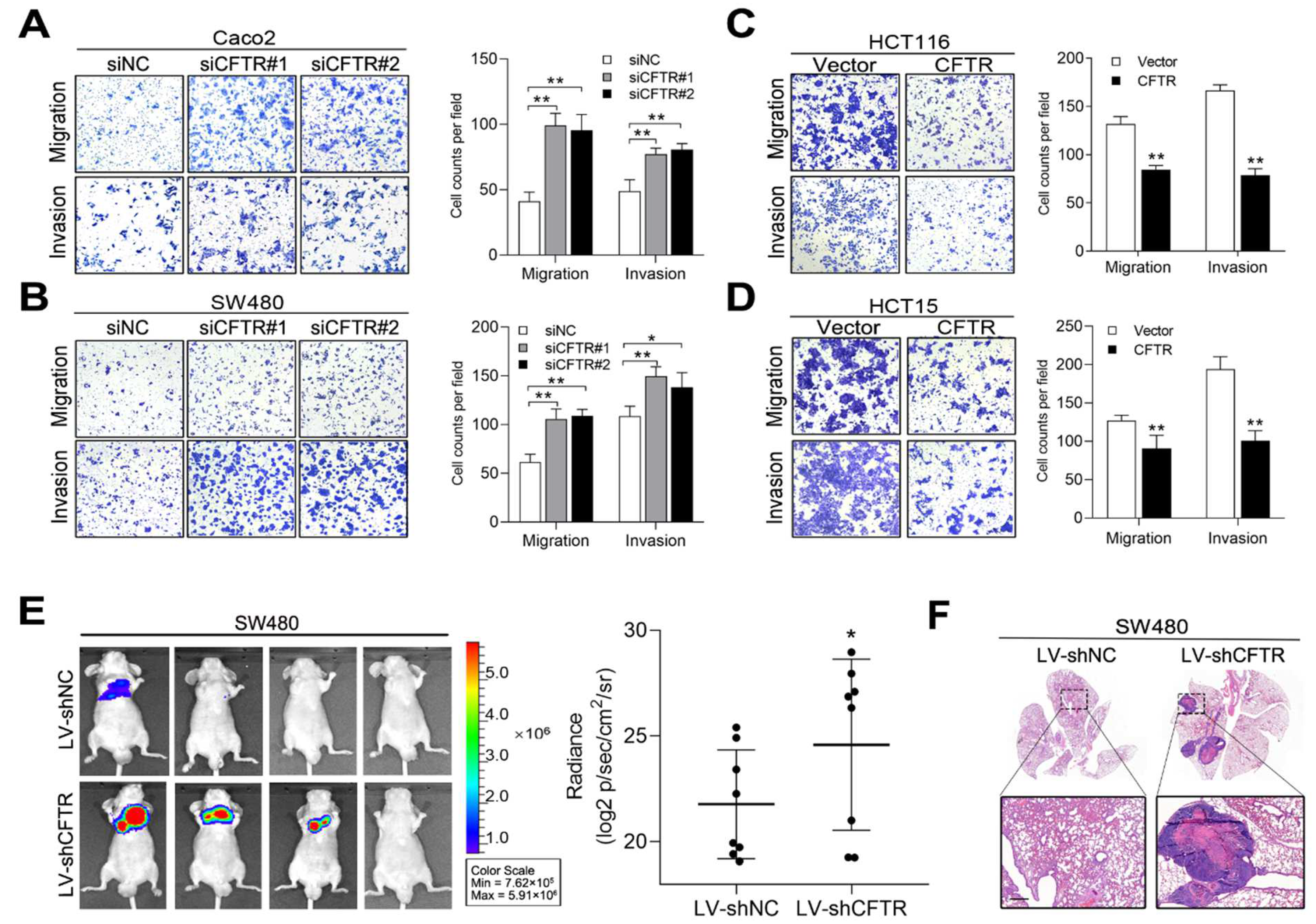
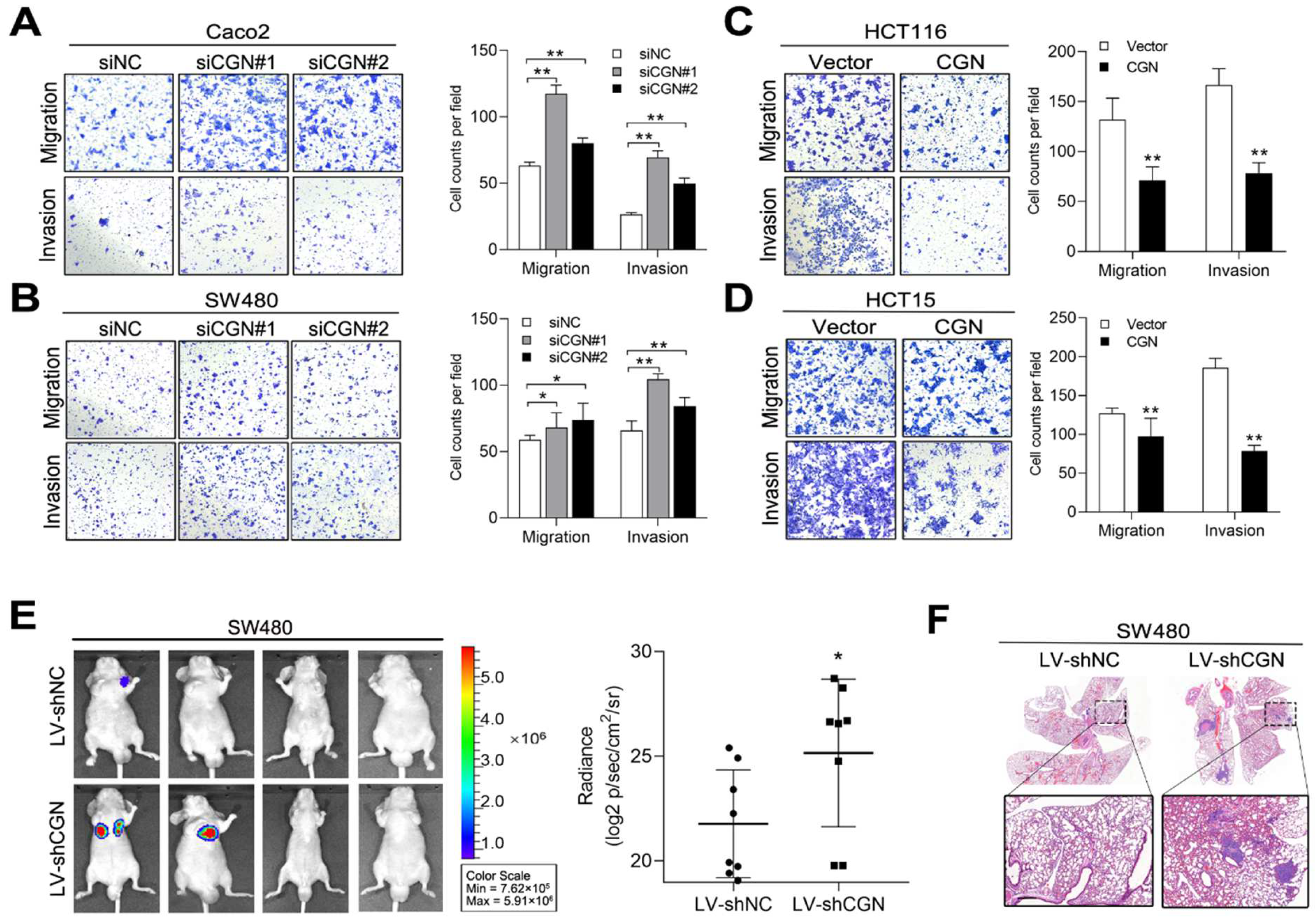
2.4. CFTR and CGN Inhibited CRC Metastasis In Vitro and In Vivo
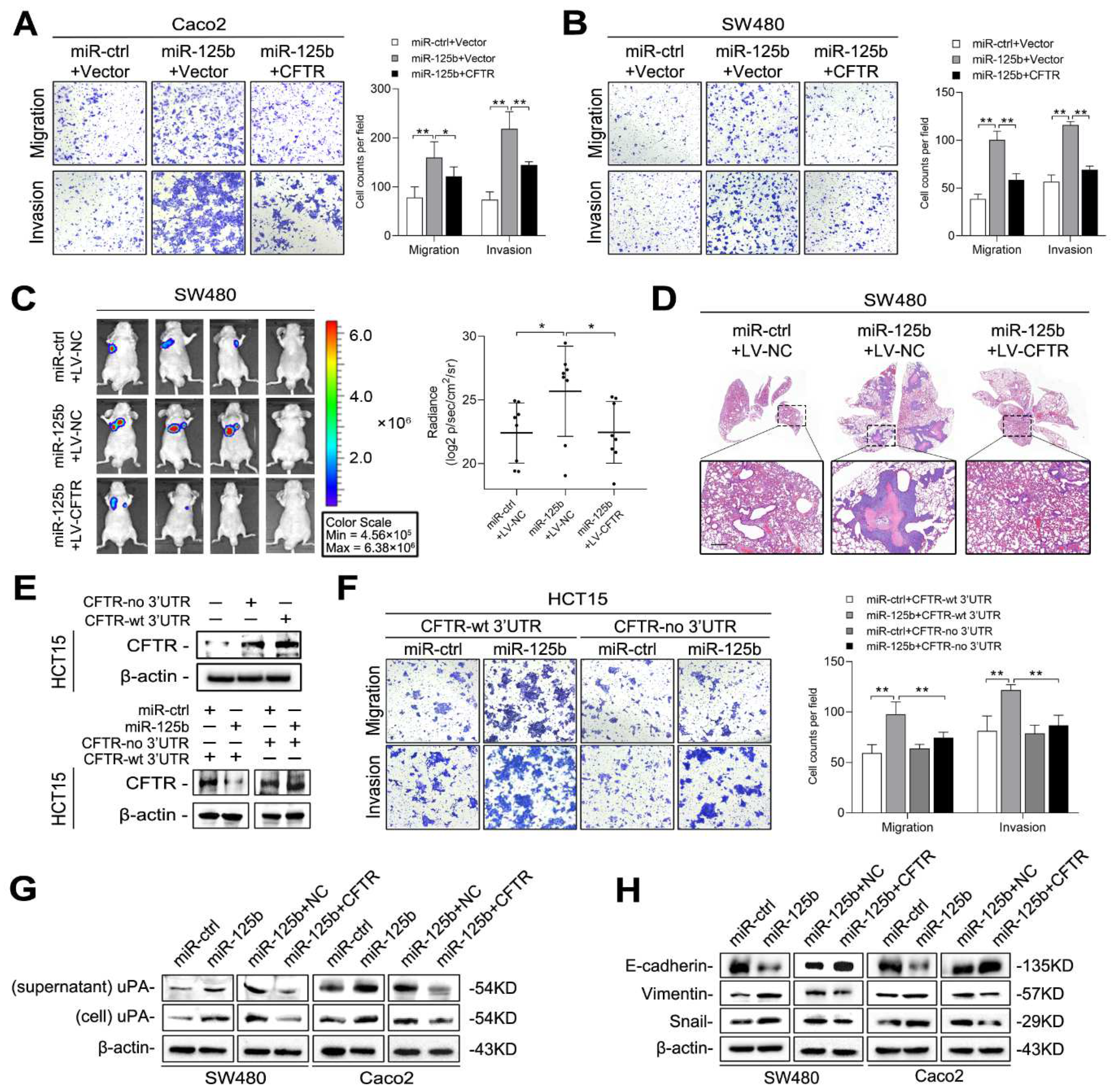
2.5. miR-125b Induces uPA Expression and EMT by Inhibiting CFTR
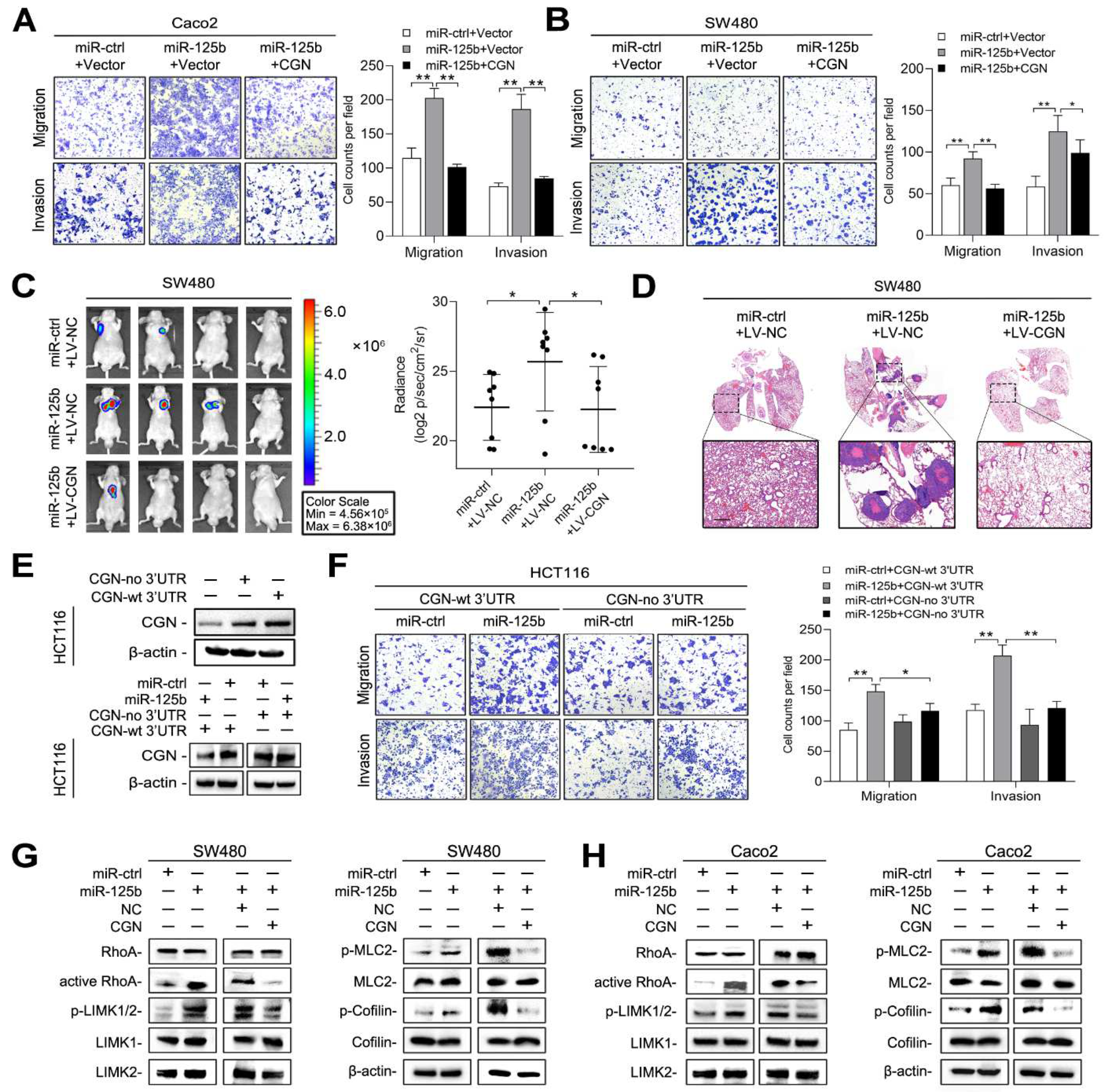
2.6. miR-125b Activates RhoA/ROCK Signaling by Targeting CGN
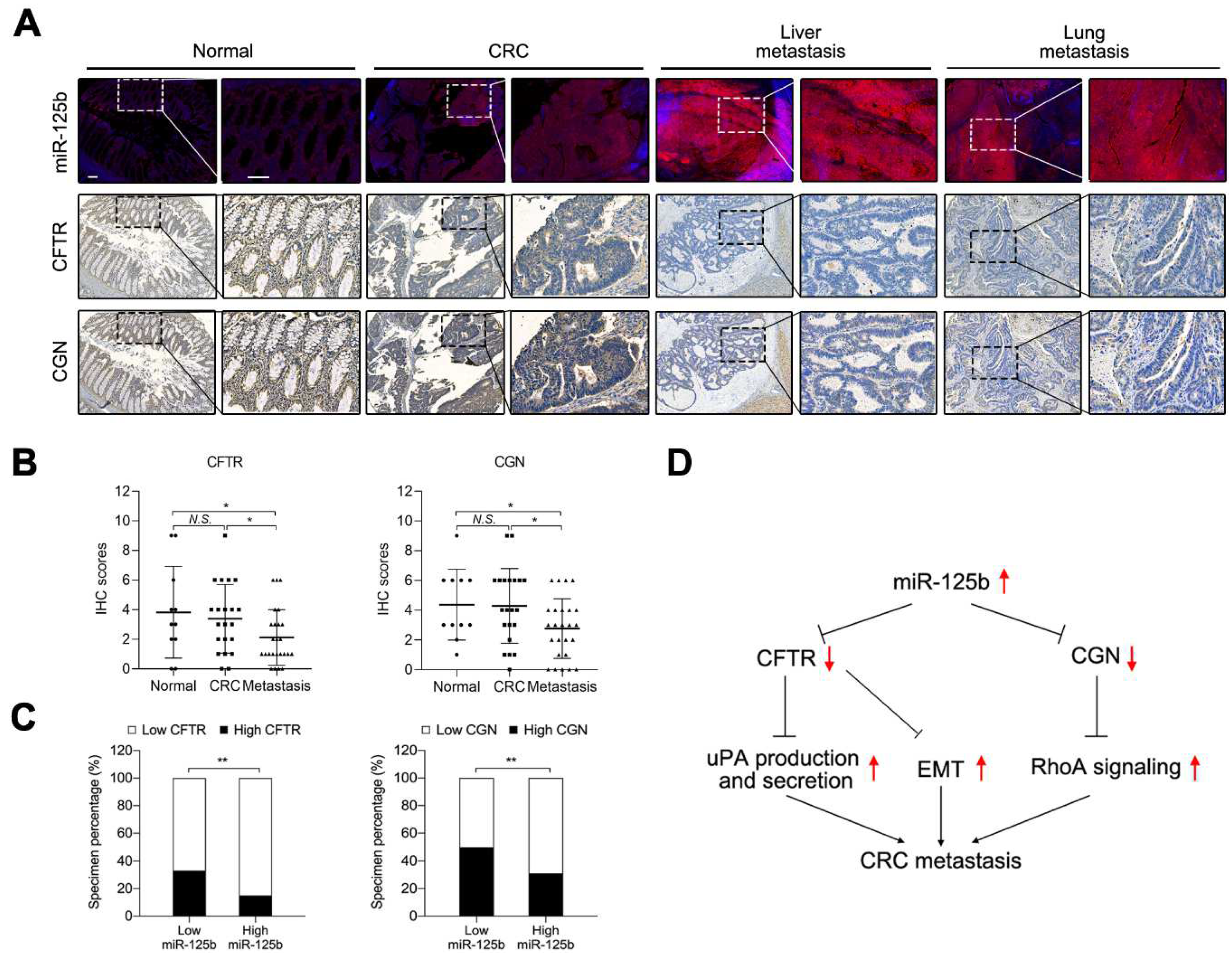
2.7. miR-125b Was Negatively Correlated with CFTR and CGN in CRC Specimens
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Tissue Collection and Tissue Microarrays
4.3. Protein Extraction and Western Blot Analysis
4.4. RNA Extraction and Real-Time PCR
4.5. FISH
4.6. Immunohistochemistry (IHC)
4.7. Transwell Invasion and Migration Assay
4.8. In Vivo Metastasis Assay
4.9. Dual-Luciferase Reporter Assay
4.10. Constructs, Oligonucleotides, Infection and Transfection
4.11. RhoA Activity Assay
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhao, X.; Liu, Q.; Mingli, Y.; Graves-Deal, R.; Cao, Z.; Singh, B.; Franklin, J.L.; Wang, J.; Bhuminder, S.; et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat. Med. 2017, 23, 1331–1341. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, X.; An, H.; Xu, C.; Cao, W.; Yuan, W.; Ma, J. Upregulation of microRNA-125b by G-CSF promotes metastasis in colorectal cancer. Oncotarget 2017, 8, 50642–50654. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Shi, W.; Zhang, Y.; Wang, X.; Sun, S.; Song, Z.; Qu, X.; Liu, M.; Zeng, Q.; Cui, S. CXCL12/CXCR4 axis induced miR-125b promotes invasion and confers 5-fluorouracil resistance through enhancing autophagy in colorectal cancer. Sci. Rep. 2017, 7, 42226. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Fan, H.; Zhang, Z.; Li, N. miR-125b-5p inhibits breast cancer cell proliferation, migration and invasion by targeting KIAA1522. Biochem. Biophys. Res. Commun. 2018, 504, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Tian, K.; Liu, W.; Zhang, J.; Fan, X.; Liu, J.; Zhao, N.; Yao, C.; Miao, G. MicroRNA-125b exerts antitumor functions in cutaneous squamous cell carcinoma by targeting the STAT3 pathway. Cell. Mol. Biol. Lett. 2020, 25, 12. [Google Scholar] [CrossRef]
- Csanády, L.; Vergani, P.; Gadsby, D.C. Structure, Gating, and Regulation of the CFTR Anion Channel. Physiol. Rev. 2019, 99, 707–738. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.C.; Mall, M.A.; Gutierrez, H.; Macek, M.; Madge, S.; Davies, J.C.; Burgel, P.-R.; Tullis, E.; Castaños, C.; Castellani, C.; et al. The future of cystic fibrosis care: A global perspective. Lancet Respir. Med. 2020, 8, 65–124. [Google Scholar] [CrossRef]
- Rowe, S.M.; Miller, S.; Sorscher, E.J. Cystic fibrosis. N. Engl. J. Med. 2005, 352, 1992–2001. [Google Scholar] [CrossRef]
- Fink, A.K.; Yanik, E.; Marshall, B.C.; Wilschanski, M.; Lynch, C.F.; Austin, A.A.; Copeland, G.; Safaeian, M.; Engels, E.A. Cancer risk among lung transplant recipients with cystic fibrosis. J. Cyst. Fibros. 2016, 16, 91–97. [Google Scholar] [CrossRef]
- Maisonneuve, P.; Marshall, B.C.; Knapp, E.A.; Lowenfels, A.B. Cancer Risk in Cystic Fibrosis: A 20-Year Nationwide Study From the United States. J. Natl. Cancer Inst. 2012, 105, 122–129. [Google Scholar] [CrossRef]
- Sadanandam, A.; Lyssiotis, C.; Homicsko, K.; Collisson, E.A.; Gibb, W.J.; Wullschleger, S.; Ostos, L.C.G.; Lannon, W.A.; Grötzinger, C.; Del Rio, M.; et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med. 2013, 19, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.T.; Wang, Y.; Cheng, H.; Zhang, X.H.; Xiang, J.J.; Zhang, J.T.; Yu, S.B.S.; Martin, T.A.; Ye, L.; Tsang, L.L.; et al. Disrupted interaction between CFTR and AF-6/afadin aggravates malignant phenotypes of colon cancer. Biochim. Biophys. Acta Bioenerg. 2013, 1843, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Than, B.L.N.; Linnekamp, J.F.; Starr, T.; Largaespada, D.A.; Rod, A.; Zhang, Y.; Bruner, V.; Abrahante, J.; Schumann, A.; Luczak, T.; et al. CFTR is a tumor suppressor gene in murine and human intestinal cancer. Oncogene 2016, 35, 4191–4199. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, J.T.; Jiang, X.; Shi, X.; Shen, J.; Feng, F.; Chan, H.C. The cystic fibrosis transmembrane conductance regulator as a biomarker in non-small cell lung cancer. Int. J. Oncol. 2015, 46, 2107–2115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.T.; Jiang, X.H.; Xie, C.; Cheng, H.; Dong, J.D.; Wang, Y.; Fok, K.L.; Zhang, X.H.; Sun, T.T.; Tsang, L.L.; et al. Downregulation of CFTR promotes epithelial-to-mesenchymal transition and is associated with poor prognosis of breast cancer. Biochim. Biophys. Acta 2013, 1833, 2961–2969. [Google Scholar] [CrossRef]
- Tu, Z.; Chen, Q.; Zhang, J.T.; Jiang, C.; Xiaohua, J.; Chan, H.C. CFTR is a potential marker for nasopharyngeal carcinoma prognosis and metastasis. Oncotarget 2016, 7, 76955–76965. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xie, C.; Jiang, X.H.; Zhang, J.T.; Sun, T.T.; Dong, J.; Sanders, A.J.; Diao, R.Y.; Wang, Y.; Fok, E.; Tsang, L.L.; et al. CFTR suppresses tumor progression through miR-193b targeting urokinase plasminogen activator (uPA) in prostate cancer. Oncogene 2012, 32, 2282–2291. [Google Scholar] [CrossRef]
- Cordenonsi, M.; D’Atri, F.; Hammar, E.; Parry, D.A.; Kendrick-Jones, J.; Shore, D.; Citi, S. Cingulin Contains Globular and Coiled-Coil Domains and Interacts with Zo-1, Zo-2, Zo-3, and Myosin. J. Cell Biol. 1999, 147, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, H.; Nakahara, T.; Furuse, K.; Sasaki, H.; Tsukita, S.; Furuse, M. JACOP, a novel plaque protein localizing at the apical junctional complex with sequence similarity to cingulin. J. Biol. Chem. 2004, 279, 46014–46022. [Google Scholar] [CrossRef]
- Mandai, K.; Nakanishi, H.; Satoh, A.; Obaishi, H.; Wada, M.; Nishioka, H.; Itoh, M.; Mizoguchi, A.; Aoki, T.; Fujimoto, T.; et al. Afadin: A Novel Actin Filament–Binding Protein with One PDZ Domain Localized at Cadherin-based Cell-to-Cell Adherens Junction. J. Cell Biol. 1997, 139, 517–528. [Google Scholar] [CrossRef]
- Citi, S.; Sabanay, H.; Kendrick-Jones, J.; Geiger, B. Cingulin: Characterization and localization. J. Cell Sci. 1989, 93, 107–122. [Google Scholar] [CrossRef]
- Schossleitner, K.; Rauscher, S.; Gröger, M.; Friedl, H.P.; Finsterwalder, R.; Habertheuer, A.; Sibilia, M.; Brostjan, C.; Födinger, D.; Citi, S.; et al. Evidence That Cingulin Regulates Endothelial Barrier Function In Vitro and In Vivo. Arter. Thromb. Vasc. Biol. 2016, 36, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, B.; Davis, T. The Blood-Brain Barrier/Neurovascular Unit in Health and Disease. Pharmacol. Rev. 2005, 57, 173–185. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Jhingory, S.; Taneyhill, L.A. The tight junction scaffolding protein cingulin regulates neural crest cell migration. Dev. Dyn. 2011, 240, 2309–2323. [Google Scholar] [CrossRef]
- Guillemot, L.; Citi, S. Cingulin Regulates Claudin-2 Expression and Cell Proliferation through the Small GTPase RhoA. Mol. Biol. Cell 2006, 17, 3569–3577. [Google Scholar] [CrossRef] [PubMed]
- Mangan, A.J.; Sietsema, D.V.; Li, D.; Moore, J.K.; Citi, S.; Prekeris, R. Cingulin and actin mediate midbody-dependent apical lumen formation during polarization of epithelial cells. Nat. Commun. 2016, 7, 12426. [Google Scholar] [CrossRef]
- Guillemot, L.; Spadaro, D.; Citi, S. The Junctional Proteins Cingulin and Paracingulin Modulate the Expression of Tight Junction Protein Genes through GATA-4. PLoS ONE 2013, 8, e55873. [Google Scholar] [CrossRef]
- Oliveto, S.; Alfieri, R.; Miluzio, A.; Scagliola, A.; Seclì, R.S.; Gasparini, P.; Grosso, S.; Cascione, L.; Mutti, L.; Biffo, S. A Polysome-Based microRNA Screen Identifies miR-24-3p as a Novel Promigratory miRNA in Mesothelioma. Cancer Res. 2018, 78, 5741–5753. [Google Scholar] [CrossRef]
- Sidenius, N.; Blasi, F. The urokinase plasminogen activator system in cancer: Recent advances and implication for prognosis and therapy. Cancer Metastasis Rev. 2003, 22, 205–222. [Google Scholar] [CrossRef]
- Rath, N.; Olson, M.F. Rho-associated kinases in tumorigenesis: Reconsidering ROCK inhibition for cancer therapy. EMBO Rep. 2012, 13, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Peverelli, E.; Catalano, R.; Giardino, E.; Treppiedi, D.; Morelli, V.; Ronchi, C.L.; Mantovani, G.; Fusco, N.; Ferrero, S.; Spada, A.; et al. Cofilin is a cAMP effector in mediating actin cytoskeleton reorganization and steroidogenesis in mouse and human adrenocortical tumor cells. Cancer Lett. 2017, 406, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Heasman, S.J.; Carlin, L.M.; Cox, S.; Ng, T.; Ridley, A.J. Coordinated RhoA signaling at the leading edge and uropod is required for T cell transendothelial migration. J. Cell Biol. 2010, 190, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhu, X.; Yu, Y.; Zhu, W.; Jin, L.; Zhang, X.; Li, S.; Zou, P.; Xie, C.; Cui, R. Dissecting miRNA signature in colorectal cancer progression and metastasis. Cancer Lett. 2020, 501, 66–82. [Google Scholar] [CrossRef]
- Zhou, X.-G.; Huang, X.-L.; Liang, S.-Y.; Tang, S.-M.; Wu, S.-K.; Huang, T.-T.; Mo, Z.-N.; Wang, Q.-Y. Identifying miRNA and gene modules of colon cancer associated with pathological stage by weighted gene co-expression network analysis. OncoTargets Ther. 2018, 11, 2815–2830. [Google Scholar] [CrossRef]
- Liu, H.N.; Liu, H.N.; Liu, T.T.; Wu, H.; Chen, Y.J.; Tseng, Y.J.; Dong, L.; Yao, C.; Weng, S.-Q. Serum microRNA signatures and metabolomics have high diagnostic value in colorectal cancer using two novel methods. Cancer Sci. 2018, 109, 1185–1194. [Google Scholar] [CrossRef]
- Lin, M.; Chen, W.; Huang, J.; Ye, Y.; Song, Z.; Chen, W.; Gao, H.; Shen, X. MicroRNA expression profiles in human colorectal cancers with liver metastases. Oncol. Rep. 2011, 25, 739–747. [Google Scholar] [CrossRef][Green Version]
- Pelullo, M.; Savi, D.; Quattrucci, S.; Cimino, G.; Pizzuti, A.; Screpanti, I.; Talora, C.; Cialfi, S. miR-125b/NRF2/HO-1 axis is involved in protection against oxidative stress of cystic fibrosis: A pilot study. Exp. Ther. Med. 2021, 21, 1–6. [Google Scholar] [CrossRef]
- Chen, J.; Chen, Y.; Chen, Y.; Yang, Z.; You, B.; Ruan, Y.C.; Peng, Y. Epidermal CFTR Suppresses MAPK/NF-κB to Promote Cutaneous Wound Healing. Cell Physiol. Biochem. 2016, 39, 2262–2274. [Google Scholar] [CrossRef]
- Takai, Y.; Nakanishi, H. Nectin and afadin: Novel organizers of intercellular junctions. J. Cell Sci. 2003, 116, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Yano, T.; Matsui, T.; Tamura, A.; Uji, M.; Tsukita, S. The association of microtubules with tight junctions is promoted by cingulin phosphorylation by AMPK. J. Cell Biol. 2013, 203, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Gawlak, G.; Tian, X.; Shah, A.S.; Sarich, N.; Citi, S.; Birukova, A.A. Role of Cingulin in Agonist-induced Vascular Endothelial Permeability. J. Biol. Chem. 2016, 291, 23681–23692. [Google Scholar] [CrossRef]
- Martínez, C.; Rodiño-Janeiro, B.K.; Lobo, B.; Stanifer, M.L.; Klaus, B.; Granzow, M.; González-Castro, A.M.; Salvo-Romero, E.; Alonso-Cotoner, C.; Pigrau, M.; et al. miR-16 and miR-125b are involved in barrier function dysregulation through the modulation of claudin-2 and cingulin expression in the jejunum in IBS with diarrhoea. Gut 2017, 66, 1537–1538. [Google Scholar] [CrossRef] [PubMed]
- Aijaz, S.; D’Atri, F.; Citi, S.; Balda, M.S.; Matter, K. Binding of GEF-H1 to the Tight Junction-Associated Adaptor Cingulin Results in Inhibition of Rho Signaling and G1/S Phase Transition. Dev. Cell 2005, 8, 777–786. [Google Scholar] [CrossRef]
- De Planell-Saguer, M.; Rodicio, M.C.; Mourelatos, Z. Rapid in situ codetection of noncoding RNAs and proteins in cells and formalin-fixed paraffin-embedded tissue sections without protease treatment. Nat. Protoc. 2010, 5, 1061–1073. [Google Scholar] [CrossRef]
- Li, J.; Su, W.; Zhang, S.; Hu, Y.; Liu, J.; Zhang, X.; Bai, J.; Yuan, W.; Hu, L.; Cheng, T.; et al. Epidermal growth factor receptor and AKT1 gene copy numbers by multi-gene fluorescence in situ hybridization impact on prognosis in breast cancer. Cancer Sci. 2015, 106, 642–649. [Google Scholar] [CrossRef]
- Zhao, X.; He, L.; Li, T.; Lu, Y.; Miao, Y.; Liang, S.; Guo, H.; Bai, M.; Xie, H.; Luo, G.; et al. SRF expedites metastasis and modulates the epithelial to mesenchymal transition by regulating miR-199a-5p expression in human gastric cancer. Cell Death Differ. 2014, 21, 1900–1913. [Google Scholar] [CrossRef]
| Variables | MiR-125b Expression | p -Value | ||
|---|---|---|---|---|
| All (n = 58) | Low (n = 32) | High (n = 26) | ||
| Age (years) | >0.9999 | |||
| ≤65 | 28 | 15 | 13 | |
| >65 | 30 | 17 | 13 | |
| Sex | 0.7976 | |||
| male | 30 | 16 | 14 | |
| female | 28 | 16 | 12 | |
| Tumor location | 0.3629 | |||
| ascending colon | 19 | 10 | 9 | |
| transverse colon | 9 | 5 | 4 | |
| descending colon | 7 | 6 | 1 | |
| sigmoid colon | 23 | 11 | 12 | |
| Tumor size (cm) | 0.2724 | |||
| <5 | 21 | 14 | 7 | |
| ≥5 | 37 | 18 | 19 | |
| Tumor invasion | 0.0457 | |||
| T1 | 0 | 0 | 0 | |
| T2 | 3 | 3 | 0 | |
| T3 | 44 | 26 | 18 | |
| T4 | 11 | 3 | 8 | |
| Lymph node metastasis | 0.021 | |||
| N0 | 33 | 23 | 10 | |
| N1 | 19 | 8 | 11 | |
| N2 | 6 | 1 | 5 | |
| N3 | 0 | 0 | 0 | |
| AJCC stage | 0.0207 | |||
| stage 1 | 3 | 3 | 0 | |
| stage 2 | 30 | 20 | 10 | |
| stage 3 | 25 | 9 | 16 | |
| Tumor Differentiation | 0.0453 | |||
| well | 11 | 8 | 3 | |
| moderate | 40 | 23 | 17 | |
| poor | 7 | 1 | 6 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Li, T.; Han, Y.-N.; Ge, M.; Wang, P.; Sun, L.; Liu, H.; Cao, T.; Nie, Y.; Fan, D.; et al. miR-125b Promotes Colorectal Cancer Migration and Invasion by Dual-Targeting CFTR and CGN. Cancers 2021, 13, 5710. https://doi.org/10.3390/cancers13225710
Zhang X, Li T, Han Y-N, Ge M, Wang P, Sun L, Liu H, Cao T, Nie Y, Fan D, et al. miR-125b Promotes Colorectal Cancer Migration and Invasion by Dual-Targeting CFTR and CGN. Cancers. 2021; 13(22):5710. https://doi.org/10.3390/cancers13225710
Chicago/Turabian StyleZhang, Xiaohui, Tingyu Li, Ya-Nan Han, Minghui Ge, Pei Wang, Lina Sun, Hao Liu, Tianyu Cao, Yongzhan Nie, Daiming Fan, and et al. 2021. "miR-125b Promotes Colorectal Cancer Migration and Invasion by Dual-Targeting CFTR and CGN" Cancers 13, no. 22: 5710. https://doi.org/10.3390/cancers13225710
APA StyleZhang, X., Li, T., Han, Y.-N., Ge, M., Wang, P., Sun, L., Liu, H., Cao, T., Nie, Y., Fan, D., Guo, H., Wu, K., Zhao, X., & Lu, Y. (2021). miR-125b Promotes Colorectal Cancer Migration and Invasion by Dual-Targeting CFTR and CGN. Cancers, 13(22), 5710. https://doi.org/10.3390/cancers13225710







